Abstract
Paf1 complex (Paf1C) is a transcription elongation factor whose recruitment is stimulated by Spt5 and the CDKs Kin28 and Bur1, which phosphorylate the Pol II C-terminal domain (CTD) on Serines 2, 5, and 7. Bur1 promotes Paf1C recruitment by phosphorylating C-terminal repeats (CTRs) in Spt5, and we show that Kin28 enhances Spt5 phosphorylation by promoting Bur1 recruitment. It was unclear, however, whether CTD phosphorylation by Kin28 or Bur1 also stimulates Paf1C recruitment. We find that Paf1C and its Cdc73 subunit bind diphosphorylated CTD repeats (pCTD) and phosphorylated Spt5 CTRs (pCTRs) in vitro, and that cdc73 mutations eliminating both activities reduce Paf1C recruitment in vivo. Phosphomimetic (acidic) substitutions in the Spt5 CTR sustain high-level Paf1C recruitment in otherwise wild-type cells, but not following inactivation of Bur1 or Kin28. Furthermore, inactivating the pCTD/pCTR-interaction domain (PCID) in Cdc73 decreases Paf1C-dependent histone methylation in cells containing non-phosphorylatable Spt5 CTRs. These results identify an Spt5 pCTR-independent pathway of Paf1C recruitment requiring Kin28, Bur1, and the Cdc73 PCID. We propose that pCTD repeats and Spt5 pCTRs provide separate interaction surfaces that cooperate to ensure high-level Paf1C recruitment.
Keywords: CTD, Paf1C, Spt5, transcription elongation
Introduction
The Paf1 complex (Paf1C) is an important cofactor for elongating RNA Polymerase II (Pol II), comprised of five subunits: Paf1, Rtf1, Cdc73, Ctr9, and Leo1, which is recruited to the coding sequences (CDS) of actively transcribed genes (Kim et al, 2004; Sims et al, 2004; Mayer et al, 2010). Paf1C orchestrates the co-transcriptional ubiquitylation of histone H2B on Lys-123 and methylation of histone H3 on Lys-4, which requires H2B ubiquitylation, and also the methylation of H3 on Lys-36 and Lys-79 (Sims et al, 2004; Smith and Shilatifard, 2010). Paf1C also promotes correct 3′ end formation by its role in recruiting termination factors (Penheiter et al, 2005; Sheldon et al, 2005; Nordick et al, 2008; Rozenblatt-Rosen et al, 2009; Kim and Levin, 2011), and enhances Pol II recruitment to promoters that depend on activator SBF (Swi4/Swi6) (Kim and Levin, 2011). Thus, efficient Paf1C recruitment is crucial for events occurring throughout the transcription process and spanning the entire CDS; however, the mechanism of Paf1C recruitment by elongating Pol II is not well understood.
Multiple factors have been implicated in Paf1C recruitment, including the FACT, Ccr4-Not, and Spt4/Spt5 (DSIF) complexes, and the cyclin-dependent kinases (Cdks) Kin28 (cyclin Ccl1) and Bur1 (cyclin Bur2) (Laribee et al, 2005; Wood et al, 2005; Pavri et al, 2006; Qiu et al, 2006; Mulder et al, 2007; Liu et al, 2009; Zhou et al, 2009). Kin28 phosphorylates the heptad repeats (Y1S2P3T4S5P6S7) in the C-terminal domain (CTD) of Pol II subunit Rpb1 on Ser5 and Ser7 (Akhtar et al, 2009; Glover-Cutter et al, 2009; Kim et al, 2009), and this activity stimulates recruitment of mRNA processing factors, histone modifying enzymes and elongation factors near the promoter (Phatnani and Greenleaf, 2006; Buratowski, 2009; Govind et al, 2010). As inhibiting Kin28 does not impair Spt4 recruitment, it appears that Spt4/Spt5 acts directly in Paf1C recruitment (Qiu et al, 2006). Indeed, strong evidence was presented that Bur1 stimulates Paf1C recruitment by phosphorylating the C-terminal repeats (CTRs) of Spt5 (Liu et al, 2009; Zhou et al, 2009). Similar to the role of the phosphorylated Pol II CTD in cofactor recruitment, the hexad repeats in Spt5, with consensus sequence S1A2W3G4G5Q6, could provide a platform for Paf1C recruitment when phosphorylated by Bur1. However, it was reported that Paf1C does not bind directly to phosphorylated Spt4/Spt5 complex (Liu et al, 2009), making it unclear how Bur1 and Spt5 promote Paf1C recruitment. Moreover, the reduction in Paf1C recruitment evoked by inhibiting Bur1 exceeded that of Ala substitutions of the Ser1 residue in all 15 hexad repeats of Spt5 (Liu et al, 2009), raising the possibility that Bur1 promotes Paf1C recruitment by an Spt5-independent pathway.
In fact, we and others have shown that Bur1 also phosphorylates Ser2 of the Pol II CTD (S2P) in vivo, augmenting the activity of the major, but non-essential, S2P kinase Ctk1 (Liu et al, 2009; Qiu et al, 2009). We found that Bur1 is recruited to the 5′ end of the CDS at ARG1 in a manner stimulated by Kin28 and the pCTD-interaction domain (PCID) in the Bur1 C-terminus. Moreover, Bur1 contributed significantly to S2P formation near the ARG1 promoter and in the fraction of bulk elongating Pol II highly enriched in Ser5P (S5P), while Ctk1 was responsible for the majority of S2P at promoter-distal ARG1 sequences and in bulk Pol II (Qiu et al, 2009). An auxiliary role for Bur1 in S2P formation in the CDS was also detected by genome-wide measurements of S2P occupancy (Tietjen et al, 2010; Bataille et al, 2012). Interestingly, Tietjen et al (2010) further implicated Bur1 in S7P formation at sites distal from the promoter; although Bataille et al (2012) found that Bur1’s contribution to S7P was most pronounced under conditions of Kin28 inhibition. Thus, it appears that Bur1 contributes to the formation of both Ser2P and Ser7P in elongating Pol II.
Considering our previous finding that Kin28 promotes Bur1 recruitment (Qiu et al, 2009), it was possible that the stimulatory role of Kin28 in Paf1C recruitment (Qiu et al, 2006) merely reflects the positive effect of Kin28 on Bur1 recruitment and attendant phosphorylation of Spt5 CTRs. We set out to test the alternative hypothesis that Kin28 acts more directly by phosphorylating the Pol II CTD on Ser5 and Ser7 to provide an alternative platform for Paf1C recruitment. In addition to phosphorylating the CTD directly, Kin28 could also cooperate with Bur1 to produce S2,S5- or S5,S7-diphosphorylated CTD repeats, and this collaboration with Bur1 in CTD phosphorylation, as well as Spt5 CTR phosphorylation by Bur1, would be enhanced by Kin28’s stimulatory role in Bur1 recruitment. Our results indicate that Paf1C binds specifically to both types of diphosphorylated CTD repeats and to phosphorylated Spt5 CTRs (pCTRs) in vitro, and they provide strong evidence that Kin28 and Bur1 can promote Paf1C recruitment independently of Spt5 CTR phosphorylation. Accordingly, we envision a dual mechanism of Paf1C recruitment via diphosphorylated Pol II CTD repeats and Spt5 pCTRs, with both pathways stimulated by Kin28 and Bur1.
