SYNOPSIS
A critical cis-regulatory element for the cystic fibrosis transmembrane conductance regulator (CFTR) gene is located in intron 11, 100 kb distal to the promoter, with which it interacts. This sequence contains an intestine-selective enhancer and associates with enhancer signature proteins, such as p300, in addition to tissue-specific transcription factors (TFs). Here we identify critical TFs that are recruited to this element and demonstrate their importance in regulating CFTR expression. In vitro DNase I footprinting and electrophoretic mobility shift assays (EMSA) identified four cell-type selective regions that bound TFs in vitro. Chromatin immunoprecipitation (ChIP) identified forkhead box A1/A2 (FOXA1/A2), hepatocyte nuclear factor 1 (HNF1), and caudal type homeobox 2 (CDX2) as in vivo trans-interacting factors. Mutation of their binding sites in the intron 11 core compromised its enhancer activity when measured by reporter gene assay. Moreover, siRNA-mediated knockdown of CDX2 caused a significant reduction in endogenous CFTR transcription in intestinal cells, suggesting that this factor is critical for the maintenance of high levels of CFTR expression in these cells. The ChIP data also demonstrate that these TFs interact with multiple cis-regulatory elements across the CFTR locus implicating a more global role in intestinal expression of the gene.
Keywords: Gene expression, transcriptional networks, enhancer, cis-acting regulatory elements, CFTR
INTRODUCTION
The recruitment of transcription factors (TFs) and chromatin remodelers to cis-regulatory elements is fundamental to the mechanisms that control gene expression. Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) has generated specific chromatin signatures for many different types of regulatory elements found in non-coding regions of the genome [1-5]. For example, enhancer elements are often enriched for the histone acetyltransferase p300, and are marked by H3 histones mono-methylated on lysine 4 (H3K4me1), but lack H3K4me3 modified histones [3-5]. Activity of distal enhancers is often facilitated by long-range interactions between these elements and their target promoters, causing tissue-specific chromatin looping events and associated transcriptional activation (reviewed in [6]). Though the location of many tissue-specific enhancers can be identified through mapping regions of open chromatin genome-wide [7], it remains a challenge to identify trans-acting factors responsible for enhancer activity.
The 189 kb cystic fibrosis transmembrane conductance regulator (CFTR) gene shows a complex pattern of expression, driven in part by cell-type specific enhancers in non-coding regions of the locus. Mutations in CFTR cause the common autosomal recessive disorder cystic fibrosis (CF) [8]. Here, we aimed to characterize the factors that regulate the activity of a cell-type selective enhancer element located in a region of open chromatin in intron 11 of the gene [9]. CFTR is expressed in specialized epithelial cells of many endoderm-derived tissues, including the airway, pancreas, small intestine and male genital duct, and also in some non-epithelial cell types [10-13]. However, the mechanisms governing CFTR expression are cell type-dependent, and differ in the intestinal and airway epithelium [9, 14]. This selectivity is apparently achieved, at least in part, by the utilization of different cis-regulatory elements within the locus. The transcriptional pathways that coordinate these elements are not fully defined (reviewed in [15]). Here our goal was to establish the network of transcription factors that activate CFTR intronic enhancers in the intestine.
The intestine-selective enhancer in intron 11 at 1811 + 0.8 kb (where 1811 is the last coding base in exon 11) was identified by DNase-chip within a 1.5 kb DNase I hypersensitive site (DHS). This DHS contains a cis-acting element that recruits p300, and cooperates with other intestinal enhancer elements within CFTR [9]. Moreover, the intron 11 element associates closely with the CFTR promoter by direct chromatin looping, demonstrated by chromosome conformation capture (3C) [9]. Here, in vitro DNase I footprinting was used to identify sequences within the intron 11 DHS element that bound nuclear factors in a cell-type selective manner. Next, bioinformatic predictions of candidate TF interactors were evaluated by electrophoretic mobility shift assays (EMSA) in vitro, and chromatin immunoprecipitation (ChIP) in vivo. These studies reveal a TF complex interacting with the intron 11 DHS element core that includes forkhead box A1/A2 (FOXA1/A2), hepatocyte nuclear factor 1 (HNF1), and caudal type homeobox 2 (CDX2).
FOXA1/A2, important regulators of liver development and differentiation, are expressed in many endoderm-derived tissues [16, 17] and can act as pioneer factors that facilitate binding of additional TFs to enhancers (reviewed in [18]). FOXA family members remodel histones in a SWI/SNF-independent manner by interacting with H3 and H4 histones, and subsequently displacing H1-linker histones [19]. HNF1 was implicated in intestinal control of CFTR expression, through its interactions with other cis-acting regulatory elements in introns 1, 10, 17a, and 20. [20, 21]. Hnf1α is also required for normal expression of Cftr in the mouse intestine [20]. Here we show its recruitment to the intron 11 DHS element in vivo [9]. CDX2, a master regulator of gut development and differentiation [22], is known to coordinate gene expression with HNF1α [23-30]. In vitro studies previously implicated CDX2 as a candidate TF interacting with CFTR cis-regulatory elements in intestinal and pancreatic cells; however, no evidence was obtained for its role in vivo [31]. We now show interaction of CDX2 with the intron 11 DHS element in vitro and in vivo, and also demonstrate its recruitment to adjacent cis-regulatory elements. Moreover, a decrease in CFTR mRNA levels following siRNA-mediated depletion of CDX2 confirms the importance of this factor in regulating CFTR expression in vivo. Thus, activity of the CFTR locus in intestinal epithelial cells may be coordinated by the interaction of pioneer factors with the intron 11 DHS core element, and subsequent recruitment of a network of intestinal-selective TFs that bind to multiple cis-regulatory elements across the locus.
EXPERIMENTAL
Plasmids and expression vectors
Reporter constructs containing the 787 bp minimal CFTR promoter and the 1.5 kb full-length DHS11 enhancer fragment were described previously [21, 32, 33]. Mutagenesis of plasmids was performed using the QuikChange® II XL or Lightning Multi Site-Directed Mutagenesis Kits (Agilent Technologies) per the manufacturer’s protocol with primers listed in Table S1. Rat HNF1α cDNA was PCR amplified from rHNF1α-CMV4 (kind gift of Riccardo Cortese), subcloned into pRCII (Invitrogen), and cloned into pcDNA3.1(-) using BamHI and XbaI to generate the HNF1α expression plasmid. The CDX2 [31], CUX1 p110 and p200 [34] and mFOXA1 [35] expression plasmids were described previously. The rat FOXA2 expression plasmid was made by cloning the rFOXA2 cDNA [35] into pcDNA3.1(-) using XhoI and BamHI. The HOXB7 expression plasmid was purchased from Addgene (plasmid 8537) [36].
Cell culture and transient reporter assays
The human colon carcinoma cell lines Caco2 and HT29, and primary skin fibroblasts (GM08333) were grown in DMEM supplemented with 10% fetal bovine serum. Caco2 cells were co-transfected with the luciferase reporter constructs and Renilla expression vector (Promega), in 24-well plates with Lipofectin® (Invitrogen), using standard methods. Cells were lysed after 48 hours and luciferase assays performed as reported previously [21]. Data from at least two independent plasmid preparations of each construct were consistent.
