Abstract
The process of Sister Chromosome Cohesion (SCC), which holds together sister chromatids upon replication, is essential for chromosome segregation and DNA repair in eukaryotic cells. Although cohesion at the molecular level has never been described in E. coli, previous studies have reported that sister sequences remain co-localized for a period after their replication. Here, we have developed a new genetic recombination assay that probes the ability of newly replicated chromosome loci to interact physically. We show that Sister Chromatid Interaction (SCI) occurs exclusively within a limited time frame after replication. Importantly, we could differentiate sister cohesion and co-localization since factors such as MatP and MukB that reduced the co-localization of markers had no effect on molecular cohesion. The frequency of sister chromatid interactions were modulated by the activity of Topo-IV, revealing that DNA topology modulates cohesion at the molecular scale in bacteria.
Keywords: chromosome segregation, DNA topology, sister chromatid cohesion, site-specific recombination, Topo-IV
Introduction
In all living cells, the inheritance of genetic material requires the faithful segregation of duplicated chromosomes into daughter cells. In eukaryotic cells, this process involves the alignment of chromatids upon replication, a process called sister chromatid cohesion (SCC) (Nasmyth and Haering, 2009). Newly replicated chromosomes remain tightly aligned and associated until the onset of mitosis. SCC is essential for genome stability since it is required for both high fidelity chromosome segregation and DNA damage repair. Furthermore, SCC is also involved in gene expression and cell development (Nasmyth and Haering, 2009). Cohesion is mediated by cohesin, a specific multi-subunit complex composed of “structural maintenance of chromosomes” (SMC) proteins. Other SMC complexes have a DNA condensing activity (Losada et al, 1998). The establishment of SCC on newly synthesized DNA is coupled with replication fork progression (Lengronne et al, 2006), and it is thought that cohesin topologically holds sister DNA strands together by trapping the DNA inside a molecular ring (Haering et al, 2002). The dissociation of cohesin complexes precedes a bipolar migration of sister chromosomes during mitosis.
In prokaryotic cells, in contrast to eukaryotic cells, little is known about sister chromosome cohesion and its control. No molecular evidence revealing SCC at the molecular level has been reported, and no proteins with cohesin-like activity have been described. Bacteria contain SMC proteins that have been shown to play a role in chromosome organization and segregation in E. coli, C. crescentus and B. subtilis (Jensen and Shapiro, 1999; Danilova et al, 2007; Cui et al, 2008; Gruber and Errington, 2009; Sullivan et al, 2009). However, experimental evidences indicated a role in condensing-like rather than in cohesin-like activity (Petrushenko et al, 2006). SCC in bacteria has been proposed to occur based on cytological fluorescence imaging that revealed a colocalization period of newly duplicated loci, i.e., the period between the time a locus is replicated and the visual separation of the two sister loci. Depending on the growth conditions, the lifespan of loci colocalization varied (Adachi et al, 2008). Furthermore, sister chromatid co-localization is not constant along the E. coli chromosome. It was demonstrated that the region containing the migS site (200 kb from oriC) was segregated before the oriC locus while it is replicated later (Yamaichi and Niki, 2004). In the Ori macrodomain (MD), a large region of about 600 kb adjacent to oriC on the right replichore was more cohesive than the distal part of the replichore, the non-structured region (Bates and Kleckner, 2005; Espeli et al, 2008). An abrupt segregation of the region would trigger nucleoid splitting and segregation of the rest of the chromosome (Bates and Kleckner, 2005). Recently, it was shown that two particular regions inside the Ori MD, called snaps, present extensive colocalization as compared to the rest of the MD (Joshi et al, 2011). The Ter MD also displays significant sister co-localization (Espeli et al, 2008). A number of factors (MukB, MatP, Topo-IV) have been shown to modulate the period of sister loci colocalization (Sunako et al, 2001; Mercier et al, 2008; Wang et al, 2008). The effect of Topo-IV inactivation has led authors to propose that catenation might be a major contributor of SCC through colocalization of the sister loci (Wang et al, 2008).
In the present work, we set-up a system that probes the ability of sister chromatids to interact physically, thus revealing sister chromatid cohesion directly in live cells. This new genetic system reports the probability of newly replicated copies of a locus colliding with each other. We used the extensively characterized site-specific recombination system of bacteriophage P1 Cre/loxP (Hamilton and Abremski, 1984) combined with a reporter gene to disclose sister chromatid molecular interactions (SCI) along the E. coli chromosome. The assay, called LacloxP, revealed that SCI is tightly controlled and lasts for 10–30 min following replication. Beside, the Laclox assay was used to test the involvement of several candidates in SCI. We showed that factors that promote co-localization of sister foci do not necessarily increase the ability of sister loci to recombine. Topo-IV was the only factor to display strong variation in sister chromatid interactions (SCI) when altered. The decatenation activity of Topo-IV was required to limit the extent of SCI, suggesting that precatenation links modulates post-replication cohesion between sister chromatids.