Results
Paf1C can bind directly to phosphorylated Rpb1 CTD repeats
Considering that Paf1C recruitment is stimulated by both Kin28 and Bur1 (Laribee et al, 2005; Qiu et al, 2006; Liu et al, 2009), and the evidence cited above that these Cdks phosphorylate the Pol II CTD on Ser5 and Ser7 (Kin28) or Ser2 and Ser7 (Bur1), we investigated whether Paf1C can bind directly to Pol II CTD repeats in a manner stimulated by S2P, S5P, or S7P. To this end, Paf1C was purified from a CDC73-TAP strain through the first step of tandem affinity purification, that is, prior to cleavage of the calmodulin binding domain (CBD) (Puig et al, 2001; see Supplementary Figure S1A), and tested for binding to biotinylated peptides comprised of three CTD repeats, either unphosphorylated, monophosphorylated at Ser2, Ser5, or Ser7, or diphosphorylated at either Ser2,Ser5 or Ser5,Ser7, in all three repeats. (Note that S2P,S7P-diphosphorylated peptides were not successfully synthesized.) Cdc73-CBD and co-purifying Rtf1 show no binding to unphosphorylated peptides, little or no binding to S2P and S7P peptides, and relatively weak binding to S5P peptides (Figure 1A). Remarkably, the purified Paf1C binds much more strongly to both S2P,S5P and S5P,S7P peptides, with consistently slightly greater binding for the S2P,S5P peptides (Figure 1A, lanes 1–25 and data not shown). As expected if Cdc73-CBD and Rtf1 bind as subunits of the same complex, the extent of depletion of Cdc73-CBD in the supernatant (S) fraction using S2P,S5P peptides relative to the ‘mock’ no-peptide reaction was comparable to that observed for Rtf1 (Figure 1A, cf. lane 14 versus 10 and 25 versus 17). (The depletion of signal in the supernatant does not equal the increase in signal in the bound fraction (B) because a larger proportion of the latter was examined.) These findings suggest that native Paf1C interacts with CTD repeats in a manner stimulated moderately by S5P and strongly by S2,S5 or S5,S7 diphosphorylation.
Figure 1.
Paf1C binds to phosphorylated CTD peptides. (A, B) Biotinylated CTD peptides (1.5 μg) phosphorylated on Ser2 (S2P), Ser5 (S5P), Ser7 (S7P), Ser2 and Ser5 (S2P S5P), Ser5 and Ser7 (S5P S7P), unphosphorylated (CTD) were adsorbed to streptavidin-coated magnetic beads. Native Paf1C purified from a CDC73-TAP strain was incubated with beads alone (Mock) or beads bearing peptides at 4°C. 1/40 input (Inp), 1/40 supernatants (S) and bound (B) proteins were subjected to Western analysis with anti-TAP and anti-Rtf1 antibodies. For lanes 24–29, immobilized (S2P S5P) peptides were treated with CIP in the presence or absence of PhosSTOP prior to incubation with purified Paf1C. (B) As in (A) except CTD peptides with Ser2 and Ser5 substituted by Asp were also used. Lanes 1–3 and 4–7 derive from different portions of the same blot as described in Supplementary Figure S10A. (C, D) As in (A, B) except using the indicated purified recombinant GST fusion proteins (500 ng) in place of Paf1C and anti-GST antibodies in Western analysis. Also, 1/10 input (Inp) and supernatants (S) (1/20 input and supernatant in (D) for GST-Ctr9(594–855) were examined. Figure source data can be found with the Supplementary data.
To confirm that enhanced binding of Paf1C to the phosphorylated peptides is dependent on the phosphate groups, the S2P,S5P peptides were pre-treated with calf intestine phosphatase (CIP), in the absence or presence of phosphatase inhibitor PhosSTOP, before incubation with purified Paf1C. Indeed, specific binding of Cdc73-CBD and Rtf1 to S2P,S5P peptides was abolished by CIP treatment and fully recovered by inclusion of PhosSTOP (Figure 1A, lanes 24, 26, and 28). Similar results were observed for the S5P peptides (Supplementary Figure S2A). Second, we tested purified Paf1C for binding to phosphomimetic peptides containing Asp2 and Asp5 in all three repeats, and observed no detectable binding (Figure 1B). Accordingly, Paf1C binding to phosphorylated CTD repeats likely involves specific recognition of the phosphate dianion in a manner inefficiently mimicked by monoanionic carboxylate groups. The results in Figure 1A and B support the possibility that Paf1C is recruited by Pol II CTD repeats diphosphorylated on Ser5/Ser2 or Ser5/Ser7.
Cdc73, Ctr9, and Rtf1 each contains a putative PCID
In an effort to determine which Paf1C subunits can mediate binding to phosphorylated CTD peptides, GST fusions to each subunit were purified from E. coli and used in binding assays. As the full-length GST-Ctr9 fusion was not well expressed, we analysed fusions containing different segments of Ctr9. Interestingly, GST-Rtf1 binds only to the S2P,S5P peptides. By contrast, the GST fusion containing residues 594–855 of Ctr9 binds weakly to S2P and S5P, but tightly to both kinds of diphosphorylated peptides, with a slight preference for S2P,S5P over S5P,S7P (Figure 1C; Supplementary Figure S2B). The GST-Cdc73 fusion binds to all three peptides harbouring monophosphorylated serines but, similar to the above findings on other recombinant subunits and purified Paf1C, it binds much better to the S2P,S5P- and S5P,S7P-diphosphorylated peptides. Based on the fractions of input bound, and the ratios of signals in the B versus S fractions, recombinant Cdc73 binds more tightly than does Ctr9 or Rtf1 to all peptides to which the latter two subunits can bind (Figure 1C; Supplementary Figure S2B). The fusions to Paf1, Leo1, and three other segments of Ctr9 (1–491, 414–800, and 701–1077) showed no binding to any CTD peptides (Figure 1C and data not shown) using comparable levels of fusion proteins in the binding reactions (Supplementary Figure S1B). As shown above for purified Paf1C, dephosphorylating the S2P,S5P peptides greatly reduced binding by GST-Cdc73, GST-Ctr9(594–855), and GST-Rtf1 in a manner restored with PhosSTOP (Supplementary Figure S2C). Furthermore, GST-Rtf1 and GST-Ctr9(594–855) showed little or no binding to the S2D,S5D phosphomimetic peptides (Figure 1D), and GST-Cdc73 showed reduced binding to the S5D and S2D,S5D peptides compared to the cognate mono- and diphosphorylated peptides (Supplementary Figure S2D).
Together, these findings suggest that Cdc73, Rtf1, and Ctr9 each contains a PCID that binds more tightly to one or both kinds of diphosphorylated CTD repeats than to monophosphorylated repeats and that also prefers dianionic phosphate groups over monoanionic acidic residues. These results can account for the higher affinity of native Paf1C for diphosphorylated versus monophosphorylated repeats (Figure 1A), and they support our hypothesis that Bur1 stimulates Paf1C recruitment, in part, by phosphorylating Ser2 or Ser7 in CTD repeats already phosphorylated on Ser5 by Kin28.
The presence of PCIDs in Cdc73 and Rtf1 is consistent with the known requirements for these two subunits in Paf1C recruitment in vivo (Mueller et al, 2004; Nordick et al, 2008). Given the strong evidence that Rtf1 acts directly in Paf1C recruitment (Warner et al, 2007), we examined whether native Rtf1 can interact with pCTD peptides independently of the other two Paf1C subunits harbouring PCIDs by purifying Rtf1-CBD from an RTF1-TAP cdc73Δ ctr9Δ strain. As Rtf1 association with Paf1C requires Cdc73 (Nordick et al, 2008), the Rtf1-CBD preparation should also be devoid of Paf1 and Leo1. As shown in Supplementary Figure S2E, Rtf1-CBD bound only to the S2P,S5P peptides, and the proportion of input recovered in the bound fraction was considerably smaller than that seen for intact Paf1C (cf. Supplementary Figure S2E, lanes 15–16 versus Figure 1A, lanes 13–14). Thus, while native Rtf1 can interact with S2P,S5P peptides independently of Cdc73 and Ctr9, it appears that Cdc73 or Ctr9 contributes to the relatively stronger pCTD-binding activity of intact Paf1C and also to its interaction with S5P,S7P peptides. Based on the relative affinities of recombinant Cdc73 and Ctr9 for S2P,S5P peptides (Figure 1C), it is likely that Cdc73 makes the greater contribution to Paf1C interaction with this diphosphorylated species of CTD heptad.