DNase I footprinting
Overlapping 200-300 bp fragments of the DHS11 region were PCR amplified with Pfu DNA polymerase and primers in Table S1, and cloned into pSC-B in the reverse orientation using the StrataClone Blunt PCR Cloning Kit (Agilent Technologies). Probes were gel purified following excision from vectors using ClaI and SmaI or KpnI and SpeI, to label the sense or antisense strands respectively. Nuclear extracts were generated and DNase I footprinting reactions were carried out as described previously [20].
Electrophoretic mobility gel shift assay (EMSA)
EMSA reactions were done using standard protocols using double stranded probes and competitors detailed in Table S1. In vitro translated (IVT) proteins were produced using TNT® T7 or SP6 Quick Coupled Reticulocyte System (Promega), and confirmed by western blot analysis. Antibodies specific for HNF-1 (Santa Cruz, sc-8986x), FOXA2 (Santa Cruz, sc-6554x), FOXA1 (Abcam, ab5089), CDX2 (Bethyl Laboratories, A300-691A), CUX1 1300 [37], FLAG® M2 (Sigma-Aldrich, F3165), BARX2 (Santa Cruz, sc-9128X), PDX1 (Santa Cruz, sc-14662X), and PBX1 (Santa Cruz, sc-889X) were used for supershifts.
Chromatin immunoprecipitation (ChIP)
ChIP was performed by standard methods. Chromatin was sonicated using a Bioruptor® Plus (Diagenode) to an average size of 500 bp. Immunoprecipitations were performed with antibodies specific for FOXA1, FOXA2, CDX2 (Bethyl Laboratories, A300-691A), HNF1, normal goat IgG (Santa Cruz, sc-2028), or normal rabbit IgG (Millipore, 12-370). Enrichment was analyzed relative to IgG using SYBR® Green quantitative PCR (qPCR) with primers listed in Table S1.
Transient siRNA knockdown and rescue
For siRNA knockdown only, Caco2 cells were reverse transfected with Lipofectamine™ RNAiMAX (Invitrogen) as per the manufacturer’s protocol in 24-well plates using 20 nM of hCDX2 (Santa Cruz, sc-43680), hHNF1α (Santa Cruz, sc-35567), or control (Santa Cruz, sc-37007) siRNA. For rescue experiments Caco2 cells were plated 48 hours prior to forward transfection with Lipofectamine™ 2000 (Invitrogen) as per the manufacturer’s protocol using 10 nM of hCDX2 or control siRNA and 0.8 μg pRC-CMV or pRC-mCdx2 expression plasmid [31]. Mock-transfected cells were treated with transfection reagent only. Cells were lysed after 48 hours for assay of protein and mRNA expression.
Western blots
Cells were lysed by standard protocols and protein levels assayed by western blot with antibodies specific for CDX2 (Bethyl Laboratories, A300-692A), HNF1, FOXA1, FOXA2, and β-tubulin (Sigma-Aldrich, T 4026). Protein quantification was performed using ImageJ software (NIH; http://rsb.info.nih.gov/ij/).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted using TRIzol® (Invitrogen), and TaqMan® reverse-transcription reactions performed (Applied Biosytems). CFTR expression was measured using TaqMan® primer/probe set spanning CFTR exons 5 and 6, and normalized to endogenous 18S rRNA [38].
RESULTS
In vitro DNase I footprinting reveals binding of multiple nuclear proteins to the CFTR intron 11 DHS enhancer region
The intron 11 (1811 + 0.8 kb) enhancer was originally identified within a DHS that encompassed approximately 1.5 kb of genomic DNA (See Table 1 for all relevant CFTR DHS coordinates). The DHS was evident in CFTR-expressing intestinal epithelial cells (Caco2 and HT29, colon carcinoma) (Fig. 1A) and primary epididymis cells, but not in skin fibroblasts that do not express CFTR [9]. To characterize sequences within the intron 11 DHS region that contribute to enhancer activity, in vitro DNase I footprinting was performed. Four overlapping 250-300 bp fragments (C-F) were designed to span the central core of the DHS region (Fig. 1B). Sense and antisense strands of these fragments were radiolabeled and used as probes for DNase I footprinting with Caco2 nuclear extracts. These experiments identified four DNA sequences that were protected from DNase I digestion (PR), two each in fragments 11D (PR1 and PR2) and 11F (PR3 and PR4) (Fig. 1C-E). The trans-acting factors that generate the PRs show some cell-type specificity, as the PRs were not observed using nuclear extracts from 16HBE14o- cells (human bronchial epithelial cells; CFTR +) or skin fibroblasts (CFTR -) (data not shown).
Table 1.
CFTR DHS locations.
| DHS | Location in CFTR locus | hg17 coordinates |
|---|---|---|
| -20.9 kb*† | 5’ | chr7:116693001-116693396 |
| 185 + 10 kb | intron 1 | chr7:116723600–116724700 |
| 1716 + 13.2/13.7 kb | intron 10 (ab) | chr7:116806300–116807600 |
| 1716 + 23 kb | intron 10 (c) | chr7:116816300–116817000 |
| 1811 + 0.8 kb | intron 11 | chr7:116822000–116823400 |
| 3271 + 0.7 kb† | intron 17a | chr7:116844713-116845585 |
| 4005 + 3.7 kb† | intron 20 | chr7:116878911-116881521 |
| 4374 + 1.3 kb | intron 23 | chr7:116899700–116901100 |
| +6.8 kb‡ | 3’ | chr7:116907887-116908087 |
| +15.6 kb‡ | 3’ | chr7:116916600–116918100 |
Distance upstream of CFTR translational start site
DHS identified through Southern blotting, not detected by DNase-chip
Distance downstream of last coding base of CFTR transcript
Figure 1. Identification of multiple sites of DNA:protein interaction in the intron 11 DHS region of CFTR.
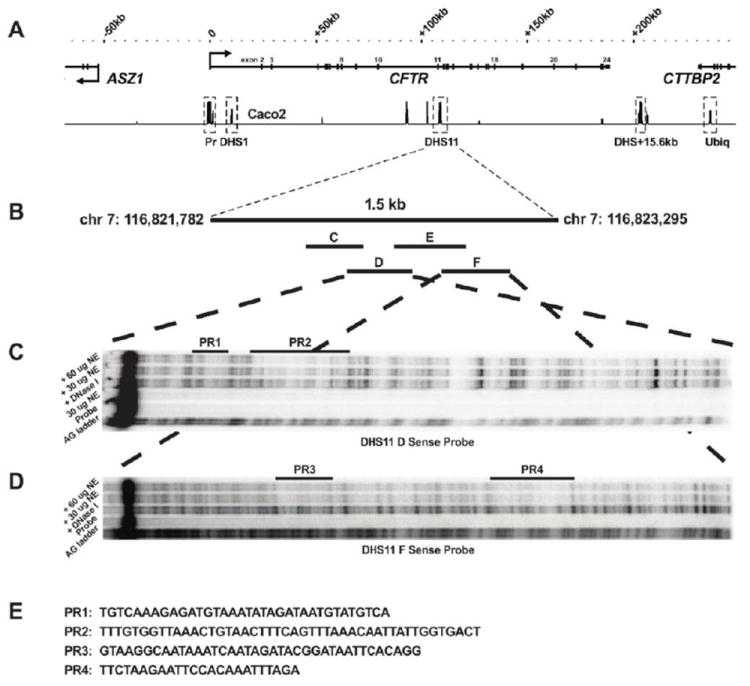
(A) DHS regions identified across the CFTR locus in Caco2 cells by DNase-chip [9] including the 1.5 kb region in intron 11 (DHS11). Distances along the locus are relative to the first base of exon 1 of CFTR. (B) Four overlapping 250-300 bp probes spanning DHS11 were used for DNase I footprinting which revealed four regions protected from DNase I (PRs 1-4) on the sense strand of DHS11 fragment D (C) and fragment F (D) probes. AG ladder shows sequence reference, and probes were incubated alone or with DNase I as controls, or with DNase I plus 30 or 60 μg of Caco2 nuclear extract (NE). (E) Sense strand sequences (5’-3’) are shown for the four PRs.