Results
The LacloxP assay to probe sister chromatid interactions
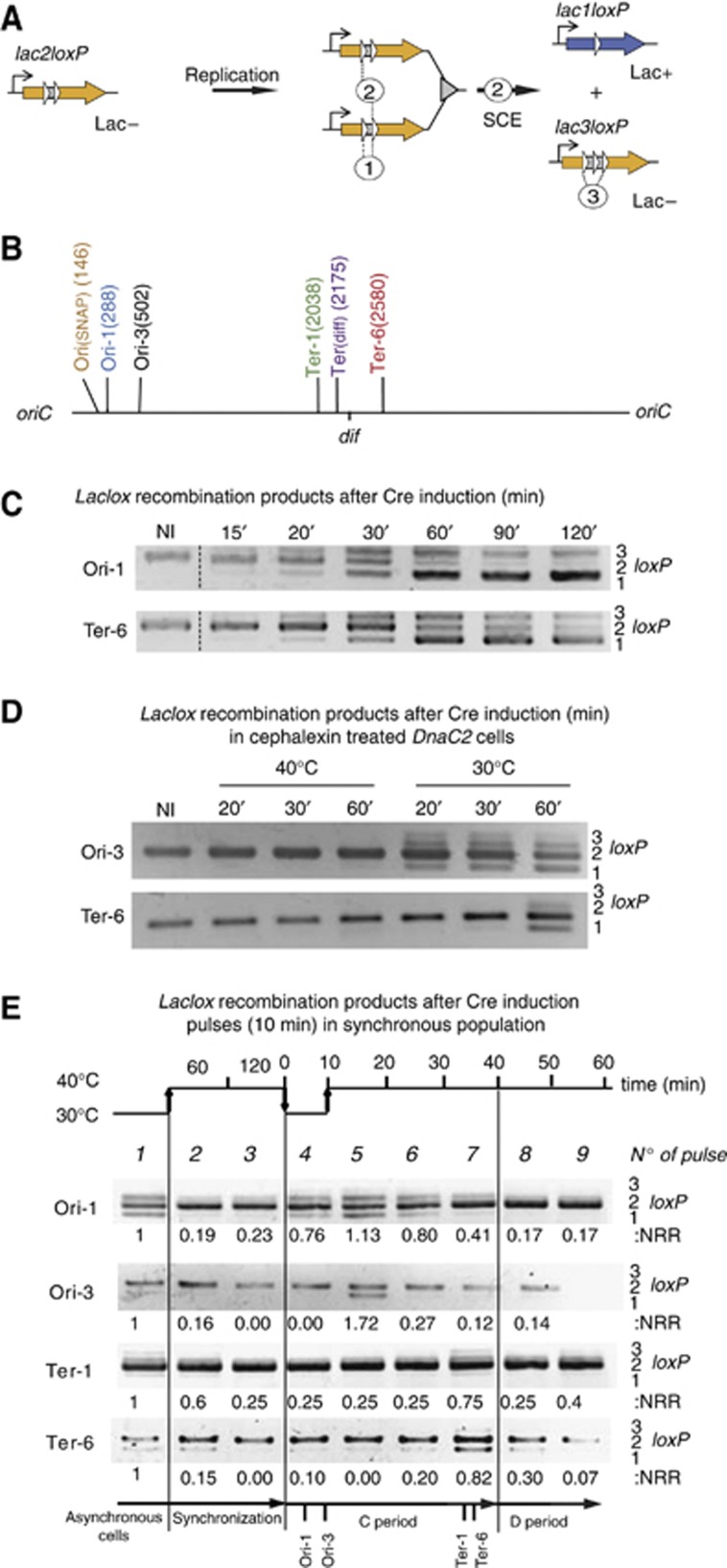
We developed a genetic tool to reveal the physical interactions between sister chromatids in vivo (Figure 1A). This LacloxP assay was based on the Cre/loxP site-specific recombination system of the bacteriophage P1. The loxP cassettes were designed to detect recombination between sister chromatids. The lacZ gene was interrupted by two directly repeated loxP sites separated by 21 bp (lac2loxP in Figure 1A). β-galactosidase activity was retained in the presence of an in-frame fusion of a single loxP in lacZ (lac1loxP). The recombination of two loxP sites carried by the same molecule, i.e., an intramolecular/intrachromatid event (event 1 Figure 1A), was prevented by the proximity of the two loxP sites. The minimal distance required between two loxP sites for them to recombine was reported to be 82 bp; no recombination was detected in vitro below this distance (Hoess and Abremski, 1985). Thus, the reconstitution of an active lacZ gene that conferred the Lac+ phenotype (lac1loxP) was predicted to require recombination between the replicated sister LacloxP cassettes, i.e., an intermolecular/interchromatid event (event 2, Figure 1A). We introduced the LacloxP recombination cassette in six different chromosome loci: three close to the oriC (Ori-1, Ori-3 and Ori(SNAP)), three in the terminus region (Ter-1, Ter(dif) and Ter-6) (Figure 1B). To control recombination, the Cre recombinase gene was carried by a pSC101 plasmid and was under the control of the PBAD promoter. Induction of cre was achieved by the addition of 0.1% L-arabinose in order to ensure the homogenous induction of the PBAD promoter in the population.
Figure 1.
The LacloxP assay revealed sister chromatid interactions. (A) The LacloxP construct consisted of the lacZ gene (orange arrow) under a constitutive promoter that was interrupted by two directly repeated loxP sites (white arrows). This construct conferred the Lac− phenotype. The intramolecular recombination event (noted 1) was prevented by the proximity between the two loxP sites. After replication, intermolecular recombination occurred between sister chromatids (noted 2) when Cre was produced. This recombination forms the lac1loxP product (blue arrow), conferring the Lac+ phenotype. The intramolecular recombination between the first and third loxP sites of lac3loxP product is noted event 3. (B) Map of the E. coli chromosome with the position of the six loci tested in this study labeled. The positions are indicated in kb from the oriC. (C) Recombination events were revealed using PCR amplification that detected the substrate, lac2loxP (406 bp), and products, lac1loxP (351 bp) and lac3loxP (460 bp). The gel shows PCR amplification obtained on total DNA sample extracted from MG1655 strains containing the LacloxP system at the Ori-1 or Ter-6 loci before (NI) and after Cre induction for the indicated time. (D) LacloxP recombination happens on replicating chromosomes but not fully segregated chromosomes. Recombination events were revealed using PCR amplification following Cre induction in the MG1655dnaC2 strain. Replication initiation was blocked by a 2-h shift to a non-permissive temperature (40°C) simultaneously cytokinesis was prohibited by the addition of cephalexin (10 μg/ml). In these conditions the cells filamented and accumulated segregated nucleoids (data not shown). Cre induction was performed at 40°C for 20, 30 or 60 min, or following replication initiation after a downshift from 40 to 30°C for 20, 30 and 60 min. (E) LacloxP recombination follows the replication forks. We used the dnaC2 thermosensitive allele to synchronize the population. Cells were grown at 30°C until OD 0.2 and were shifted to 40°C for two hours to allow the completion of ongoing replications. Replication was initiated by a 10 min shift at 30°C, and cells were returned to 40°C to avoid over-initiation. Nine pulses of 10 min Cre induction were performed and then the cells were immediately harvested and frozen in liquid nitrogen. Genomic DNA was extracted and used for PCR amplification, revealing the formation of LacloxP recombination products. The quantification of recombination is presented (NRR), it corresponds to the measure of the amount of lac1loxP plus lac3loxP compared to the amount of lac2loxP. The values were normalized to that observed for the asynchronous culture.