Evidence that Paf1C binds directly to phosphorylated Spt5 CTRs
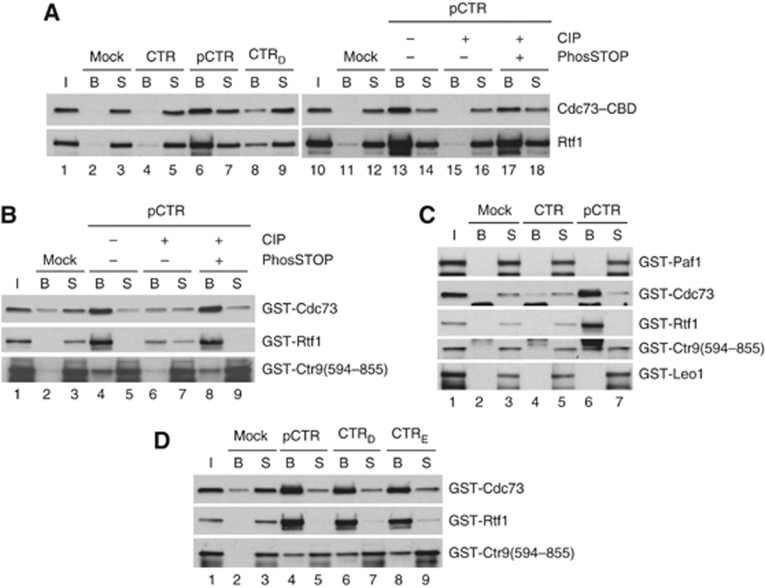
There is strong evidence that Bur1 promotes Paf1C recruitment by phosphorylating Ser1 in the 15 CTRs of Spt5 (Liu et al, 2009; Zhou et al, 2009); however, evidence was lacking for direct interaction of Paf1C with phosphorylated Spt5 (Liu et al, 2009). To determine whether Paf1C can interact with Ser1-phosphorylated CTRs, we conducted peptide binding assays with purified Paf1C and biotinylated peptides consisting of four repeats of the Spt5 consensus sequence S1A2W3G4G5Q6, synthesized with Ser1 unphosphorylated (CTR), phosphorylated (pCTR), or substituted with aspartic acid (CTRD).
The Paf1C purified from the CDC73-TAP strain described above displayed strong, phosphorylation-dependent binding to the Spt5 CTR peptides, binding at high levels to pCTR but not to CTR peptides, and binding in a manner abolished by CIP treatment of pCTR and recovered fully with PhosSTOP (Figure 2A). Moreover, purified Paf1C displayed reduced binding to CTRD versus pCTR peptides (Figure 2A). Similar to our results with Pol II CTD peptides, bacterially expressed GST-Cdc73, GST-Rtf1, and GST-Ctr9(594–855) all bind to pCTR peptides in a manner greatly diminished by CIP treatment and recovered with PhosSTOP (Figure 2B), and show no binding to unphosphorylated CTR peptides (Figure 2C). By contrast, GST-Paf1 and GST-Leo1 do not bind to either pCTR or CTR peptides (Figure 2C). Based on the proportion of input recovered with the pCTR peptides, GST-Cdc73 and GST-Rtf1 bind more tightly than GST-Ctr9(594–855) to pCTR peptides (Figure 2B and C). Consistent with this, Rtf1-CBD purified from cdc73Δ ctr9Δ cells also displays strong binding to pCTR peptides (Supplementary Figure S2F).
Figure 2.
Paf1C binds to phosphorylated Spt5 C-terminal repeats (CTR) peptides. (A–D) Peptide binding assays were conducted as described in Figure 1 using biotinylated CTR peptides (1.5 μg) with Ser1 unphosphorylated (CTR), Ser1 phosphorylated (pCTR), or Ser1 substituted with Asp (CTRD) or Glu (CTRE), and purified native Paf1C or recombinant GST fusion proteins, except using 1/5 or 1/10 input and supernatants of GST-Rtf1 and GST-Ctr9(594–855), respectively. Lanes 10–18 in (A) derive from the same experiment that produced Figure 1B, and the input and Mock lanes from the latter are reproduced here as lanes 10–12. Figure source data can be found with the Supplementary data.
Summarizing the binding data for the recombinant subunits in Figure 1C and Supplementary Figure S2B and C, Cdc73 binds tightly to phosphorylated CTR peptides and to both kinds of diphosphorylated CTD peptides, Rtf1 binds strongly to pCTR but only moderately to S2P,S5P peptides, and Ctr9 binds relatively weakly to both pCTR and both kinds of diphosphorylated CTD peptides. Unlike our findings for purified Paf1C, recombinant GST-Cdc73, GST-Rtf1, and GST-Ctr9(594–855) fragments bound to the phosphomimetic Spt5 CTRD peptides to nearly the same extent as to pCTR peptides (Figure 2D). While this last discrepancy remains to be explained, the results in Figure 2 provide strong evidence that Paf1C can interact specifically with phosphorylated Spt5 CTRs.
The Cdc73 PCID promotes Paf1C recruitment in vivo
Having found that recombinant Cdc73 displays the strongest affinity for both kinds of diphosphorylated CTD repeats, we sought to identify the PCID in Cdc73 and assess its importance in Paf1C recruitment. To this end, we tested a panel of GST-Cdc73 deletion variants for binding to the S5P-CTD peptides in vitro. As shown in Figure 3A, a C-terminal fragment of residues 201–393 was the smallest Cdc73 segment that bound S5P-CTD peptides indistinguishably from full-length Cdc73 (Figure 3A), and it showed the same relative affinities for different CTD peptides observed for full-length Cdc73, namely S2P,S5P>S5P>S2P>unphosphorylated peptides (cf. Figure 3B, top row (WT) with Figure 1C, GST-Cdc73). These results suggested that the functional PCID spans a relatively large region (of 193 residues) in the Cdc73 C-terminus.
Figure 3.
Phospho-CTD interaction domain (PCID) of Cdc73 is located in its C-terminal region. (A) GST fusions to full-length (F.L.) Cdc73 or the Cdc73 truncations shown schematically on the right were purified and used in peptide binding assays with the CTD peptide S5P as described in Figure 1. The results are summarized in the schematic. (B–D) Peptide binding assays conducted as in Figure 1 using the indicated CTD peptides (B, C) or Spt5 pCTR peptides (D) and either WT or the indicated mutant versions of GST-Cdc73(201–393). Lanes 1–3 and 4–5 of the W357A panel derive from different sections of the same blot as described in Supplementary Figure S10B. Figure source data can be found with the Supplementary data.
A multiple sequence alignment of Cdc73 from different yeasts and fungi revealed blocks of conserved residues throughout the C-terminal segment (Supplementary Figure S3), including a possible WW motif—a PCID containing two conserved Trp residues (Meinhart et al, 2005)—and other strings of conserved residues with no obvious similarity to previously established PCIDs (Li et al, 2005; Meinhart et al, 2005; Phatnani and Greenleaf, 2006; Vojnic et al, 2006; Lunde et al, 2010; Xiang et al, 2010; Ghosh et al, 2011). We introduced mutations into the GST-Cdc73(201–393) construct to generate Ala substitutions in the conserved Trp residues (W357A and W380A), or in 6–7 consecutive residues in three other conserved blocks, and tested the mutant proteins for peptide binding. As shown in Figure 3B and C, all of the mutations except W357A reduced or abolished binding of the fusion protein to pCTD peptides. Interestingly, all substitutions besides W357A also greatly impaired binding to the Spt5 pCTR peptides (Figure 3D), consistent with the possibility that the same structural elements in the Cdc73 C-terminus responsible for binding to pCTD repeats mediate binding to the Spt5 pCTRs. However, it is possible that these substitutions disrupt folding of the Cdc73 C-terminal region and impair binding to pCTD and/or pCTR peptides by an indirect mechanism.
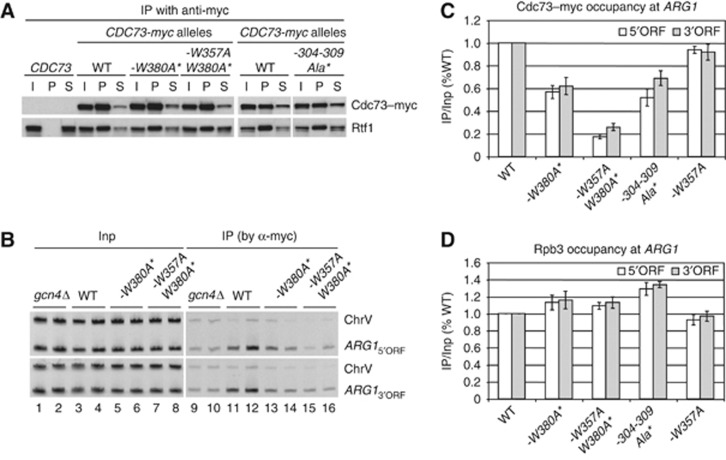
After introducing a subset of these PCID mutations into chromosomal CDC73 (along with a C-terminal myc tag), we discovered by Western analysis of yeast WCEs that they moderately reduce abundance of Cdc73–myc protein. However, for three of the mutants we could mitigate these reductions by introducing a low-copy plasmid harbouring the same mutant allele: W380A; W357AW380A; and 304-309Ala (Supplementary Figure S4A). The resulting strains also display normal levels of Paf1C, as indicted by the fact that nearly WT amounts of Rtf1 (Figure 4A) and HA-tagged Paf1 (Supplementary Figure S4B) co-immunoprecipitated with each cdc73–myc protein. Accordingly, we proceeded to evaluate the effects of the cdc73 mutations on Paf1C recruitment by chromatin immunoprecipitation (ChIP) analysis of the myc-tagged Cdc73 subunits.