Prediction of candidate transcription factors that interact with the enhancer core
In silico analysis of the PR sequences using MatInspector (www.genomatix.de) identified an extensive list of potential transcription factor binding sites (TFBS), and this was used to generate a subset of candidate TFs based on cell-specificity and likely functional relevance (Table 2). These candidates include TFs expressed in epithelial cell types that are relevant to CF, those that have previously been shown to coordinate regulation of CFTR expression, and those that recruit chromatin remodelers. Included in this list are FOXA2, FOXF2, CDX2, HNF1, MECOM, C/EBPα and HOXB7, among others (see Table 2).
Table 2.
Candidate transcription factors predicted to bind PRs 1-4.
| Protected Region 1: TGTCAAAGAGATGTAAATATAGATAATGTATGTCA | |||
|---|---|---|---|
|
| |||
| Factor Name | Matrix | Matrix Similarity | Sequence (strand)§ |
| MECOM | V$EVI1.04 | 0.734 | aagagatgtaaaTATAg (+) |
| FOXA2 | V$XFD1.01 | 0.931 | gagatgTAAAtatagat (+) |
| CUX1 | V$CLOX.01 | 0.871 | cattATCTatatttacatc (-) |
|
| |||
| Protected Region 2: TTTGTGGTTAAACTGTAACTTTCAGTTTAAACAATTATTGGTGACT | |||
|
| |||
| Factor Name | Matrix | Matrix Similarity | Sequence (strand) |
|
| |||
| HNF1α | V$HNF1.01 | 0.809 | aGTTAcagtttaaccac (-) |
| FOXF2 | V$FREAC2.01 | 0.904 | tcagttTAAAcaattat (+) |
| HOXB7 | V$HOXB7.01 | 0.841 | accaatAATTgtttaaact (-) |
| BARX2 | V$BARX2.02 | 0.929 | gtttaaacAATTattggtg (+) |
|
| |||
| Protected Region 3: GTAAGGCAATAAATCAATAGATACGGATAATTCACAGG | |||
|
| |||
| Factor Name | Matrix | Matrix Similarity | Sequence (strand) |
|
| |||
| HOXB8 | V$HOXB8.01 | 0.897 | taaggcaATAAatcaatag (+) |
| CDX2 | V$CDX2.03 | 0.995 | ctattgatTTATtgcctta (-) |
| HNF6 | V$HNF6.01 | 0.979 | gcaataaaTCAAtagat (+) |
| SOX9 | V$SOX9.01 | 0.905 | ataaatCAATagatacggataattc (+) |
| CUX1 | V$CLOX.01 | 0.865 | ataaATCAatagatacgga (+) |
| HMGA1 | V$HMGIY.01 | 0.929 | cctgtgAATTatccgtatctattga (-) |
| 0.849 | cctgtGAATtatccgtatc (-) | ||
| HOXB5 | V$HOXB5.01 | ||
| 0.843 | acggaTAATtcacaggctt (+) | ||
|
| |||
| Protected Region 4: TTCTAAGAATTCCACAAATTTAGA | |||
|
| |||
| Factor Name | Matrix | Matrix Similarity | Sequence (strand) |
|
| |||
| STAT5(α/β) | V$STAT5.01 | 0.966 | cttaTTCTaagaattccac (+) |
| C/EBPα | V$CEBPA.01 | 0.972 | aaatttgtGGAAttc (-) |
CAPITALS indicate the core sequence comprised of the most conserved consecutive positions of the matrix
As an additional approach to identify sites of functional DNA:protein interactions with the intron 11 DHS region, we analyzed the predicted TFBS in regions of high cross-species conservation within the element, as these sequences often encompass sites that are subject to positive selection. Fifteen peaks of high conservation were identified from the 17 vertebrate species multiz alignment and conservation track available on the UCSC genome browser [39-41] (Fig. S1). MatInspector analysis of the fifteen conserved regions (CRs) identified 152 potential TFBS, including general and tissue-specific TFs (Table S1). Most notably, there were additional sites for some of the candidate factors predicted to bind the PRs, including CDX2 (3 sites), HNF1 (5 sites), and FOXA2 (1 site). The interaction of these factors with the intron 11 DHS region is evaluated further below.
Identification of factors that interact in vitro with PR1 and PR2 sequences
The in vitro potential of the candidate TFs (Table 2) to bind their predicted cognate sites in DHS11 PR1 and PR2 was investigated using electrophoretic mobility shift assays (EMSAs) (Fig. 2). Double-stranded DNA probes corresponding to the individual PRs (Table S1) were radiolabeled and used in EMSA reactions with nuclear extracts from Caco2 cells. Nuclear proteins bound to both PR1 and PR2 in vitro, and the specificity of these interactions was shown by self-competition (10x and 100x molar excess of the probe) and by competition experiments with oligonucleotides corresponding to consensus binding sites of the candidate TFs.
Figure 2. In vitro interactions of FOXA1/A2, HNF1, and CDX2 with the intron 11 DHS enhancer.
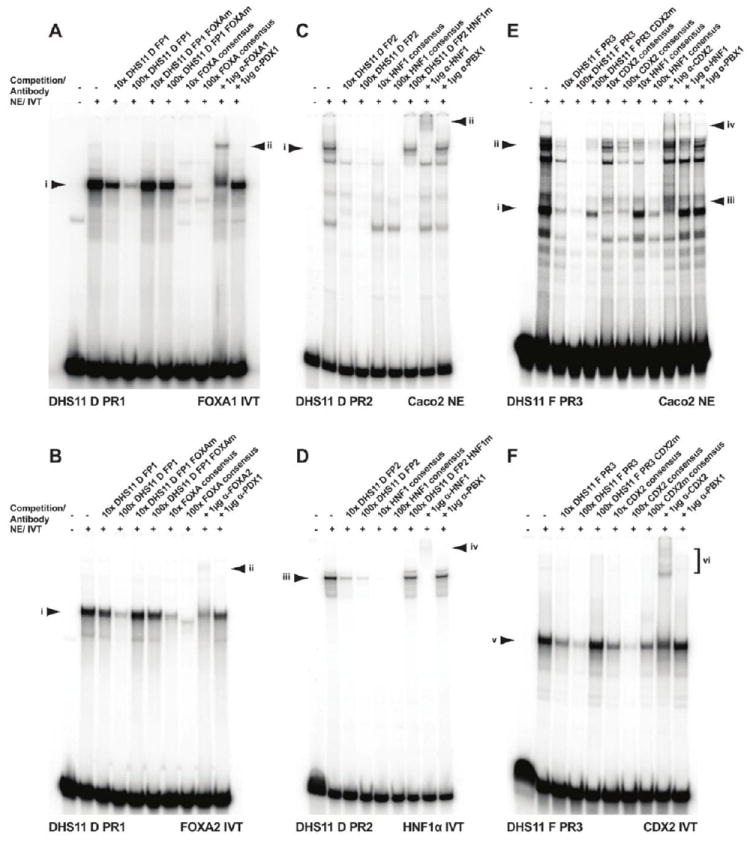
EMSAs using probes for PR1 (A-B), PR2 (C-D), or PR3 (E-F) sequences of the DHS11 enhancer. PR1 interacts with in vitro translated (IVT) FOXA1 (A) and FOXA2 (B), complex (i). Competition with 10x or 100x molar excess of unlabeled oligonucleotides are shown. A supershift is seen with FOXA1-specific (A) or FOXA2-specific (B) antibody (ii), but not with isotype matched PDX1 antibody. (C-D) PR2 generates a major complex with Caco2 nuclear extracts (i in C) or IVT HNF1α (iii in D). Competition with cold oligonucleotides is shown. A supershift is seen with HNF1-specific antibody (ii in C, iv in D), but not with isotype matched PBX1 antibody. (E-F) Protein complexes form between PR3 and Caco2 nuclear extracts (i and ii in E) or IVT CDX2 (v in F). Competition with cold oligonucleotides is shown. Supershifts are seen with CDX2-specific antibody (iii and iv in E, vi in F), and HNF1-specific antibody (iv in E), but not with isotype matched PBX1 antibody.