Recombination events between sister chromatid are rapidly detected by a PCR assay
The formation of recombinant products, lac1loxP and lac3loxP, was detected by PCR assay (Figure 1C). The lac1loxP and lac3loxP products were detected 15 min after Cre induction and accumulated to high levels after 30 and 60 min, respectively; most of the lac2lox substrate was converted into lac1loxP and lac3loxP products. After 120 min, the reciprocal recombination product lac3loxP was hardly detected, indicating that it was converted into the lac1loxP product. In lac3loxP, the external loxP core sequences were separated by 105 bp and therefore supported intramolecular recombination in vivo (event 3, Figure 1A). Because of event 3 on the lac3loxP substrate, the amount of SC in the lac1lox configuration can nearly reach 100% upon a long cre induction.
We first tested if lac2loxP recombination could occur between fully segregated chromosomes. We used an E. coli strain where replication initiation was under the control of a dnaC2, a thermosensitive allele of the DnaC initiation protein. At a non-permissive temperature (40°C), replication initiation was blocked, but ongoing rounds of replication were completed (Withers and Bernander, 1998). In the presence of cephalexin, which inhibits cytokinesis, the cells grew into filaments that harbored 4–8 fully replicated and segregated nucleoids. In these conditions, the induction of the Cre recombinase (for 20–60 min) did not result in the formation of any of the recombinant product (Figure 1D). This showed that, according to predictions, recombination did not occur between the directly repeated loxP sites that were spaced 21 bp apart on the same chromosome. Furthermore, recombination did not occur between fully segregated chromosomes. In contrast, when the temperature was shifted to 30°C concomitantly with cre induction, recombination products were observed after 20 min for the Ori-3 locus and after 60 min for the Ter-1 locus, consistent with their relative position on the replichores. This suggested that lac2loxP recombination was strictly dependant on replication of the locus.
Sister chromatid cohesion during a limited time window after their replication
We designed an experiment to determine the period of the replication cycle that is permissive for SCI (Figure 1E). To do so, we synchronized the cells using a thermosensitive allele of DnaC (dnaC2) that blocks initiation of replication at non-permissive temperature (40°C). The synchronization was achieved through three successive temperature shifts: (i) a 30–40°C shift for 2 h to terminate the ongoing round of replication; (ii) a 40–30°C shift for 6 min to initiate replication; (iii) a 30–40°C shift to inhibit further initiation of chromosome replication. We used flow cytometry to control the synchrony and measure the replication rate at 40°C (Supplementary Figure S1A); replication appeared to be completed in about than 40–50 min and half of the population had achieved cell division after 70 min (Supplementary Figure S1B). Assuming a linear progression of the forks, we approximated the times of replication of the Ori-1, Ori-3, Ter-1 and Ter-6 loci to be 5, 8, 35 and 36 min after initiation of replication, respectively.. The expression of cre was induced by 10 min pulses: one at 30°C before synchronization, two during the period of synchronization at 40°C (after 60 and 120 min), one during the temperature downshift at 30°C that triggered the initiation of replication, and finally five more during the subsequent incubation at 40°C. Cells were immediately frozen in liquid nitrogen after each pulse and genomic DNA was extracted. PCR amplification was performed to reveal the formation of lac1loxP recombination products (Figure 1E). Recombination was observed for every locus in the asynchronous cultures (pulse number 1). The amount of recombinant product lac1loxP in the asynchronous culture was used as a reference and normalized to 1 for the quantification of the recombination in the synchronized cells. The recombinant products were not detected after the inhibition of replication initiation at 40°C (pulses no 2 and no 3 in Figure 1E). Recombinant products were detected at early time points (10 and 20 min) following the synchronous replication initiation at the Ori-1 locus (no 4 and no 5 in Figure 1E) and were still detectable 30 and 40 min after replication initiation (no 6 and no 7). For the Ori-3 locus, recombinant products were detected 20 min after initiation of replication (pulse no 5) and were hardly visible 30 min after replication initiation (pulse No 6). For the Ter-1 andf Ter-6 loci, recombinant products were detected 40 and 50 mintes after replication initiation. These results indicated that recombination between sister chromatids can occur immediately after replication and only for a limited period of time, about 30 min for Ori-1 and 10–20 min for Ori-3, Ter-1 and Ter-6. Note that the amount of lac1loxP product detected by PCR was relatively low. This result suggested either that only a small fraction of the sister chromatids was maintained in a sufficient proximity to allow recombination or that the level of recombinant product was underestimated, perhaps because part of the long-lived Cre/loxP synapse intermediate was not a suitable template for PCR amplification (see below).