Figure 4.
Substitutions in the Cdc73 PCID eliminating binding to pCTD peptides reduce Cdc73–myc occupancies at ARG1. (A) Untagged WT strain BY4741 and the following CDC73-myc13 strains were cultured in synthetic complete medium lacking leucine (SC-Leu) to log phase (A600 of ∼0.6): HQY1147 (CDC73-myc13); HQY1468 (cdc73-W380A-myc13 harbouring low-copy plasmid pHQ1862 with cdc73-W380A-myc13, henceforth abbreviated as -W380A*); HQY1469 (cdc73-W357A,W380A-myc13 harbouring pHQ1861, abbreviated as -W357A,W380A*); and HQY1471(cdc73-6Ala(304-309)-myc13 harbouring pHQ1952, abbreviated as -304-309Ala*). WCEs were prepared and immunoprecipitated with anti-myc antibodies and subjected to Western analysis with anti-myc and anti-Rtf1 antibodies. The last six lanes derive from a separate blot as described in Supplementary Figure S10C. (B–D) ChIP analysis of Cdc73–myc binding at ARG1. The CDC73-myc13 strains described in (A) and CDC73-myc13 gcn4Δ strain HQY1148 were cultured in SC lacking Ile, Val, and Leu (SC-Ilv-Leu) and treated with sulfometuron (SM) for 30 min to induce Gcn4. ChIP analysis was conducted using anti-myc antibodies for Cdc73–myc (B, C) or anti-Rpb3 antibodies for Rpb3 (D). DNA extracted from immunoprecipitates (IP) and input chromatin (Input) samples was subjected to PCR in the presence of [33P]-dATP with the appropriate primers to amplify radiolabelled ARG1 5′ORF or 3′ORF sequences, or sequences from a chromosome V intergenic region (ChrV) examined as a control. PCR products were resolved by PAGE and visualized by autography, with representative results shown for two biological replicates in adjacent lanes of (B), and quantified with a phosphorimager. The resulting values were expressed as ratios of IP to Input signals and normalized to the corresponding ratio for WT CDC73-myc cells. Error bars correspond to standard errors of the mean derived from four or more independent cultures and two independent immunoprecipitations for each culture. (See Materials and methods in Supplementary data for further details on quantification of ChIP data.) Figure source data can be found with the Supplementary data.
We showed previously that the occupancy of myc-tagged Paf1 in ARG1 chromatin is increased on induction of transcriptional activator Gcn4 (by starvation for isoleucine and valine) to levels ∼6-fold above background in GCN4 cells but remains at background levels in gcn4Δ cells (Qiu et al, 2006). As shown in Figure 4B, Cdc73–myc occupancy in the ARG1 CDS is similarly induced by Gcn4, showing much higher levels in GCN4 (WT) versus gcn4Δ cells (lanes 11–12 versus 9–10). Importantly, the W380 and 304-309Ala mutations significantly reduced Cdc73–myc occupancy under inducing conditions (Figure 4B and C). An even stronger reduction was observed for the W357A,W380A double mutant, although the W357A single mutation had little or no effect on its own. The reductions in Cdc73 occupancy occurred without significant decreases in Pol II (Rpb3) occupancy at ARG1 (Figure 4D). These results suggest that the Cdc73 PCID is crucial for efficient Paf1C recruitment at induced ARG1.
We also measured Cdc73–myc occupancies at the GAL1 gene under conditions of galactose induction, and at the constitutively expressed genes PMA1 and ADH1. As shown in Supplementary Figure S5A–F, the W380A, W357A, W380A and 304-309Ala mutations all produced significant reductions in Cdc73–myc occupancies, but not Pol II (Rpb3) occupancies, in the coding sequences of these three genes, consistent with a general requirement for the Cdc73 PCID in Paf1C recruitment.
Bur1 promotes Paf1C recruitment by Spt5 pCTR dependent and independent pathways
The results above indicate that the Cdc73 PCID is required for robust Paf1C recruitment in vivo, but do not reveal whether it mediates interaction with Spt5 pCTRs, Pol II pCTD repeats, or both, to promote Paf1C recruitment. It was shown previously that substituting Ser1 with Ala in all 15 CTRs of Spt5 (spt5-S1-15A) reduced Paf1–myc occupancy at the 5′ ends of ARG1 and PYK1 in vivo (Liu et al, 2009), consistent with the idea that Spt5 phosphorylation by Bur1 promotes Paf1C recruitment. To provide additional evidence supporting this interpretation, we examined whether the phosphomimetic Asp and Glu substitutions in the Spt5 CTRs would have less effect than Ala substitutions on Paf1C recruitment to ARG1. Indeed, Paf1–myc recruitment was reduced by a factor of 3–4 in the non-phosphorylatable spt5-S1-15A mutant, but only by 20–30% in the phosphomimetic mutants spt5-S1-15D and spt5-S1-15E, at the 5′ and 3′ ends of the ARG1 CDS (Figure 5A and B). Thus, even though the Spt5 CTRD and CTRE peptides were inferior to the pCTR peptides in binding Paf1C in vitro (Figure 2), the phosphomimetic Spt5 substitutions can sustain a substantial level of Paf1C recruitment in vivo, supporting the conclusion that phosphorylation of Ser1 in the Spt5 CTRs by Bur1 stimulates Paf1C recruitment (Liu et al, 2009). We speculate that phosphomimetic Spt5 CTRs are highly effective in Paf1C recruitment in vivo because they contain an immutable negative charge, whereas the phosphorylation level of the WT CTRs is likely modulated by protein phosphatases.
Figure 5.
Inactivating bur1-as reduces Paf1–myc occupancy at ARG1 in spt5 phosphomimetic and non-phosphorylatable CTR mutants. (A, B) Phosphomimetic substitutions in Spt5 CTRs sustain Paf1 recruitment at ARG1. PAF1-myc13 strains with the indicated WT or mutant SPT5-HA3 alleles (HQY1379, HQY1414, HQY1441, and HQY1447) were subjected to ChIP analysis with anti-myc antibodies as described in Figure 4. Representative results are shown in (A), with results from independent cultures displayed in adjacent lanes, and the quantification of results from four or more biological replicates is summarized in (B) as percentages of the values measured in the WT SPT5 strain. (C) Inactivating bur1-as reduces Paf1–myc occupancy at ARG1 in phosphomimetic spt5 mutants. PAF1-myc13 bur1-as strains with the indicated SPT5-HA3 alleles (HQY1379, HQY1416, HQY1444, Y1449, and HQY1419)were cultured in SC-Ilv to A600 of ∼0.4 and half of each culture was treated with 6 μM 3MB-PP1 (+3 MB) for 20 min while the other half was left untreated (−3 MB); and all cultures were then treated with sulfometuron (SM) for 30 min to induce Gcn4. ChIP analysis was conducted using anti-myc or anti-Rpb3 antibodies. Paf1–myc occupancies at ARG1 were calculated as described in Figure 4, normalized for the corresponding Rpb3 levels and the resulting Paf1–myc:Rpb3 ratios are presented in (C) relative to those observed for the WT SPT5-HA3 strain. The Paf1–myc and Rpb3 occupancies used to calculate the ratios in (C) are presented in Supplementary Figure S6.