FOXA1/A2 interact in vitro with PR1
As FOXA1 and FOXA2 have identical consensus binding sequences, we investigated the in vitro potential of both of these factors to bind PR1. A shift in the migration of PR1 oligonucleotide was observed upon incubation with in vitro translated (IVT) FOXA1 or FOXA2 protein (Fig. 2A-B, lanes 2, complex i). Self-competition with 10x and 100x molar excess of PR1 or with a FOXA consensus oligonucleotide inhibited formation of this complex (Fig. 2A-B, self, lanes 3-4, consensus, lanes 7-8). Competition with a PR1 oligonucleotide containing a mutation in the predicted FOXA binding site did not inhibit complex i (Fig. 2A-B, lanes 5-6). Moreover, inclusion of an antibody specific for either FOXA1 or FOXA2 in the EMSA reaction resulted in a supershift of the PR1 probe (Fig. 2A-B, lane 9, complex ii). Though these results demonstrate an interaction of IVT FOXA1/A2 with PR1, equivalent data were not obtained with nuclear extracts from Caco2 cells, which also express both FOXA1 and FOXA2 as shown by western blot (Fig. S2A). This suggests that in the complex mixture of nuclear proteins in Caco2 cells, there may be other factors that compete with FOXA1/A2 at this site.
HNF1 interacts in vitro with PR2
EMSA experiments using PR2 as probe and Caco2 nuclear extracts generated multiple complexes (Fig. 2C). Though these were all specific, as shown by their complete loss upon competition with 100x excess of unlabeled PR2, only the slowest mobility complex (i) apparently contains HNF1 protein. Loss of complex i occurs upon competition with HNF1 consensus oligonucleotide (Fig. 2C, lanes 5-6), but not with unlabeled PR2 oligonucleotide that contains a mutation in the predicted HNF1 binding site (Fig. 2C, lane 7). Co-incubation of Caco2 nuclear extracts, PR2 probe, and an antibody specific for HNF1 results in a supershift of complex i (Fig. 2C, lane 8, complex ii). Equivalent EMSA experiments with IVT HNF1α confirmed the in vitro binding of this factor to PR2 (Fig. 2D). Thus, HNF1α is a major factor in the complex that interacts with PR2 in vitro. Though additional proteins appear to interact specifically with this probe, as demonstrated by the complexes of different mobility seen in Fig. 2C, at present these have not been characterized.
Other candidate factors
The other candidate TFBS in PR1 (MECOM and CUX1) and PR2 (FOXF2, HOXB7, and BARX2) were also investigated by EMSA using Caco2 nuclear extracts (data not shown). Competition with MECOM consensus oligonucleotide did not compete with protein complexes formed with PR1. While these complexes partially competed with CUX1 consensus oligonucleotide, IVT p110 or p200 CUX1 isoforms did not interact strongly with PR1 probe. Protein complexes generated with PR2 were not destabilized with FOXF2 consensus oligonucleotide. Both HOXB7 and BARX2 consensus oligonucleotides partially competed PR2 protein complexes, however, no interaction was observed upon addition of BARX2-specific antibody and IVT HOXB7 did not complex with PR2 probe. These data suggested that further investigation of the interaction of these factors with PR1 and PR2 was not warranted.
An in vitro interaction of CDX2 with PR3
Of the seven factors predicted to bind PR3 (Table 2), CDX2 was pursued due to its probable functional importance, and our previous work showing an in vitro interaction of CDX2 with other cis-regulatory elements in CFTR [31]. EMSA experiments using PR3 as a probe and Caco2 nuclear extracts demonstrated multiple interacting proteins (Fig. 2E). Competition with excess PR3 showed these complexes were specific (Fig. 2E, lanes 3-4). A CDX2 consensus oligonucleotide was used in competition experiments to reveal which complex, if any, included CDX2 protein. A fast migrating complex, was specifically competed in these experiments (Fig. 2E, lanes 6-7, complex i). Competition with excess unlabeled PR3 oligonucleotide containing a mutation in the CDX2 consensus binding site did not destabilize complex i (Fig. 2E, lane 5). Moreover, addition of an antibody specific for CDX2 to the EMSA reaction caused a supershift of complex i (Fig. 2E, lane 10, complex iii). IVT CDX2 generated an EMSA profile with PR3 that supported the characterization of complex i (Fig. 2F): a single complex was formed (v) at the mobility predicted from complex i (Fig. 2E), and this supershifted with CDX2-specific antibody (Fig. 2F lane 9, complex vi).
Since multiple protein complexes were seen with PR3 and Caco2 nuclear extracts, and it is known that HNF1 and CDX2 co-regulate the transcription of many genes [23-30], we next evaluated the potential contribution of HNF1 to these complexes, despite lack of a predicted binding site for this factor in PR3. Interestingly, upon competition with excess HNF1 consensus oligonucleotide, complex ii is competed more effectively than with the CDX2 consensus (Fig. 2E, lane 9 compared to lane 7), while complex i is only partially inhibited (Fig. 2E, lanes 8-9). Additionally, the supershift that is caused by interaction of an antibody specific to HNF1 appears to be mainly from complex ii (Fig. 2E, lane 11, complex iv). Interestingly IVT HNF1α, alone or in combination with IVT CDX2, did not generate protein complexes with PR3 (data not shown), suggesting that though HNF1 contributes to the complex binding at PR3 in vivo, it requires additional partners in a multi-protein complex to form stable interactions.
Mutation of FOXA, HNF1, or CDX2 consensus binding sites reduces the activity of the intron 11 DHS enhancer element
Using reporter constructs in which the CFTR basal promoter drives luciferase expression, we evaluated the contribution of the FOXA, HNF1 and CDX2 motifs in PRs 1-3 to the enhancer activity of the intron 11 DHS element reported previously [9]. The consensus motifs in the full-length DHS 11 element were destroyed by site-directed mutagenesis, constructs were transiently transfected into Caco2 cells, and luciferase activity was measured (Fig. 3). Disruption of the FOXA consensus site core sequence in PR1 reduced enhancer activity by 34%, while mutation of the predicted HNF1 and CDX2 site cores, in PR2 and PR3 respectively, reduced enhancer activity by over 60%. When both the HNF1 and CDX2 sites were mutated in the same construct, an 87% reduction of activity was observed. These data suggest that FOXA1/A2, HNF1, and CDX2 contribute to the enhancer activity of the DHS11 element.