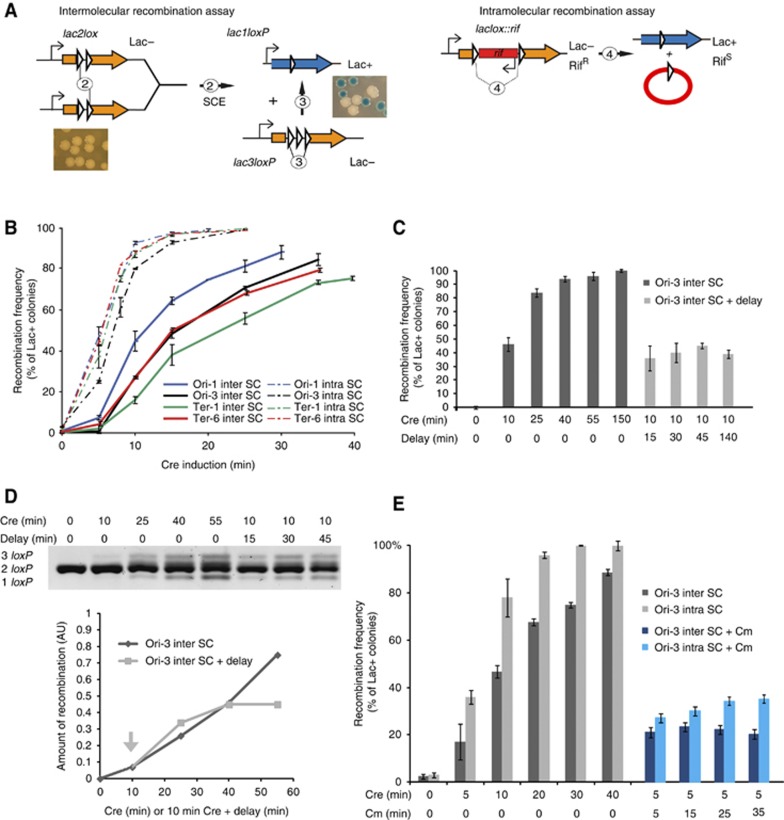
Lac+ colonies formation is an accurate measure of SCI
While the direct detection of recombinant products was enlightening the temporal control of SCI, because of the strong constraints imposed by the synchronization procedure, this approach was not appropriate to quantify SCI and analyze parameters that may modulate SCC. We used the same intermolecular recombination assay described above but the frequency of recombination (forming lac1loxP product) was measured according to the amount of Lac+ colonies observed on X-Gal plates (Figure 2). To assess the level of SSC in different regions of the chromosome, we performed the recombination assay with lac2loxP cassettes inserted at six different positions (Ori-1, Ori-3, Orisnap, Ter-1, Ter-6 and Terdif). Differential abilities of chromosome regions to promote LacloxP recombination could reflect either differential abilities for sister chromatids to interact or the difficulty for Cre to form synapsis between loxP sites in some regions of the chromosome. To discriminate between these two possibilities, we also measured for the same loci the capacity of intramolecular loxP/Cre recombination to estimate the relative access of Cre to different chromosomal regions (Figure 2A). The intramolecular recombination cassette contains two directly repeated loxP sites spaced by the rifampicin resistance gene: the 1 kb intervening segment separating the two loxP sites ensured efficient synapse formation in cis (Figure 2A). We recorded the number of recombinants as a function of Cre induction time for the six loci carrying cassettes for the intermolecular or the intramolecular assay (Figure 2B and Supplementary Figure S2C). The curves for the intramolecular recombination followed simple enzymatic rules: a short lag is observed (<1 min), then a robust and linear recombination rate is observed for every locus (∼10–13%/min). After 10 min of induction, a plateau is reached at approximately 100%. The curves for the intermolecular recombination differed slightly. We observed a longer lag (∼3 min) followed by a linear recombination rate (∼3–7%/min). A plateau (70–80% of Lac+ colonies) is reached after 25–40 min of induction (Figure 2B).
Figure 2.
Lac+colonies formation is an accurate measure of SCI. (A) Description of the inter and intramolecular recombinations. The intermolecular reaction, as described on Figure 1A, involves sister chromatid exchange between loxP sites (SCE). The intramolecular recombination between loxP sites distant from 1 kb can be monitored at every loci tested by the deletion of a Rif cassette and the reconstitution of the lacZ gene. It is used to control Cre accessibility at the different loci tested and the influence of the genetic background on Cre reactivity. Both inter and intramolecular recombination frequencies can be measured on plate by the formation of blue colonies. (B) Measurement of the frequency of SCI (inter-SC) and intramolecular recombination (intra-SC) according to the extent of Cre induction. The curves represent the average of four experiments. (C) Measure of the frequency of SCI by Lac+ colonies counting at the Ori-3 locus (inter SC). Following Cre inductions for 10–150 min after induction the cells were diluted 104 fold and immediately plated (dark gray). After the 10 min induction, cells were diluted 104 fold and kept in liquid culture with shaking for a delay up to 140 min before plating (light gray). (D) PCR analysis of the recombination products observed for the Ori-3 locus in the same induction conditions than on Figure 2C. Genomic DNA was extracted immediately after induction or after the indicated dilution delay. The amount of recombined products (3 loxP+1 loxP) was measured by density scanning with a Typhoon after electrophoresis (AU arbitrary densitometry unit). The arrow indicates the timing of the dilution. (E) Measure of the ability of Cre to produce recombined colonies following translation inhibition by the addition of chloramphenicol (Cm). The recombination frequency at the Ori-3 inter SC and Ori-3 intra SC cassette was measured by Lac+ colonies counting in the absence of Cm (gray boxes) or in the presence of Cm (120 μg/ml) 5 min after the addition of arabinose (blue boxes), the cultures were incubated at 30°C for the indicated times, then diluted 104 fold and immediately plated.
In order to quantitatively relate the level of recombination to the molecular cohesion, we tested if the rate of recombination of the Ori-3 locus was dependent on the time left after induction and before plating. Following cre induction, cultures were diluted to stop the induction, and either immediately plated or kept at 30°C for 15–140 min before plating (Figure 2C). When the cultures were immediately diluted and plated, the frequency of Lac+ colonies observed on plate raised quickly to 100%. Strikingly, increasing the delays before plating did not change the frequency of Lac+ colonies. Similar results were observed with the the Ori-1 and Ter-1 loci (Supplementary Figure S2A). These observations suggest that most of the recombination took place during the induction pulse. The amount of recombined DNA was also directly monitored by PCR analysis (Figure 2D) or Southern blot (Supplementary Figure S2B); they revealed that recombined Ori-3 products were not efficiently detected immediately after the recombination pulse, they accumulated during the dilution step to reach a plateau (∼40% for a 10 min pulse, i.e. a similar amount than Lac+ colonies). These experiments indicated that recombination intermediates formed at the end of the induction period were converted into recombinant products during the 30 min that followed cre induction. These observations imply a rapid inactivation of the Cre recombinase following dilution of the arabinose inductor. We confirmed that the formation of Lac+ colonies did not rise after induction by using chloramphenicol to block translation at the end of the induction pulse (Figure 2E). The intramolecular recombination cassette was used at the Ori-3 position to measure recombination in the presence of chloramphenicol (for 5–35 min after induction). As observed for the dilution experiments, the amount of Lac+ colonies did not increase after inhibition of translation. These experiments indicated that the amount of Cre able to perform new recombination on SC formed in the minutes following the induction pulse is very limited. Therefore the number of recombinant resulted from the number of loxP synapses that formed during the induction time or shortly after; discrepancies between the PCR and the plating assays likely originated from a slow resolution of the synapse into recombination products. The amount of recombinant colonies on plate is likely the best quantification of the number of cells presenting molecular cohesion for the defined locus in the population for a given time interval, i.e. sister chromatids involved in molecular cohesion.