The phosphomimetic substitutions should render the contribution of Spt5 CTRs to Paf1C recruitment independent of Bur1. Hence, if Bur1 stimulates Paf1C recruitment solely by phosphorylating Spt5, then inhibiting an analogue-sensitive Bur1 variant (encoded by bur1-as) should not affect Paf1C recruitment in the spt5 phosphomimetic mutants. Consistent with previous results (Qiu et al, 2006; Liu et al, 2009), treatment of the bur1-as strain with the inhibitory ATP analogue 3MB-PP1 dramatically reduced Gcn4-dependent Paf1–myc occupancy at ARG1 (Supplementary Figure S6A, bur1-as, +MB versus −MB). Neither inhibition of bur1-as with 3MB-PP1 nor the spt5-S1-15A mutation reduces Pol II (Rpb3) occupancy at ARG1 (Supplementary Figure S6B). Accordingly, the ratio of Paf1–myc to Rpb3 occupancies was strongly reduced by inhibition of bur1-as (Figure 5C, bur1-as, +MB versus −MB). The Paf1–myc:Rpb3 ratio was also reduced in the spt5-S1-15A bur1-as double mutant in the absence of bur1-as inhibition by 3MB-PP1, but not to the extent observed in the bur1-as single mutant treated with 3MB-PP1 (Figure 5C, bur1-as spt5-S1-15A, −MB versus bur1-as, +MB). Importantly, analogue treatment of all three bur1-as spt5 double mutants evoked reductions in the Paf1–myc:Rpb3 ratios at ARG1 compared to the levels seen without 3MB-PP1 (Figure 5C and Supplementary Figure S6A and B, bur1-as spt5 double mutants, +MB versus −MB). These results indicate that Bur1 contributes to Paf1C recruitment at ARG1 by a mechanism independent of Ser1 phosphorylation of the Spt5 CTRs. ChIP analysis of Paf1–myc recruitment at the GAL1, PMA1, and ADH1 genes in the same mutants gave qualitatively similar findings, with inhibition of bur1-as evoking significant reductions in Paf1–myc occupancies in both the bur1-as spt5-S1-15D and bur1-as spt5-S1-15E double mutants (Supplementary Figure S7A–C). These findings support the possibility that diphosphorylated CTD repeats produced by Bur1 and Kin28 act together with the Spt5 pCTRs to promote Paf1C recruitment.
Paf1C recruitment also requires Kin28 kinase activity in phosphomimetic spt5 mutants
We showed previously that robust Paf1 recruitment is dependent on Kin28 (Qiu et al, 2006), and that Kin28 promotes Bur1 recruitment to promoter-proximal CDS (Qiu et al, 2009). Hence, we predicted that Kin28 should promote Paf1C recruitment, at least in part, by enhancing Bur1 recruitment and attendant phosphorylation of Spt5 CTRs. Supporting this prediction, inhibiting the analogue-sensitive variant of Kin28 (kin28-as) with NA-PP1 strongly reduces phosphorylation of the Spt5 CTRs, as judged by Western analysis of bulk Spt5 with phosphospecific antibodies (Liu et al, 2009; Figure 6A, lanes 3–4). Strikingly, phosphorylated Spt5 was eliminated on removal of the PCID in the C-terminus of Bur1 by the bur1-ΔC mutation (Figure 6A, lanes 5–6). Given that bur1-ΔC reduces Bur1 binding to S5P CTD repeats in vitro and decreases Bur1 recruitment to ARG1 in vivo (Qiu et al, 2009), these findings strongly support the idea that Kin28 promotes Spt5 CTR phosphorylation by enhancing co-transcriptional recruitment of Bur1.
Figure 6.
Paf1 recruitment in spt5 phosphomimetic mutants requires Kin28 activity. (A) Inactivating kin28-as, or eliminating the Bur1 PCID, reduces phosphorylation of Spt5 CTRs in vivo. WCEs were prepared by the denaturing method of TCA precipitation from strains BY4741 (WT) and HQY1455 (kin28-as) cultured in SC-Ilv medium to A600 of ∼0.35, untreated (−) or treated (+) with 48 μM NA-PP1 for 42 min, and from HQY1223 (WT) and HQY1280 (bur1-CΔ) untreated with NA-PP1, and subjected to Western analysis with antibodies against phosphorylated Spt5 (Spt5-P) or total Spt5. (B–D) Inactivating kin28-as reduces S5P and decreases the Paf1-myc:Rpb3 ratio in phosphomimetic spt5 mutants without diminishing Spt5 occupancies at ARG1. PAF1-myc13 strains HQY1379 (WT SPT5-HA3), HQY1414 (spt5-S1-15A-HA3), HQY1441 (spt5-S1-15D-HA3), HQY1447 (spt5-S1-15E-HA3), HQY1465 (kin28-as SPT5-HA3), HQY1575 (kin28-as spt5-S1-15A-HA3), HQY1466 (kin28-as spt5-S1-15D-HA3), and HQY1467 (kin28-as spt5-S1-15E-HA3) were cultured in SC-Ilv and treated with NA-PP1 at 12 μM for 12 min and then with sulfometuron for 30 min. ChIP analysis was conducted as described in Figures 4 and 5 using H14 antibodies against S5P (B), anti-myc antibodies and anti-Rpb3 antibodies (C), and HA antibodies against Spt5-HA (D). Figure source data can be found with the Supplementary data.
We asked next whether the contribution of Kin28 to recruitment of Paf1C is limited to its role in stimulating Spt5 phosphorylation by Bur1. If so, then analogue inhibition of kin28-as should not reduce Paf1C recruitment in strains harbouring the Spt5 variants with non-phosphorylatable CTRs. Consistent with our previous results (Qiu et al, 2006), ChIP analysis showed that treatment of kin28-as cells with ATP analogue NA-PP1 lowers the occupancy of the S5P form of Rpb1 at the promoter and 5′ORF regions of ARG1 (Figure 6B, kin28-as). Furthermore, inhibition of kin28-as by NA-PP1 evokes reduced Paf1–myc:Rpb3 ratios in the ARG1 coding sequences (Figure 6C, kin28-as). Importantly, inhibition of kin28-as also reduced Paf1 recruitment in cells harbouring the phosphomimetic variants spt5-S1-15D and spt5-S1-15E, and also in the strain containing the non-phosphorylatable mutant spt5-S1-15A (Figure 6C). ChIP analysis of Spt5 in the same strains insured that inhibition of kin28-as by NA-PP1 does not unexpectedly reduce the occupancies of WT or mutant forms of Spt5 at ARG1 (Figure 6D). Similar results were obtained for the induced GAL1 gene, at least for the 5′ end of the CDS, where inhibition of kin28-as reduced the Paf1:Rpb3 ratio in cells harbouring the phosphomimetic spt5 mutations (Supplementary Figure S8C). Although Paf1 occupancies were likewise reduced at the 3′ end of GAL1 on kin28-as inhibition (Supplementary Figure S8A), this result is complicated by the similar reductions in Rpb3 occupancies that occur at the GAL1 3′ end (Supplementary Figure S8B). Nevertheless, the bulk of the data in Figure 6 and Supplementary Figure S8 indicate that cells harbouring Spt5 variants that cannot be phosphorylated by Bur1 are still dependent on Kin28 activity for efficient Paf1C recruitment. This implies that Kin28 promotes Paf1C recruitment by a mechanism distinct from its stimulation of Spt5 phosphorylation by Bur1, consistent with the notion that CTD repeats diphosphorylated by Kin28 and Bur1 provide an alternative platform for Paf1C recruitment.
The fact that the kin28-as mutation alone does not evoke a more dramatic reduction in Paf1C recruitment (Figure 6C) might be explained by noting that inhibition of kin28-as alone reduces, but does not abolish, Spt5 CTR phosphorylation (Figure 6A), and previous results indicating that Bur1 and Srb10 contribute to Ser5P and that Bur1 promotes Ser7P formation in vivo (Liu et al, 2004; Kanin et al, 2007; Tietjen et al, 2010; Bataille et al, 2012). Thus, the residual Spt5 CTR and Pol II CTD phosphorylation occurring in analogue-inhibited SPT5 kin28-as cells could be sufficient for appreciable Paf1C recruitment by both partially phosphorylated scaffolds.