Figure 3. FOXA, HNF1, and CDX2 sites contribute to the enhancer activity of the DHS11 region in vitro.
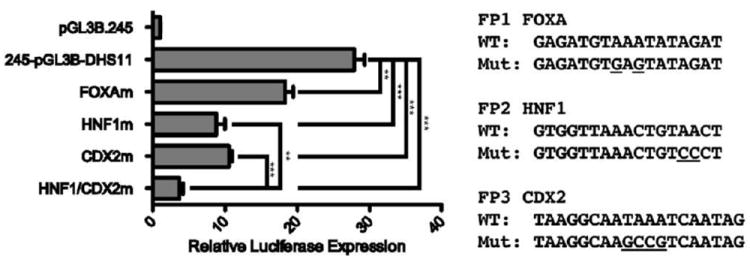
Caco2 cells were transfected with pGL3B luciferase reporter vectors containing the 787 bp basal CFTR promoter (PROM) and the 1.5 kb DHS11 region cloned into the enhancer site of the vector. Mutations were made in FOXA, HNF1, or CDX2 predicted consensus sequences, alone or in combination. Renilla luciferase vector was used as a transfection efficiency control. Luciferase expression is shown relative to the CFTR basal promoter-only vector. Error bars represent SEM (n=9). ** denotes P<0.01; *** denotes P<0.001 comparing the mutant vectors to the wild-type DHS11-containing vector using an unpaired t-test.
FOXA1/A2 interact in vivo with CFTR cis-regulatory elements
The interaction of FOXA1 and FOXA2 with the CFTR locus in vivo was investigated by chromatin immunoprecipitation (ChIP), followed by SYBR Green qPCR (Fig. 4). PCR primer sets were within multiple defined cis-regulatory enhancer elements across the CFTR locus and other non-regulatory regions (Table S1). In Caco2 cells, the DHS element in intron 11 was the highly enriched for both FOXA1 and FOXA2 (~4-8-fold relative to IgG). These factors were also similarly enriched at nearby cis-regulatory elements that lack enhancer activity at DHS in intron 10 (DHS10a,b, 1716 + 13.2 kb/13.7 kb) [9], and slightly enriched (~2-fold) at other CFTR cis-regulatory elements, including those in introns 1 (185 + 10 kb) and 23 (4374 + 1.3 kb) (FOXA2 only) [14, 42]. FOXA2 was enriched at many of the same regions in HT29 cells, an additional intestinal cell line that expresses similar amounts of CFTR [9] and FOXA2 (Fig. S2A-B). No FOXA2 enrichment was observed in the CFTR (-) skin fibroblasts (Fig. S2C). These data indicate that FOXA proteins interact directly and/or indirectly at many locations across the CFTR locus in intestinal cells. The most prominent interactions are at regions of open chromatin in introns 10 and 11, close to the center of the locus.
Figure 4. FOXA1/A2 interact with CFTR regulatory elements in vivo.
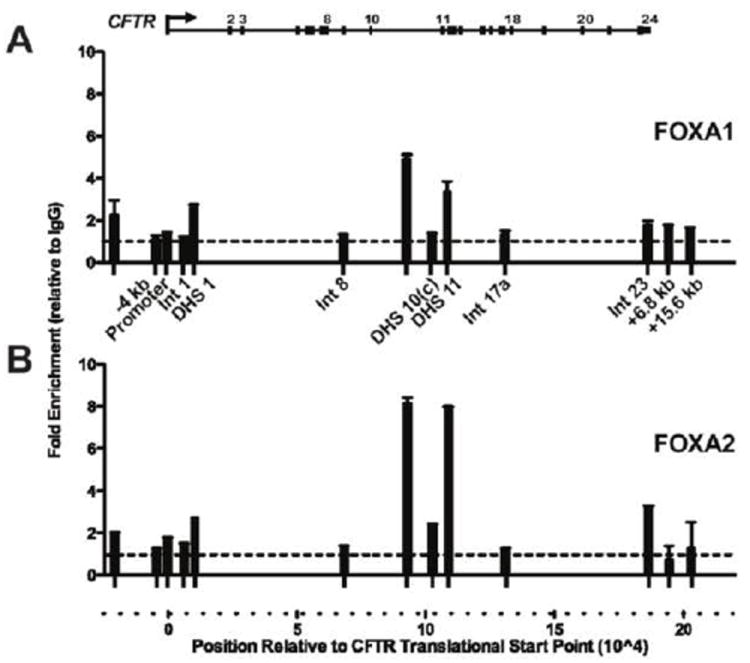
ChIP with FOXA1 (A) or FOXA2 (B) specific antibodies and Caco2 chromatin. Primer sets spanning the CFTR locus (Table S1) were used to assay enrichment by SYBR Green qPCR analysis. Results were normalized to 18s rRNA levels, and compared to goat IgG enrichment at the same site (dotted line). Error bars represent SEM of two PCR reactions per primer set. Data are shown from a representative ChIP experiment though consistent results were obtained in three independent experiments.
HNF1 interacts in vivo with CFTR regulatory elements
We previously showed that HNF1 is necessary for normal CFTR expression levels in the mouse intestinal epithelium [20] and interacts with multiple cis-regulatory elements across the CFTR locus [9]. These interactions were confirmed by ChIP in the current series of experiments (Fig. S3A). We next used a siRNA approach to efficiently knockdown 90% of endogenous HNF1α protein in Caco2 cells, but this did not impact levels of CFTR mRNA expression as measured by qRT-PCR (Fig. S3B-D).
CDX2 interacts in vivo with CFTR regulatory elements, and contributes to CFTR expression in intestinal epithelial cells
Next, we used ChIP followed by qPCR to investigate the interaction of CDX2 with the CFTR locus in Caco2 cells (Fig. 5A). Enrichment (~ 9 fold) of CDX2 was seen at the intron 11 element, and at equivalent levels at DHS elements in intron 1 and intron 10 (DHS10c, 1716 + 23 kb). A much higher enrichment (~23-fold) of CDX2 was seen at the DHS10a,b cis-elements. Moreover, slight enrichment of CDX2 (4-9 fold) was also evident at well characterized enhancer-blocking insulator elements associated with the -20.9 kb DHS upstream of the CFTR translational start site and two others at +6.8 kb and +15.6 kb downstream from the CFTR translational stop site and at the CFTR promoter. Thus, CDX2 associates with CFTR in vivo at multiple genomic locations.
Figure 5. CDX2 interacts with CFTR regulatory elements and contributes to CFTR expression in vivo.