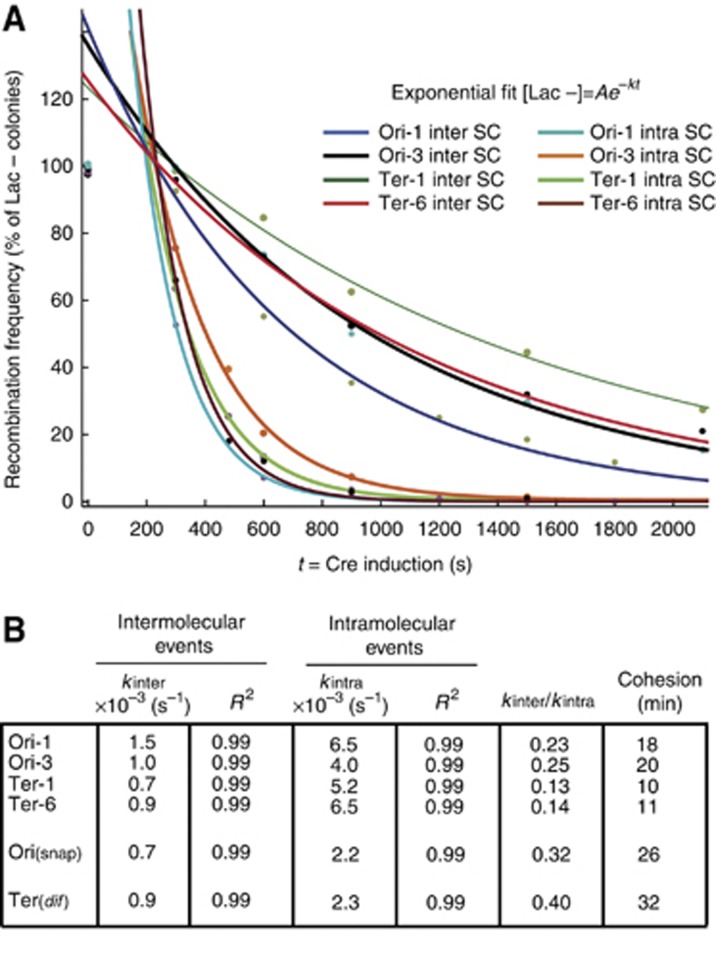
LacloxP recombination reveals variability in sister chromatid cohesion along the chromosome
Considering that intramolecular recombination is possible at any step of the E. coli cell cycle, we postulated that the intermolecular recombination rate relates to the fraction of the cell cycle when SC are available for recombination, i.e. the cohesion period. After the initial lag, both intermolecular and intramolecular reaction were following a single exponential decay (Amount of Lac- colonies)=Ae−kt. A is a constant that is close to 150% for the intermolecular reaction and close to 300% for the intramolecular reaction, t is the length of the induction period and k is catalytic constant of each reaction (sec−1) (Figure 3A and Supplementary Figure S2D). For the intramolecular reaction, k varies from 2 to 6.5 × 10−3 s−1 suggesting that Cre recombination is not equivalent in every region of the chromosome. These values are in the same range as that measured in vitro (3–7 min per event) confirming that Cre recombination is not a fast reaction (Fan, 2012). For the intermolecular reaction, k varies from 0.7 to 1.5 × 10−3 s−1 (Figure 3B). Considering that when the SC are in close contact the inter and intramolecular reactions are equally efficient, the ratio kinter/kintra gives an estimation of the portion of the cell cycle allowing inter SC recombination. We estimated that SC were in contact for 18 min (23% of the 80 min cell cycle) at the Ori-1 locus, 20 min at the Ori-3 (25% of 80 min) locus, 10 min (13% of 80 min) at the Ter-1 locus and 11 min (14% of 80 min) at the Ter-6 locus (Figure 3B). Since intramolecular recombination efficiency were comparable for the Ori and Ter loci, it was unlikely that the difference in the estimated cohesion would be caused by the higher level of recombination resulting from a higher copy number of the Ori loci compared to the Ter loci. We measured the kinetic parameter of SCI for two other loci known for an extended colocalization of newly replicated sister chromatid, in the snap region (Orisnap) (Joshi et al, 2011) and near the dif site (Terdif) (Espeli et al, 2008). SC at the Orisnap and Terdif loci, respectively, support intermolecular recombination for 32% of the cell cycle (26 min) and 40% of the cell cycle (32 min) (Figure 3B, Supplementary Figure S2B and C). SCI was high for the snap and dif regions; it suggested that the LacloxP assay and the colocalization of FROS tags were able to catch similar constraints on the SC. These observations suggested that the frequency of LacloxP recombination is an accurate tool to monitor SC proximity.
Figure 3.
Sister chromatid interaction varied according to the locus considered. (A) Cre recombination follows single exponential decay kinetics for the inter and the intramolecular recombination reactions. The data from Figure 2B were plotted according to the disappearing of Lac− colonies versus the length of the induction. Single exponential fits are represented. To avoid taking into consideration the lag observed during the first 5 min, the fits exclude the time 0 and 2.5 min of induction. (B) Kinetic constants, kinter and kintra were presented for every reaction. The estimation of the Cohesion Period was given by CP=τ × (kinter/kintra), where τ is the generation time (80 min).