The Cdc73 CID is required for robust histone H3 methylation in the absence of Ser1 phosphorylation of Spt5 CTRs
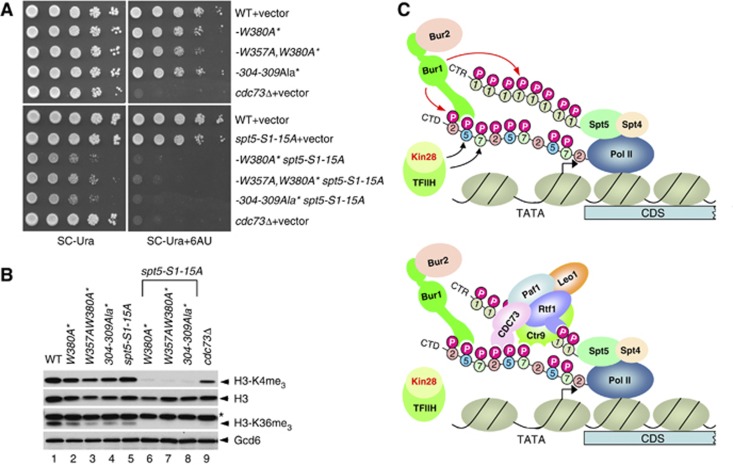
Finally, we reasoned that if the Cdc73 PCID mediates Paf1C binding to both the Spt5 pCTRs and Pol II pCTD, then the cdc73 PCID substitutions that impair binding to phosphorylated CTD repeats should impair Paf1C recruitment even in cells where the Spt5 CTRs are rendered non-phosphorylatable by S1A substitutions. To test this prediction, we first exploited the fact that elimination of Cdc73 in cdc73Δ cells confers strong 6-azauracil (6-AU) sensitivity, a phenotype conferred by various mutants with defects in Pol II elongation, owing to reduced nucleotide pools evoked by 6-AU (Riles et al, 2004). Importantly, we found that all three double mutants harbouring the spt5-S1-15A allele combined with one of the three cdc73 PCID mutations display slow-growth phenotypes on complete medium and also much greater sensitivity to 6-AU than observed in the corresponding spt5-S1-15A or cdc73 single mutants (Figure 7A). Employing a second in-vivo assay for Paf1C function, we measured the levels of Lys4 and Lys36 trimethylation in bulk histone H3 in the same set of strains. As expected (Laribee et al, 2005; Chu et al, 2007), these two modifications were greatly reduced in cdc73Δ cells (Figure 7B, lane 9 versus 1). The spt5-S1-15A and cdc73 PCID mutations produced slight to moderate reductions in the modifications (Figure 7B, lanes 1–5), with the strongest effects observed for the W357A,W380A* mutant (lane 3). Importantly, however, both H3-K4me3 and H3-K36me3 were nearly undetectable in all three cdc73 spt5-S1-15A double mutants (Figure 7B, lanes 6–8).
Figure 7.
The Cdc73 PCID promotes Paf1C function independently of phosphorylated Spt5 CTRs, and a model for Paf1C recruitment via the Spt5-pCTR and Pol II pCTD. (A) cdc73 PCID mutations confer synthetic sensitivity to 6-AU and slow growth in combination with spt5-S1-15A. Strains HQY1468 (-W380A), HQY1469 (-W357AW380A), HQY1471 (-304-309Ala), HQY1509 (-W380A spt5-S1-15A), HQY1510 (-W357AW380A spt5-S1-15A), and HQY1507 (-304-309Ala spt5-S1-15A) were transformed with a low-copy URA3 plasmid containing the cognate chromosomal cdc73 allele present in that strain (designated as cdc73* alleles, as described in Figure 4), whereas strains HQY1147 (WT), HQY1508 (spt5-S1-15A), and 5326 (cdc73Δ) were transformed with an empty URA3 vector. The transformants were grown to saturation and serial 10-fold dilutions were spotted on SC-Ura and SC-Ura+6AU (with 6-azauracil at 100 μg/ml) and incubated at 30°C for 3 or 4 days, respectively. (B) WCEs prepared from strains in (A) were subjected to Western analysis with antibodies against total H3, the indicated methylated forms of H3, or Gcd6 (as loading control). Non-specific bands are marked by an asterisk. (C) Model for dual pathway of Paf1C recruitment by the pCTR and pCTD. (Upper panel) Kin28 phosphorylates Ser5 and Ser7 in the CTD and the S5P stimulates Bur1/Bur2 recruitment to the CTD via the Bur1 PCID. Spt5/Spt4 associates with Pol II via the ‘clamp domain’ of Pol II (Martinez-Rucobo et al, 2011), independently of CTD phosphorylation. Recruited Bur1/Bur2 phosphorylates Ser2 (and possibly Ser7) in CTD repeats harbouring S5P to produce diphosphorylated repeats, and also phosphorylates the Spt5 CTRs (upper panel). (Lower panel) Paf1C recruitment occurs through cooperation between interactions of the PCIDs in Cdc73, Rtf1 and Ctr9 with diphosphorylated CTD repeats and the Spt5 pCTR. Figure source data can be found with the Supplementary data.
The findings that the cdc73 PCID mutations exacerbate the growth phenotypes (Figure 7A) and impair H3 methylation (Figure 7B) in spt5-S1-15A cells, where the Spt5 pCTRs cannot contribute to Paf1C recruitment, strongly supports our hypothesis that the Cdc73 PCID mediates an important interaction of Paf1C with Pol II pCTD repeats in addition to Spt5 pCTRs. This model can also explain why the -W357A,W380A* single mutation confers 6AU sensitivity and reduces H3-K36me3 abundance while the spt5-S1-15A mutation provokes neither phenotype, as the -W357A,W380A* mutation is expected to reduce Paf1C recruitment by both the pCTD and pCTR, whereas spt5-S1-15A impairs only the pCTR-dependent mechanism. Presumably, the spt5-S1-15A, -W357A,W380A* double mutation reduces Paf1C occupancy below the level conferred by -W357A,W380A* alone because spt5-S1-15A also decreases the ability of the Rtf1 and Ctr9 subunits to support Paf1C recruitment via the pCTR. In addition, although W357A,W380A* reduces Cdc73 occupancy at ARG1 to the background level seen in gcn4Δ cells (Figure 4B), it clearly does not abolish Cdc73 occupancy at the other genes we tested (Supplementary Figure S5), and it may reduce Cdc73 levels at ARG1 only to the basal levels observed in gcn4Δ cells. Hence, the spt5-S1-15A mutation likely also eliminates the residual contribution of the W357A,W380A* variant of the Cdc73 PCID to Paf1C recruitment by the pCTR.
Discussion
Paf1C is recruited directly by phosphorylated Spt5 CTRs in a manner stimulated by Kin28
Previous work established that phosphorylation of the Spt5 CTRs by Bur1 is a key pathway for stimulating Paf1C recruitment by elongating Pol II (Liu et al, 2009; Zhou et al, 2009), but it was unclear whether the Spt5 pCTRs interact directly with Paf1C. We observed here that purified Paf1C and recombinant forms of three Paf1C subunits, Cdc73, Rtf1, and Ctr9, bind specifically to Ser1-phosphorylated Spt5 CTR peptides in vitro. The pCTR-binding activity of Cdc73 was mapped to its C-terminus and we demonstrated that substitution mutations in this PCID that inactivate pCTR binding in vitro reduce Paf1C recruitment to ARG1, GAL1, ADH1, and PMA1 CDS in vivo. These findings provide the first evidence for direct binding of Paf1C to the Bur1-phosphorylated form of the Spt5 CTRs.
We showed previously that Paf1C recruitment is also enhanced by Kin28 (Qiu et al, 2006), but the underlying mechanism was unknown. Consistent with the fact that Kin28 promotes Bur1 recruitment near the promoter via the PCID in the Bur1 C-terminus (Qiu et al, 2009), we showed here that inactivating kin28-as, or deleting the Bur1 PCID, reduces phosphorylation of the Spt5 CTRs in vivo. This finding implies that Kin28 promotes Paf1C recruitment, at least partly, by stimulating Bur1 recruitment and attendant Ser1 phosphorylation of the Spt5 CTRs.