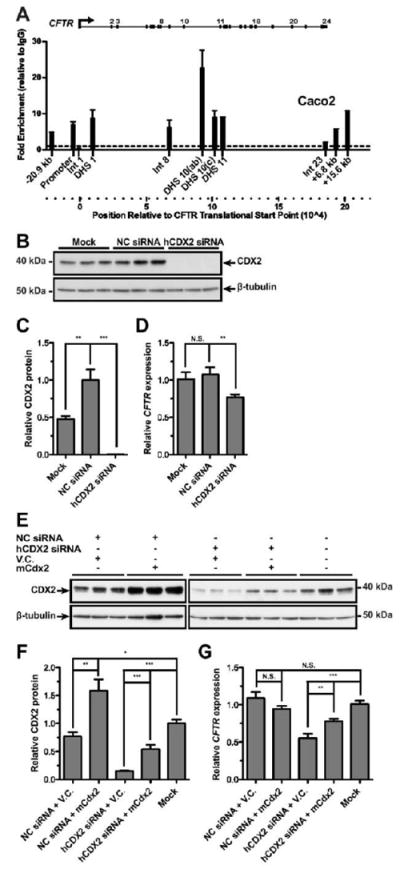
(A) Caco2 ChIP with CDX2-specific antibody; enrichment detected and calculated as in Fig. 4, relative to rabbit IgG. (B-D) Caco2 cells were reverse transfected for 48h with negative control (NC) siRNA, siRNA targeting human CDX2, or transfection reagent only (mock). (E-G) Caco2 cells were forward transfected for 48h with combinations of NC siRNA, hCDX2 siRNA, empty pRC-CMV vector (V.C.), or pRC-mCdx2 vector (mCdx2). (B and E) Western blots of whole cell lysate for total CDX2, or β-tubulin for loading control. CDX2 protein levels were quantified from three independent experiments; results were normalized to β-tubulin, and are shown relative to the NC sample (C) or mock transfected (F). CFTR mRNA levels measured by Taqman qRT-PCR from total RNA. Results were normalized to 18s rRNA levels, and are shown relative to the NC siRNA transfected (D) or mock transfected (G) cells. Error bars represent SEM, n=8 or 9; *** denotes P<0.001, ** denotes P<0.01, N.S. denotes not significant, using an unpaired t-test.
To determine the functional relevance of these CDX2 interactions within the CFTR locus we used a pool of three siRNAs to knockdown CDX2 protein. Fig. 5B-C show that the CDX2-targeted siRNAs efficiently depleted CDX2 in Caco2 cells. Knockdown of CDX2 caused CFTR expression levels to fall by approximately 25% in the same cells (Fig. 5D). Overexpression of mouse Cdx2 upon knockdown of endogenous CDX2 protein in Caco2 cells partially restored CDX2 protein levels (Fig. 5E-F). This resulted in a comparable increase in CFTR expression (Fig. 5G). These data confirm the important role of CDX2 in achieving maximal CFTR expression in Caco2 intestinal epithelial cells.
DISCUSSION
The ability to map regions of open chromatin genome-wide has the potential to rapidly identify novel regulatory cis-acting regulatory elements. However, confirming the function of these elements and identifying trans-acting factors remains challenging. We previously identified multiple cis-acting regulatory elements for the CFTR gene, located within open chromatin. In an effort to identify transcriptional networks that coordinate expression of this epithelial chloride channel in the intestine, we now investigate the transcription factors that bind to an intestine-selective enhancer in intron 11 of the gene. We identify FOXA1/A2, HNF1, and CDX2 as important factors that interact with this element, and other cis-regulatory sequences across the locus. Moreover, we show by mutation of their predicted binding sites and siRNA knockdown of CDX2 that these factors are critical for the maintenance of high levels of CFTR mRNA in intestinal epithelial cells.
FOXA1/A2, HNF1, and CDX2 are key regulators of gene expression in endoderm-derived tissues [16, 17, 22, 43], including many that express CFTR, such as the intestine, pancreas and airway. These factors, and others such as GATAs 4-6, HNF4α and CDX1, comprise a transcriptional network that co-regulates the expression of many genes in the intestinal epithelium [23-30]. In the case of CFTR, the recruitment of FOXA1/A2, HNF1, and CDX2 to multiple intestinal cis-regulatory elements may coordinate their interaction with the gene promoter by altering local chromatin structure. TF binding and subsequent chromatin modifications may facilitate looping of the locus [9] either directly, or indirectly through recruitment of additional chromatin remodelers such as p300 [44-46].
Members of the forkhead box (FOX) family of TFs regulate chromatin remodeling through their activity as pioneer factors and classic TFs [47]. To date, FOXA, FOXE, and FOXO classes have been described as pioneer factors due to their ability to bind within condensed chromatin, an environment from which many other TFs are excluded. The bound FOX factors locally remodel chromatin to provide access to other TFs, for example by their SWI/SNF-independent histone remodeling activity, which displaces H1-linker histones (reviewed in [18]). Though not yet identified as a pioneer factor, FOXI1 repressed the CFTR promoter in a transgene construct introduced into a vas deferens cell line, and this repression was greater in CFTR promoter variants associated with congenital bilateral absence of the vas deferens (CBAVD) [48]. In contrast, our data suggest that FOXA factors positively regulate CFTR expression through binding to intronic enhancers, including the one in intron 11. FOXA proteins may remodel chromatin at these sites, facilitating the recruitment of other transcription factors and co-factors.
Our data also demonstrate that HNF1 is one of the TFs that populate the remodeled chromatin, along with p300 with which it interacts [44]. Though we previously showed enrichment of both factors in vivo at sites across the CFTR locus in intestinal cells [9], we now identify a critical HNF1 binding site in the intron 11 enhancer. To further examine the in vivo role of HNF1, we used a pool of three siRNAs to knockdown HNF1α in Caco2 cells. Though 90% knockdown was achieved as estimated by western blot, this had no impact on CFTR mRNA expression levels. This observation is consistent with our previous data showing that an antisense HNF1α ribozyme, which stably depleted HNF1α was ineffective in reducing CFTR expression in Caco2 cells but inhibited CFTR in Capan-1 pancreatic adenocarcinoma cells, where endogenous levels of HNF1α are much lower [20]. In Caco2 cells the small amount of HNF1α protein persisting after knockdown may be sufficient to support HNF1-mediated CFTR expression. Moreover, the low levels of HNF1β in Caco2 cells may compensate for the depleted HNF1α, since both factors bind the same consensus sequence.
CDX2 can interact with different classes of the SWI/SNF chromatin-remodeling complex to regulate expression of target genes. In mouse blastocysts, Cdx2 and Brg1 physically interact to repress Oct4 expression [49]. In contrast, villin expression in human intestinal cells is dependent on CDX-mediated targeting of the Brm-type SWI/SNF complex to the promoter [50]. We demonstrate here that CDX2 is enriched in vivo at the intron 11 enhancer and at other cis-regulatory elements across the CFTR locus in Caco2 cells. Moreover, effective siRNA inhibition of CDX2 significantly reduced CFTR mRNA expression levels in these cells. ChIP-chip, ChIP-seq and further data from other groups support an important role for CDX2 in cis-regulatory element-mediated control of CFTR in Caco2 cells [51-54]. Though the siRNA depleted more than 95% of CDX2 protein, we only observed a 25% reduction in CFTR expression. However, as multiple cis-regulatory elements interact with the CFTR promoter and recruit diverse factors, loss of a single factor would be unlikely to totally destroy CFTR promoter activity. Moreover, while the intron 11 enhancer and other cis-regulatory elements contribute to the high level of CFTR expression observed in the intestinal epithelium, previous evidence suggests that loss of a single enhancer does not abolish CFTR expression [55].