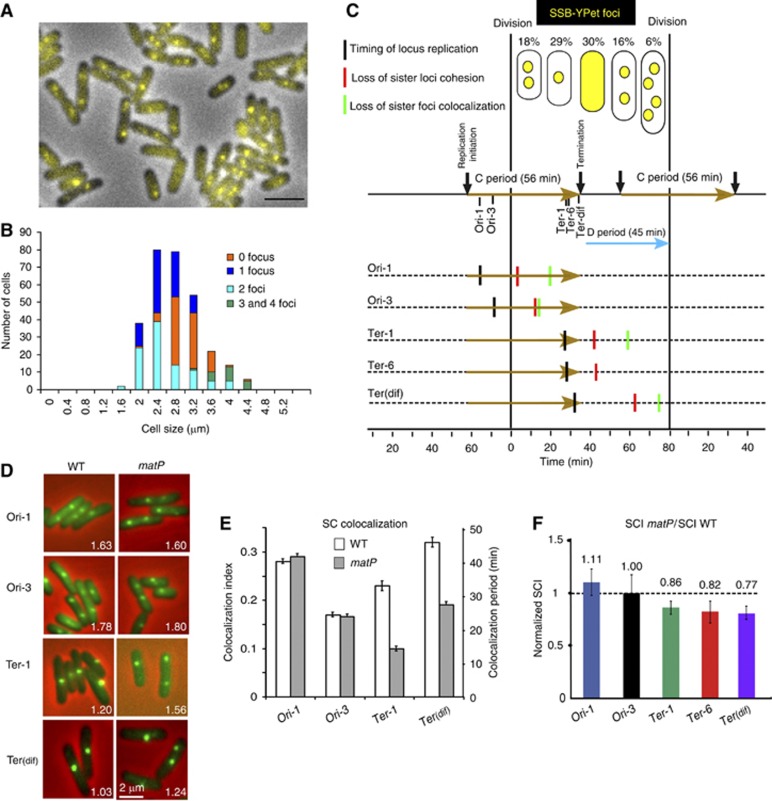
Distinction of SCI and colocalization of loci
Sister loci colocalization has been extensively used as an indirect measure of the cohesion period in both eukaryotic (Straight et al, 1996) and bacterial cells (Sunako et al, 2001; Bates and Kleckner, 2005; Espeli et al, 2008). In bacteria, the FROS systems, the parS/ParB-GFP systems and the FISH technique have been used to monitor SC colocalization. However, these approaches remain unsatisfactory and controversial because colocalization varies according to the growth rate (Adachi et al, 2008), it does not necessarily imply that sister loci can interact physically and it relies on a difficult determination of the cell cycle parameters, C and D periods, in bacteria, (Nielsen et al, 2007; Adachi et al, 2008; Espeli et al, 2008; Lesterlin et al, 2008). The quantification of the number of SSB-Ypet foci and their repartition according to the cell size is a precise way to define the C and D period (Reyes-Lamothe et al, 2008; Espeli et al, 2012). The characterization of the cell cycle parameters according to the SSB-Ypet pattern (Figure 4A and B) is presented on Figure 4C. New-born cells were finishing the replication rounds initiated in the mother cell; they presented two SSB-YPet foci corresponding to the forks that replicated the right and left replichores. Later, the two foci merged at mid-cell for the replication of the terminus region. After a period without replication (cells lacking SSB-YPet focus), the replication was initiated simultaneously on two chromosomes (large cells with two SSB-YPet foci). A few minutes before division, the replication forks separated, giving rise to cells with 4 SSB-Ypet foci. The amount of cells without SSB-YPet foci (30%) allows the direct estimation of the replication period C (56 min; ∼70% of 80 min), The amount of large cells with 2 and 4 foci (22%) allows the determination of the timing of initiation (22 min before division) and therefore of the time between termination and cell division, the D period (46 min).
Figure 4.
Sister chromatid colocalization and SCI are not equivalent measures of sister chromatid cohesion. (A) The SSB-Ypet fusion was used to monitor replication parameters. A representative picture of the MG1655-SSB-YPet strain grown in Minimal Medium A+casamino acids+glycerol. (B) histogram of the amount of cells present in each cell size category according to the number of SSB-YPet foci observed in the cells. (C) Representation of the cell cycle in the MG1655 strain grown in Minimal Medium A+casamino acids+glycerol at 30°C. The percentage of cells in the population according to the number of SSB-Ypet foci is indicated. The cells with two foci were split into two categories: the small cells represent a size below the median size of the cells with 1 focus, and the large cells represent a size above the median size of the cells with 1 focus. The generation time (τ) is 80 min, the C period is 56 min (brown arrows) and the D period is 45 min (cyan arrow). The timing of replication (black vertical bars), the estimated timing of the loss of SC cohesion (red vertical bars) and the estimated timing of the loss of SC colocalization (green vertical bars) are indicated for each locus. (D) Representative pictures of the parS/ParBP1-GFP tag inserted at the Ori-1, Ori-3 Ter-1 and Terdif locus. The MG1655 and MG1655matP cells were grown in minimal medium A supplemented with casamino acids and glycerol (Green parS/ParBP1-GFP, Red Phase contrast microscopy). For each strain the average number of foci per cell is indicated. (E) Measure of the colocalization of the sister parS/ParBP1GFP foci. The cell cycle parameters (generation time, length of the C period and D period) measured in Figure 4A–C were used to estimate the average number of copies of each locus in the cells (Nbloc). The average number of foci per cell (Nbfoc) was measured from (Figure 4D). The left axis represents the colocalization index, defined as (Nbloc−Nbfoc)/Nbloc. The right axis represents the estimated lifespan of SC colocalization. (F) Measure of SCI with the LacloxP system for the Ori-1, Ori-3, Ter-1, Terdif and Ter-6 loci in the MG1655matP strain. The relative normalized recombination frequency is defined as (Inter SC matP/Intra SC matP)/(Inter SC wt/Intra SC wt).
We compared the extent of molecular cohesion observed with the LacloxP assay (Figure 3) and the extent of sister loci colocalization observed by the parS/ParB-GFP FROS (Figures 4C and D). We used the cell cycle parameters to calculate the timing of replication and the extent of the colocalization period for each locus (Figure 4E). We estimated that sister Ori-1, Ori-3, Ter-1 and Terdif loci were colocalized for 37, 22, 31 and 45 min, respectively, following their replication and cohesive at the molecular level for 18, 20, 10 and 32 min, respectively (Figure 3B). For most of the tested loci, the estimated period of colocalization was longer to that of molecular cohesion (Figure 4C). The LacloxP assay only picks up a fraction of the colocalized SC; this suggests that colocalized SC correspond at least to two states: one where SC are close enough to recombine frequently, i.e., the SCC step, and one where the sister loxP are not able to recombine. In order to get more insight into the relationship between SCI and colocalization, we measured SCI in genetic backgrounds that have been previously reported to modulate the duration of chromosome loci colocalization.