Evidence that Paf1C is recruited by diphosphorylated CTD repeats generated by Kin28 and Bur1 in addition to Spt5 pCTRs
We obtained evidence that Kin28, and possibly Bur1, also promote Paf1C recruitment more directly through Paf1C binding to the phosphorylated Pol II CTD. This idea was stimulated by the fact that Kin28, a component of TFIIH recruited to promoters (Mayer et al 2010), phosphorylates the CTD on Ser5 and Ser7 (Akhtar et al, 2009; Glover-Cutter et al, 2009; Kim et al, 2009) at promoter-proximal sites genomewide (Kim et al, 2010; Mayer et al, 2010; Tietjen et al, 2010; Bataille et al, 2012). Furthermore, because Kin28 promotes Bur1 recruitment (Qiu et al, 2009), and Bur1 also contributes to CTD phosphorylation on Ser2 and Ser7 (Liu et al, 2009; Qiu et al, 2009; Tietjen et al, 2010; Bataille et al, 2012), Kin28-stimulated Bur1 recruitment should augment formation of S5P,S7-CTD repeats at promoter-distal sites and also produce S2P,S5P-CTD repeats near the promoter by phosphorylating Ser7 and Ser2, respectively, in repeats already phosphorylated on Ser5 by Kin28. Importantly, we found that purified Paf1C can interact directly with pCTD peptides in vitro with a strong preference for S2P,S5P- and S5P,S7P-CTD repeats, and the same three Paf1C subunits that interact with Spt5 pCTR peptides also interact tightly with one or both of diphosphorylated CTD peptides. Moreover, point mutations in the Cdc73 C-terminus that reduce Paf1C recruitment in vivo abolish Cdc73 binding to both diphosphorylated CTD and pCTR peptides. Thus, Paf1C could be recruited by the Pol II pCTD in addition to the Spt5 pCTRs.
Strong support for this hypothesis came from examining the effects of inhibiting bur1-as or kin28-as in cells harbouring phosphomimetic Spt5 CTRs, where Ser1 in all 15 CTR repeats is substituted with Asp or Glu. These substitutions sustain efficient Paf1C recruitment in otherwise WT cells, whereas Spt5 substitutions with non-phosphorylatable Ala do not, supporting the model that phosphorylated Spt5 CTRs are crucial for Paf1C recruitment. Although Bur1 cannot regulate Ser1 phosphorylation in the Spt5 CTRs of these mutants, inhibiting bur1-as still reduced Paf1C recruitment in these strains, thus indicating that another Bur1 substrate is important for Paf1C recruitment. There is evidence that Bur1 phosphorylates Rad6 (Wood et al, 2005); however, we showed previously that eliminating Rad6 does not affect Paf1C recruitment at ARG1 (Qiu et al, 2006). Hence, we favour the model that the Spt5 pCTR-independent pathway of Paf1C recruitment involves Bur1’s other known substrates, Ser2 and Ser7 of the Pol II CTD, and proceeds via direct binding of Paf1C to S2,S5- and S5,S7-diphosphorylated CTD repeats. This model can also account for the fact that inhibiting kin28-as in the mutants harbouring phosphomimetic Spt5 CTRs also reduces Paf1C recruitment, as expected from the reduction in S2,S5- and S5,S7-diphosphorylated CTD repeats produced by diminished Kin28 activity. Accordingly, we propose that Kin28 enables direct binding of PCIDs in Paf1C subunits to diphosphorylated CTD repeats in addition to stimulating Spt5 CTR phosphorylation by Bur1. Finally, the fact that the cdc73 mutations that impair Cdc73 binding to pCTD peptides confer synthetic 6-AU sensitivity and strong reductions in Paf1C-dependent H3 methylation when combined with non-phosphorylatable spt5-S1-15A further supports the notion that the Cdc73 PCID mediates Paf1C binding to the pCTD in addition to the Spt5 pCTRs.
A model for dual pathways of Paf1C recruitment via Pol II pCTD and Spt5 pCTRs
Considering all of our findings and those described previously (Liu et al, 2009; Qiu et al, 2009; Zhou et al, 2009), we propose that Ser5 CTD phosphorylation by Kin28 at the promoter stimulates Bur1 recruitment with attendant phosphorylation of the Spt5 CTRs, and that Kin28 and Bur1 also collaborate to produce S2,S5-diphosphorylated CTD repeats at the promoter and 5′ end of the CDS and S5,S7-diphosphorylated repeats further downstream in the CDS. The resulting pCTRs and pCTD repeats cooperate to enable robust recruitment of Paf1C via the multiple PCIDs in Paf1C subunits Cdc73, Rtf1, and Ctf9 (Figure 7C). Leo1 interaction with the nascent transcript likely also promotes Paf1C recruitment (Dermody and Buratowski, 2010), reminiscent of the finding that co-transcriptional recruitment of yeast Yra1 depends on an RNA recognition motif (RRM) that binds both RNA and the pCTD via distinct residues (Mackellar and Greenleaf, 2011). Additional experiments will be required to determine whether Paf1C binds with thermodynamic cooperativity to the pCTD and pCTRs in the Pol II elongation complex. However, recent structural analysis of the Pol II:Spt5/Spt4 complex is consistent with this idea in suggesting that the CTRs and CTD extend for comparable distances from the same side of Pol II (Martinez-Rucobo et al, 2011) and, thus, could interact simultaneously with different Paf1C subunits (Figure 7C).
Our proposal that Ser2 CTD phosphorylation promotes Paf1C recruitment might seem at odds with the previous finding that eliminating Ctk1, the major Ser2 CTD kinase, does not reduce Paf1C recruitment at ARG1 (Qiu et al, 2006). However, we showed previously that Ctk1 and Bur1 make similar contributions to S2P in bulk elongating Pol II containing S5P, and in promoter-proximal Rpb1 at ARG1 (Qiu et al, 2009). Thus, in ctk1Δ cells, the residual S2P generated by Bur1 in promoter-proximal Rpb1 could be sufficient for robust Paf1C recruitment by the pCTD early in the elongation cycle in combination with the Bur1-dependent Spt5 pCTR recruitment pathway. By contrast, eliminating Bur1 activity affects both pathways, abrogating Paf1C recruitment via Spt5 pCTRs in addition to reducing S2P levels in the CTD. It is also possible that the known recruitment of Bur1 by CTD repeats phosphorylated on Ser5 by Kin28 (Qiu et al, 2009) makes Bur1 more efficient than Ctk1 in generating the S2,S5-diphosphorylated repeats that would constitute the Paf1C binding site near the promoter. Tethering of Bur1 to S5P repeats could also enhance S2P formation in clusters of CTD repeats and thereby facilitate binding of the different Paf1C subunits harbouring PCIDs to adjacent S2P,S5P-CTD repeats (Lunde et al, 2010). Finally, it is possible that the role of Bur1 in replacing Ser7P at promoter distal sites, which is apparently not shared by Ctk1 (Tietjen et al, 2010), is more important than its contribution to Ser2 CTD phosphorylation in Paf1C recruitment.
Although we consider the scheme in Figure 7C to be the simplest interpretation of the data, we cannot exclude the possibility that Bur1 has another unknown substrate in the elongation complex, not recognized by Ctk1, that enhances Paf1C recruitment, and that the Spt5 pCTR-independent role of Kin28 in Paf1C recruitment is limited to promoting Bur1 recruitment and attendant phosphorylation of this unknown Bur1 substrate rather than the formation of diphosphorylated CTD repeats to which Paf1C can bind. This explanation would require that the ability of Paf1C subunits to bind diphosphorylated CTD repeats in vitro is completely incidental to their physiologically relevant interactions with Spt5 pCTRs. However, this alternative model seems inconsistent with the synthetic growth defects and reductions in Paf1C-dependent histone H3 methylation events we observed in the cdc73 spt5-S1-15A double mutants, which are more readily explained by simultaneous impairment of Paf1C binding to both pCTD and pCTR repeats.