The more global role for transcriptional networks that are coordinated by FOXA1/A2, HNF1 and CDX2 in epithelial function warrants further discussion. Foxa1 was recently implicated in the regulation of genes encoding other ion transporters in the sweat glands of mice [56]. These transporters, Na-K-Cl cotransporter 1 (Nkcc1/Slc12a2) and Bestrophin 2 (Best2) are also expressed in the intestine and colon, where the Best2 anion channel has been shown to regulate bicarbonate transport [57, 58]. In Caco2 cells, multiple sites of CDX2 enrichment are evident across SLC12A2 and the SLC26A3 locus [51-53], which encodes a Cl-/HCO3- exchanger. Moreover, HNF1 regulates the expression of SLC26A3 in the intestinal epithelium [59] and CFTR and SLC26A3 are thought to functionally interact to regulate HCO3- secretion [60]. These data together with ours reported here suggest that this group of transcription factors may play a more general role in regulating expression of ion transporters in the intestine. It is probable that other TFs such as GATAs 4-6 and HNF4α play a critical role in these networks [52].
The involvement of multiple transcriptional networks in coordinating the expression of ion channel/transporter genes has implications for the nuclear organization on which this depends. It is possible that the critical TFs reside in specialized transcription factories that efficiently coordinate high levels of gene expression. However, the mechanism whereby genes would be selected for targeting to these sites and migrate there remains unclear (reviewed in [61, 62]). It is known that CFTR associates with different nuclear regions depending on its expression status [63]. The locus moves to the nuclear interior when it is expressed and this relocation might enable interaction with specialized transcription factories. In intestinal cells these transcription factories would probably be enriched for HNF1, CDX2, FOXA1/A2, and other members of the same networks.
Supplementary Material
Acknowledgments
We thank Drs. R. Cortese (CEINGE, Naples, Italy), R. Matusik (Vanderbilt University, Nashville, TN, U.S.A.), and A. Nepveu (McGill University, Montreal, QC, Canada) for their kind donation of plasmids and antibody reagents, Dr. C. Ott for generating the HNF1α expression plasmid.
FUNDING
This work was supported by the Cystic Fibrosis Foundation Harris11G0; National Institutes of Health [R01HL094585 and R01HD68901 to A.H.]; and the Northwestern University Cellular and Molecular Basis of Disease Training Grant to J.L.K.
Abbreviations used
- 3C
chromosome conformation capture
- Best
bestrophin 1
- CBAVD
congenital bilateral absence of the vas deferens
- CDX2
caudal type homeobox 2
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- ChIP
chromatin immunoprecipitation
- CR
conserved region
- DHS
DNase I hypersensitive site
- EMSA
electrophoretic mobility shift assay
- FOXA1/A2
forkhead box A1/A2
- HNF1
hepatocyte nuclear factor 1
- IVT
in vitro translated
- NC
negative control
- NE
nuclear extract
- NkccI/Slc12a2
Na-K-Cl cotransporter 1
- PR
protected region
- qPCR
quantitative PCR
- qRT-PCR
quantitative reverse transcription PCR
- TF
transcription factor
- TFBS
transcription factor binding site
References
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 3.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, Liu Z, London D, McDaniell RM, Shibata Y, Showers KA, Simon JM, Vales T, Wang T, Winter D, Zhang Z, Clarke ND, Birney E, Iyer VR, Crawford GE, Lieb JD, Furey TS. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 9.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci U S A. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy VA, Harris A. The CFTR gene and regulation of its expression. Pediatr Pulmonol. 2005;40:1–8. doi: 10.1002/ppul.20199. [DOI] [PubMed] [Google Scholar]
- 11.Mulberg AE, Weyler RT, Altschuler SM, Hyde TM. Cystic fibrosis transmembrane conductance regulator expression in human hypothalamus. Neuroreport. 1998;9:141–144. doi: 10.1097/00001756-199801050-00028. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimura K, Nakamura H, Trapnell BC, Chu CS, Dalemans W, Pavirani A, Lecocq JP, Crystal RG. Expression of the cystic fibrosis transmembrane conductance regulator gene in cells of non-epithelial origin. Nucleic Acids Res. 1991;19:5417–5423. doi: 10.1093/nar/19.19.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies WL, Vandenberg JI, Sayeed RA, Trezise AE. Cardiac expression of the cystic fibrosis transmembrane conductance regulator involves novel exon 1 usage to produce a unique amino-terminal protein. J Biol Chem. 2004;279:15877–15887. doi: 10.1074/jbc.M313628200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Ott CJ, Lewandowska MA, Leir SH, Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J Cell Mol Med. 2011 doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillen AE, Harris A. Transcriptional regulation of CFTR gene expression. Front Biosci (Elite Ed) 2012;4:587–592. doi: 10.2741/401. [DOI] [PubMed] [Google Scholar]
- 16.Lai E, Prezioso VR, Smith E, Litvin O, Costa RH, Darnell JE., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- 17.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 20.Mouchel N, Henstra SA, McCarthy VA, Williams SH, Phylactides M, Harris A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 23.Balakrishnan A, Stearns AT, Rhoads DB, Ashley SW, Tavakkolizadeh A. Defining the transcriptional regulation of the intestinal sodium-glucose cotransporter using RNA-interference mediated gene silencing. Surgery. 2008;144:168–173. doi: 10.1016/j.surg.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudreau F, Rings EH, van Wering HM, Kim RK, Swain GP, Krasinski SD, Moffett J, Grand RJ, Suh ER, Traber PG. Hepatocyte nuclear factor-1 alpha, GATA-4, and caudal related homeodomain protein Cdx2 interact functionally to modulate intestinal gene transcription. Implication for the developmental regulation of the sucrase-isomaltase gene. J Biol Chem. 2002;277:31909–31917. doi: 10.1074/jbc.M204622200. [DOI] [PubMed] [Google Scholar]
- 25.Escaffit F, Boudreau F, Beaulieu JF. Differential expression of claudin-2 along the human intestine: Implication of GATA-4 in the maintenance of claudin-2 in differentiating cells. J Cell Physiol. 2005;203:15–26. doi: 10.1002/jcp.20189. [DOI] [PubMed] [Google Scholar]
- 26.Mitchelmore C, Troelsen JT, Spodsberg N, Sjostrom H, Noren O. Interaction between the homeodomain proteins Cdx2 and HNF1alpha mediates expression of the lactase-phlorizin hydrolase gene. Biochem J. 2000;346(Pt 2):529–535. [PMC free article] [PubMed] [Google Scholar]
- 27.Valente AJ, Zhou Q, Lu Z, He W, Qiang M, Ma W, Li G, Wang L, Banfi B, Steger K, Krause KH, Clark RA, Li S. Regulation of NOX1 expression by GATA, HNF-1alpha, and Cdx transcription factors. Free Radic Biol Med. 2008;44:430–443. doi: 10.1016/j.freeradbiomed.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 28.Fang R, Olds LC, Sibley E. Spatio-temporal patterns of intestine-specific transcription factor expression during postnatal mouse gut development. Gene Expr Patterns. 2006;6:426–432. doi: 10.1016/j.modgep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Klopot A, Freund JN, Dowling LN, Krasinski SD, Fleet JC. Control of differentiation-induced calbindin-D9k gene expression in Caco-2 cells by cdx-2 and HNF-1alpha. Am J Physiol Gastrointest Liver Physiol. 2004;287:G943–953. doi: 10.1152/ajpgi.00121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonckheere N, Vincent A, Perrais M, Ducourouble MP, Male AK, Aubert JP, Pigny P, Carraway KL, Freund JN, Renes IB, Van Seuningen I. The human mucin MUC4 is transcriptionally regulated by caudal-related homeobox, hepatocyte nuclear factors, forkhead box A, and GATA endodermal transcription factors in epithelial cancer cells. J Biol Chem. 2007;282:22638–22650. doi: 10.1074/jbc.M700905200. [DOI] [PubMed] [Google Scholar]