MatP influenced loci colocalization but not molecular cohesion
MatP, the structuring factor of Ter MD is required for the colocalization of the replicated Ter MDs to hold them together in close spatial proximity before bipolar segregation (Mercier et al, 2008). It is therefore possible to use matP mutant cells to investigate the interdependence between the level of SCI and the colocalization lifespan of the structured Ter MD. The matP deletion specifically altered colocalization (50% reduction) of the sister foci in the Ter MD under growth conditions used for the recombination assays (Figures 4D and E). Ter-1 and Terdif loci, colocalization was observed for 31 and 45 min in wt cells and only for 13 and 24 min in the absence of MatP. In the matP mutant, the level of SCI was only slightly decreased compared to the wild type: a 14, 18 and 23% decrease, respectively, for the Ter-1 Ter-6 and Terdif loci (Figure 4F and Supplementary Figure S3). This result shows that most of the SCI was retained in a context where colocalization was profoundly altered. Therefore, MatP-mediated colocalization does not enhance the interactions between sister Ter loci, indicating that SCI is established by another factor or mechanism. Strikingly, in the Ter domain, the remaining colocalization observed in the absence of MatP is close to the extent of molecular cohesion determined with the LacloxP assay, it suggests that, in the Ter MD, sister chromatid colocalization is the consequence of two event: (i) a SCI step independent on MatP; (ii) a SC colocalization step dependent on MatP. These results showed that colocalization revealed by fluorescence imaging should not be associated to SCC since the duration of spatial proximity between sister loci does not reveal their ability to interact physically.
Topo-IV activity modulated sister chromatid cohesion and colocalization
Topo-IV is an important player in sister loci segregation (Wang et al, 2008) and we investigated the effect of modulating Topo-IV activity on the level of SCI. We tested the effect of transient inactivation of Topo-IV on the frequency of LacloxP recombination. The recombination frequencies of LacloxP cassettes inserted at four different positions were measured in wt and parC1215ts (Topo-IVTS) strains. The wt and Topo-IVTS strains exhibited the same frequency of intramolecular recombination under permissive and after a 20 min shift to non-permissive temperatures Supplementary Figure S4A). By contrast, intermolecular recombination was enhanced for all loci at permissive and non-permissive temperatures (Figure 5A). We also tested the effect of Topo-IV overproduction using a plasmid that encoded ParE and ParC, the two subunits of the Topo-IV. Unfortunately, upon IPTG induction, Topo-IV overexpression had detrimental consequences on cell physiology (Mossessova et al, 2000) and perturb greatly both inter and intra molecular recombination (data not shown); it limited the quantitative analysis, however intermolecular recombination was still detectable at high level in these conditions. In the absence of IPTG, conditions where a leak of the inducible promoter produced a small increase of Topo-IV amount, we observed a small reduction of SCI for the Ori-3 and Ter-1 respectively, 0.72±0.05 and 0.76±0.04 of the SCI observed with the empty vector.
Figure 5.
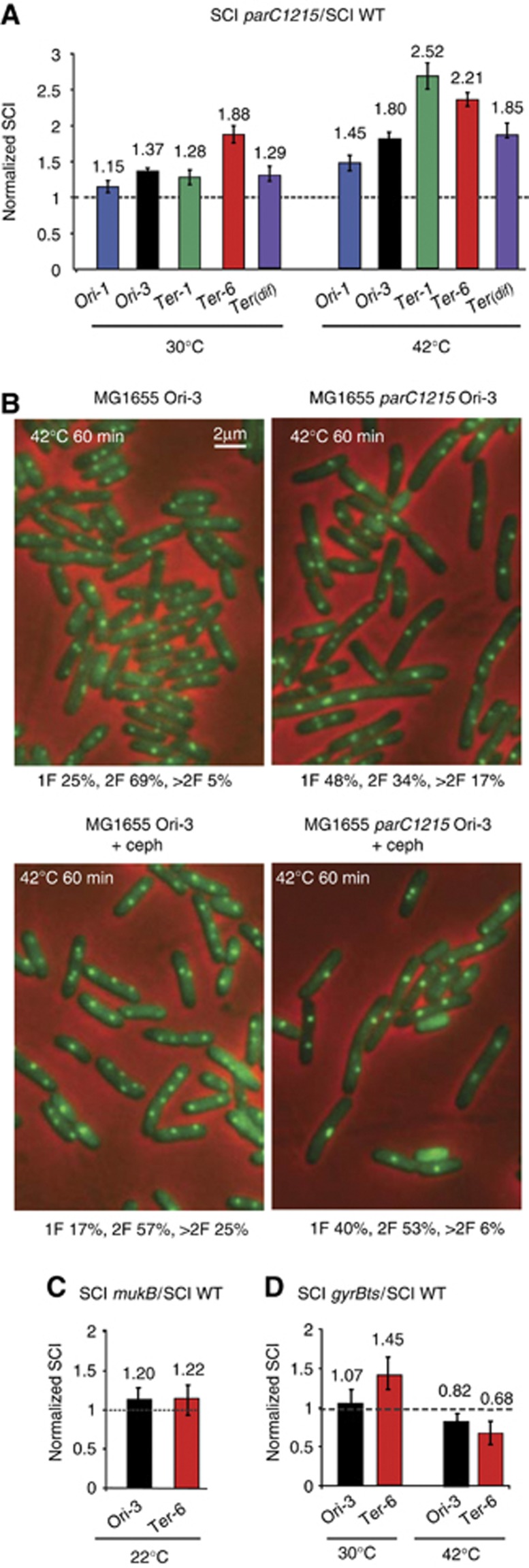
Topoisomerase-IV (Topo-IV) controls the extent of SCI. (A) The measure of SCI using the LacloxP system for the Ori-1, Ori-3, Ter-1, Ter-6 and Terdif loci in the MG1655parC1215 strain at a permissive temperature of 30°C, followed by a 20 min shift to a non-permissive temperature at 42°C. The relative normalized recombination frequency was defined as (Inter SC parC1215/Intra SC parC1215)/(Inter SC wt/Intra SC wt). (B) The inactivation of Topo-IV provokes a reduction in the number of MG1655parC1215 cells segregating the sister Ori-3 parS/ParBP1 foci. A representative picture of the Ori-3 parS/ParBP1-GFP tag in the MG1655 and MG1655parC1215 cells after a 20 min shift at 42°C in the absence or presence of cephalexin to inhibit cell division. The percentage of cells with 1 focus (1F), 2 foci (2F) or more than 2 foci (>2F) are indicated. (C) The measure of SCI using the LacloxP system for the Ori-3 and Ter-6 loci in the MG1655mukB strain at 22°C. Relative normalized recombination was defined as (Inter SC mukB/Intra SC mukB)/(Inter SC wt/Intra SC wt). (D) The measure of SCI using the LacloxP system for the Ori-3 and Ter-6 loci in the MG1655gyrBts strain after a 60 min shift at 42°C. The relative normalized recombination was defined as (Inter SC gyrBts/Intra SC gyrBts)/(Inter SC wt/Intra SC wt).