The significance of pCTD and pCTR interactions with multiple Paf1C subunits
The results of our peptide binding assays indicated that purified Paf1C binds with comparable affinities to S2P,S5P- and S5P,S7P pCTD and monophosphorylated pCTR repeats. These interactions are quite specific for the phosphorylated ligands, as purified Paf1C does not interact stably with phosphomimetic S2D,S5D peptides and also binds less tightly to CTRD versus pCTR peptides. Thus, tight interactions of Paf1C with both pCTD and pCTR repeats seem to require dianionic –PO3= groups, and presumably their greater potential for forming ionic interactions or H bonds, compared to carboxylate groups of Asp or Glu side chains. In this view, the weaker binding seen for the CTRD peptide presumably reflects its ability to make only a subset of the interactions afforded by the Ser1–PO3= group with the PCID binding pocket. This specificity makes it unlikely that the pCTD- and pCTR-binding activities of Paf1C merely reflect an RNA binding activity of the complex that would involve interactions with monoanionic phosphate groups in the backbone of the nascent transcript. Indeed, the Leo1 and Rtf1 subunits were shown to bind RNA in vitro (Dermody and Buratowski, 2010), yet we found that Leo1 does not interact with pCTD or pCTR peptides, and that Rtf1 is exquisitely specific in its binding to S2P,S5P versus S5P,S7P pCTD peptides. Thus, the ability to bind RNA does not automatically endow a protein with the ability to bind different kinds of diphosphorylated pCTD repeats or pCTR repeats. Indeed, Mackellar and Greenleaf (2011) showed that the RNA binding and S2P,S5P-CTD binding activities of Yra1 require distinct residues in the protein.
The recombinant Cdc73 and Ctr9 proteins most closely resemble the purified Paf1C complex in pCTD-binding specificity, but Cdc73 binds much more tightly than does the Ctr9 C-terminal segment to pCTD peptides. Hence, Cdc73 probably makes the greatest contribution to pCTD binding by Paf1C. By contrast, recombinant Rtf1 displays the strongest binding to pCTR peptides and could make the greater contribution to pCTR binding by Paf1C. As noted above, both Cdc73 and Rtf1 have been implicated in Paf1C recruitment in vivo, and while this manuscript was under revision, VanDemark, Arndt, and colleagues reported that truncation of nearly the entire Cdc73 PCID (as defined here) impaired Paf1C recruitment without affecting Paf1C integrity (Amrich et al, 2012), in accordance with our findings. They also found that removing the Cdc73 PCID reduced the H3-K4me3 level only in cells lacking Rtf1, which is consistent with our findings that cdc73 PCID substitutions reduce H3-K4me3 only in combination with spt5-S1-S15A, which eliminates contacts between Paf1C subunits (likely Rtf1) and the Spt5 pCTR. Interestingly, the crystal structure of this Cdc73 segment reveals a Ras-like domain lacking key elements required for GTP binding but containing several clusters of conserved surface-exposed residues that could comprise one or more binding sites for CTD or CTR repeats (Amrich et al, 2012). Certain of our clustered Ala substitutions that eliminate GST-Cdc73 binding to both pCTD- and pCTR peptides affect a subset of these conserved surface residues; however, it is possible that they alter the folding of the PCID in a way that disrupts overlapping, or even separate, pCTD- and pCTR-binding surfaces. Assuming that a single surface is responsible for both binding activities of Cdc73, a hypothetical scenario can be proposed to account for this promiscuity based on known principles of pCTD interaction derived from structural analyses of PCID-pCTD complexes (see Supplementary Figure S9). Obviously, further work is required to determine whether Cdc73, Rtf1, and Ctr9 interact with only pCTD or pCTR repeats when present as subunits of intact Paf1C; and in the event that recognition of both ligands occurs, whether single or multiple binding pockets are involved.
In conclusion, there is a large body of evidence indicating that the phosphorylated CTD provides a scaffold for recruitment of mRNA processing and histone modification enzymes, and of factors that regulate elongation or termination by Pol II (Phatnani and Greenleaf, 2006; Govind et al, 2010). Our results point to cooperation between the pCTD and Spt5 pCTR pathways for efficient Paf1C recruitment and Paf1C-dependent H3 methylation events. It is tempting to speculate that other factors recruited by the pCTD might also take advantage of the phosphorylated scaffold presented by the Spt5 pCTRs to enhance their recruitment by elongating Pol II.
Materials and methods
Yeast strains and plasmids employed are listed in Supplementary Tables S1 and S2, respectively, and are described in Supplementary data. Synthetic biotin-conjugated peptides purchased from AnaSpec are listed in Supplementary Table S4. Peptide binding assays and ChIP experiments were conducted as described in Supplementary data. Briefly, for ChIP assays, chromatin was prepared from formaldehyde-crosslinked cells, immunoprecipitated with the appropriate antibodies, and the DNA was extracted and subjected to PCR amplification in the presence of [33P]-dATP using primers listed in Supplementary Table S3. PCR conditions were optimized for each primer set to ensure that the amounts of amplified 33P-labelled products are proportional to amounts of input DNA over the range of concentrations of the relevant sequences in total or immunoprecipitated chromatin.
Co-immunoprecipitation assays using WCEs, and Western analysis of WCEs prepared under denaturing conditions using trichloroacetic acid, were conducted as described in Supplementary Data, as was the purification of GST fusions from bacteria and native Paf1C from yeast.
Supplementary Material
Acknowledgments
We thank Tom Dever for excellent suggestions and advice; Fred Winston and Steven Hahn for plasmids; and Karen Arndt, Grant Hartzog, and Steven Hahn for antibodies. We also appreciate Kevan Shokat’s kind gift of 3MB-PP1 at the outset of this study. We thank Andy VanDemark and Karen Arndt for critical reading of the manuscript and for communicating results prior to publication. This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development (NICHD), National Institutes of Health (NIH).
Author contributions: HQ conceived and executed experiments, analysed the data and assisted in writing the manuscript. CU and NG assisted in executing experiments. AGH conceived experiments, analysed the data, and assisted in writing the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ (2009) TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell 34: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrich CG, Davis CP, Rogal WP, Shirra MK, Heroux A, Gardner RG, Arndt KM, Vandemark AP (2012) Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin. J Biol Chem 287: 10863–10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F (2012) A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 45: 158–170 [DOI] [PubMed] [Google Scholar]
- Buratowski S (2009) Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Simic R, Warner MH, Arndt KM, Prelich G (2007) Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J 26: 4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody JL, Buratowski S (2010) Leo1 subunit of the yeast paf1 complex binds RNA and contributes to complex recruitment. J Biol Chem 285: 33671–33679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Shuman S, Lima CD (2011) Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell 43: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL (2009) TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol 29: 5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG (2010) Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell 39: 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanin EI, Kipp RT, Kung C, Slattery M, Viale A, Hahn S, Shokat KM, Ansari AZ (2007) Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci USA 104: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Erickson B, Luo W, Seward D, Graber JH, Pollock DD, Megee PC, Bentley DL (2010) Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol 17: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Levin DE (2011) Mpk1 MAPK association with the paf1 complex blocks sen1-mediated premature transcription termination. Cell 144: 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J 23: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Suh H, Cho EJ, Buratowski S (2009) Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem 284: 26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD (2005) BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr Biol 15: 1487–1493 [DOI] [PubMed] [Google Scholar]
- Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P (2005) Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proc Natl Acad Sci USA 102: 17636–17641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S (2004) Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol 24: 1721–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S (2009) Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29: 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G (2010) Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol 17: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackellar AL, Greenleaf AL (2011) Cotranscriptional association of mRNA export factor Yra1 with the C-terminal domain of RNA polymerase II: a mechanism for cotranscriptional recruitment. J Biol Chem 286: 36385–36395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P (2011) Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J 30: 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P (2005) A structural perspective of CTD function. Genes Dev 19: 1401–1415 [DOI] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA (2004) The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 14: 447–456 [DOI] [PubMed] [Google Scholar]
- Mulder KW, Brenkman AB, Inagaki A, van den Broek NJ, Timmers HT (2007) Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res 35: 2428–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordick K, Hoffman MG, Betz JL, Jaehning JA (2008) Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell 7: 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA (2005) A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell 20: 213–223 [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The Tandem Affinity Purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Qiu H, Hu C, Hinnebusch AG (2009) Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol Cell 33: 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Hu C, Wong CM, Hinnebusch AG (2006) The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol Cell Biol 26: 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L, Shaw RJ, Johnston M, Reines D (2004) Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M (2009) The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci USA 106: 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM (2005) A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol Cell 20: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A (2010) The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ (2010) Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P (2006) Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J Biol Chem 281: 13–15 [DOI] [PubMed] [Google Scholar]
- Warner MH, Roinick KL, Arndt KM (2007) Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 27: 6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A (2005) The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol Cell 20: 589–599 [DOI] [PubMed] [Google Scholar]
- Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L (2010) Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467: 729–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Kuo WH, Fillingham J, Greenblatt JF (2009) Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc Natl Acad Sci USA 106: 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.