- 31.McCarthy VA, Ott CJ, Phylactides M, Harris A. Interaction of intestinal and pancreatic transcription factors in the regulation of CFTR gene expression. Biochim Biophys Acta. 2009;1789:709–718. doi: 10.1016/j.bbagrm.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AN, Barth ML, McDowell TL, Moulin DS, Nuthall HN, Hollingsworth MA, Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J Biol Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 33.Phylactides M, Rowntree R, Nuthall H, Ussery D, Wheeler A, Harris A. Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. Eur J Biochem. 2002;269:553–559. doi: 10.1046/j.0014-2956.2001.02679.x. [DOI] [PubMed] [Google Scholar]
- 34.Truscott M, Denault JB, Goulet B, Leduy L, Salvesen GS, Nepveu A. Carboxyl-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J Biol Chem. 2007;282:30216–30226. doi: 10.1074/jbc.M702328200. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, Gupta A, Wang Y, Suzuki K, Mirosevich J, Orgebin-Crist MC, Matusik RJ. Foxa1 and Foxa2 interact with the androgen receptor to regulate prostate and epididymal genes differentially. Ann N Y Acad Sci. 2005;1061:77–93. doi: 10.1196/annals.1336.009. [DOI] [PubMed] [Google Scholar]
- 36.Shen WF, Largman C, Lowney P, Corral JC, Detmer K, Hauser CA, Simonitch TA, Hack FM, Lawrence HJ. Lineage-restricted expression of homeobox-containing genes in human hematopoietic cell lines. Proc Natl Acad Sci U S A. 1989;86:8536–8540. doi: 10.1073/pnas.86.21.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon NS, Premdas P, Truscott M, Leduy L, Berube G, Nepveu A. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol Cell Biol. 2001;21:6332–6345. doi: 10.1128/MCB.21.18.6332-6345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mouchel N, Broackes-Carter F, Harris A. Alternative 5’ exons of the CFTR gene show developmental regulation. Hum Mol Genet. 2003;12:759–769. doi: 10.1093/hmg/ddg079. [DOI] [PubMed] [Google Scholar]
- 39.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 42.Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo CJ, Conley PB, Hsieh CL, Francke U, Crabtree GR. Molecular cloning, functional expression, and chromosomal localization of mouse hepatocyte nuclear factor 1. Proc Natl Acad Sci U S A. 1990;87:9838–9842. doi: 10.1073/pnas.87.24.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ban N, Yamada Y, Someya Y, Miyawaki K, Ihara Y, Hosokawa M, Toyokuni S, Tsuda K, Seino Y. Hepatocyte nuclear factor-1alpha recruits the transcriptional co-activator p300 on the GLUT2 gene promoter. Diabetes. 2002;51:1409–1418. doi: 10.2337/diabetes.51.5.1409. [DOI] [PubMed] [Google Scholar]
- 45.Hussain MA, Habener JF. Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J Biol Chem. 1999;274:28950–28957. doi: 10.1074/jbc.274.41.28950. [DOI] [PubMed] [Google Scholar]
- 46.Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene. 2005;24:8277–8290. doi: 10.1038/sj.onc.1208991. [DOI] [PubMed] [Google Scholar]
- 47.Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 48.Lopez E, Viart V, Guittard C, Templin C, Rene C, Mechin D, Des Georges M, Claustres M, Romey-Chatelain MC, Taulan M. Variants in CFTR untranslated regions are associated with congenital bilateral absence of the vas deferens. J Med Genet. 2011;48:152–159. doi: 10.1136/jmg.2010.081851. [DOI] [PubMed] [Google Scholar]
- 49.Wang K, Sengupta S, Magnani L, Wilson CA, Henry RW, Knott JG. Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS One. 2010;5:e10622. doi: 10.1371/journal.pone.0010622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamichi N, Inada K, Furukawa C, Sakurai K, Tando T, Ishizaka A, Haraguchi T, Mizutani T, Fujishiro M, Shimomura R, Oka M, Ichinose M, Tsutsumi Y, Omata M, Iba H. Cdx2 and the Brm-type SWI/SNF complex cooperatively regulate villin expression in gastrointestinal cells. Exp Cell Res. 2009;315:1779–1789. doi: 10.1016/j.yexcr.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Verzi MP, Hatzis P, Sulahian R, Philips J, Schuijers J, Shin H, Freed E, Lynch JP, Dang DT, Brown M, Clevers H, Liu XS, Shivdasani RA. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc Natl Acad Sci U S A. 2010;107:15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyd M, Hansen M, Jensen TG, Perearnau A, Olsen AK, Bram LL, Bak M, Tommerup N, Olsen J, Troelsen JT. Genome-wide analysis of CDX2 binding in intestinal epithelial cells (Caco-2) J Biol Chem. 2010;285:25115–25125. doi: 10.1074/jbc.M109.089516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paul T, Li S, Khurana S, Leleiko NS, Walsh MJ. The epigenetic signature of CFTR expression is co-ordinated via chromatin acetylation through a complex intronic element. Biochem J. 2007;408:317–326. doi: 10.1042/BJ20070282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rowntree RK, Vassaux G, McDowell TL, Howe S, McGuigan A, Phylactides M, Huxley C, Harris A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum Mol Genet. 2001;10:1455–1464. doi: 10.1093/hmg/10.14.1455. [DOI] [PubMed] [Google Scholar]
- 56.Cui CY, Childress V, Piao Y, Michel M, Johnson AA, Kunisada M, Ko MS, Kaestner KH, Marmorstein AD, Schlessinger D. Forkhead transcription factor FoxA1 regulates sweat secretion through Bestrophin 2 anion channel and Na-K-Cl cotransporter 1. Proc Natl Acad Sci U S A. 2012;109:1199–1203. doi: 10.1073/pnas.1117213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Payne JA, Xu JC, Haas M, Lytle CY, Ward D, Forbush B., 3rd Primary structure, functional expression, and chromosomal localization of the bumetanide-sensitive Na-K-Cl cotransporter in human colon. J Biol Chem. 1995;270:17977–17985. doi: 10.1074/jbc.270.30.17977. [DOI] [PubMed] [Google Scholar]
- 58.Qu Z, Hartzell HC. Bestrophin Cl- channels are highly permeable to HCO3. Am J Physiol Cell Physiol. 2008;294:C1371–1377. doi: 10.1152/ajpcell.00398.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Angelo A, Bluteau O, Garcia-Gonzalez MA, Gresh L, Doyen A, Garbay S, Robine S, Pontoglio M. Hepatocyte nuclear factor 1alpha and beta control terminal differentiation and cell fate commitment in the gut epithelium. Development. 2010;137:1573–1582. doi: 10.1242/dev.044420. [DOI] [PubMed] [Google Scholar]
- 60.Greeley T, Shumaker H, Wang Z, Schweinfest CW, Soleimani M. Downregulated in adenoma and putative anion transporter are regulated by CFTR in cultured pancreatic duct cells. Am J Physiol Gastrointest Liver Physiol. 2001;281:G1301–1308. doi: 10.1152/ajpgi.2001.281.5.G1301. [DOI] [PubMed] [Google Scholar]
- 61.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Razin SV, Gavrilov AA, Pichugin A, Lipinski M, Iarovaia OV, Vassetzky YS. Transcription factories in the context of the nuclear and genome organization. Nucleic Acids Res. 2011;39:9085–9092. doi: 10.1093/nar/gkr683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