Using a parS/parBP1-GFP system at the Ori-3 locus, we observed that under the conditions of the LacloxP assay, Topo-IV alteration affected sister chromatid segregation (Figure 5B). Twenty minutes after Topo-IV inactivation, the number of cells with only one Ori-3 locus increased two-fold compared to the wt cells. Due to the concomitant increase in the number of cells with more than 2 foci, the average number of foci per cell remained constant at 1.83 and 1.84 in the WT and Topo-IVTS strains, respectively. Since the average number of foci is the value taken into account in the original estimation of colocalization, this result suggested that SC colocalization might not be changed by Topo-IV inactivation. However, we observed a delay in cell division in the Topo-IVTS strain at 42°C that could be the reason for the increased number of cells with more than two foci. We added cephalexin to block division in both strains. In the presence of cephalexin, the number of cells with only one focus increased and the average number of foci decreased in the Topo-IVTS strain (2.31 in the WT and 1.7 in the Topo-IVts). Therefore, the segregation of sister Ori-3 loci was severely impaired in the absence of active Topo-IV. We performed the same experiment with a parS/parBP1-GFP tag inserted at the Ter-1 locus, SC segregation was also impaired in the absence of Topo-IV (2.1 in the WT and 1.7 the Topo-IVts) (Supplementary Figure S4B).
Regulation of DNA topology around the replication fork controls SCI
The strong influence of Topo-IV transient inactivation on intermolecular recombination and sister foci segregation suggested that destabilizing the topological equilibrium toward the accumulation of positive supercoils and/or precatenanes could trigger a prolonged cohesion period, within which the sister chromatids were prone to interact. It has been shown recently that the ability of Topo-IV to remove positive supercoils, but not its activity on catenanes, was enhanced by an interaction with the SMC MukB protein (Hayama and Marians, 2010). In addition, MukB has been proposed to enhance the colocalization period of sister loci near the oriC (Sunako et al, 2001). We deleted mukB from the cells, which is only tolerated at low temperature (22°C), and performed the recombination assay. A slight increase in the intermolecular recombination rates was observed in the absence of MukB (Figure 5C and Supplementary Figure S5). This showed that the MukBEF complex in E. coli was not directly responsible for the establishment of the cohesion of the Ori-3 and Ter-6 loci. This result is consistent with the fact that the alteration of the decatenating activity of Topo-IV, rather than the positive supercoil removal, was involved in the modulation of recombination between sister chromatids. These results, presented in Figure 5, suggested that precatenanes enhanced sister-chromatid cohesion.
Topological management of the positive supercoils generated by the replication fork was controlled by the activity of the DNA gyrase and eventually Topo-IV downstream of the fork. On plasmid DNA, the excess positive supercoils were transferred to the replicated region and took the form of precatenanes (Peter et al, 1998). Precatenanes have not yet been directly observed on chromosomes. It has been proposed that precatenanes can form in response to an excess of positive supercoils that DNA gyrase could not remove (Postow et al, 2001). Therefore, we tested if alteration of the gyrase activity in the gyrB203ts allele led to an increase in the intermolecular recombination rate. The gyrB203ts allele produce a nearly wt gyrase at permissive temperature (30°C) and a completely deficient enzyme at non permissive temperature (42°C). Compared to the wt strain at a permissive temperature, we observed 7 and 45% increases for the Ori-3 and Ter-6 loci, respectively, in intermolecular recombination, in the strain containing the gyrB203ts allele (Figure 5D and Supplementary Figure S5). After a 20 min shift to a non-permissive temperature (42°C), a significant decrease in the intermolecular rate was observed (Figure 4D). We propose that a slight decrease in the DNA gyrase activity at a permissive temperature was sufficient to increase precatenation without impeding replication (Grompone et al, 2003). At a non-permissive temperature, excess positive supercoils ahead of the fork trigger rapid replication fork blockage, and ongoing decatenation by Topo-IV reduces the sister chromatid interaction frequencies.
Discussion
Sister chromatid cohesion at the molecular level in E. coli
Here, we report the construction of a new molecular probe that quantitatively revealed the level of sister chromatid physical interactions (SCI) in live cells. The reconstitution of the lacZ reporter involved intermolecular recombination between newly replicated sister loxP sites. Strikingly, recombination was dramatically higher between SC of replicating chromosomes compared to fully segregated chromosomes (Figure 1C), suggesting that a particular structure is linking the two SC. The ability to recombine loxP sites suggests that portions of sister chromatids are kept aligned for several minutes. These characteristics made the LacloxP genetic assay a valuable and appropriate tool to investigate the interaction ability of the newly replicated sister loci and reveal their close proximity. The rate of Lac+ colonies after cre induction appears to be a reliable and accurate method to quantify the relative molecular cohesion at a defined locus.
The lifespan of chromosome cohesion has been previously inferred from sister loci colocalization studies (Sunako et al, 2001; Bates and Kleckner, 2005; Espeli et al, 2008). This indirect approach based on fluorescent microscopy was intrinsically controversial since sister loci colocalisation does not necessarily implicates or reflects their ability to cohere at the molecular scale. Furthermore, no consensus view emerged as these studies led to different estimates of the colocalisation periods. Our study provided evidence that the level of colocalization is not always reflected in the level of sister chromatid cohesion. Sister loci were consistently involved in a cohesion step following their replication. SCI were detected at all loci tested between 10 and 32 min. These cohesion estimates are always shorter than the extent of sister chromatid colocalization under the same conditions (Figure 4C), suggesting that part of the colocalization period is not giving rise to sister loci interactions (Figure 6). For instance, the early stages of sister loci separation would still exhibit colocalisation without being able to interact at the molecular scale (see yellow zone in Figure 6). Indeed, the alteration of sister colocalization in Ter MD, following the deletion of matP, did not correlate with a comparable reduction of SCI. Therefore, as revealed in the matP strain, colocalization involved mechanisms that were superimposed to the cohesion itself. Remarkably, loci in the snap regions and near the dif site that presented long colocalization periods also presented a strong SSC suggesting that the two events could be correlated. Further studies using the LacloxP assay would allow performing a quantitative analysis of the SCI extent at the genome scale. Furthermore, this genetic assay can theoretically be adapted to a variety of organisms and be used to measure the ability of two homologous or heterologous DNA pieces to interact physically in live cells.
Figure 6.
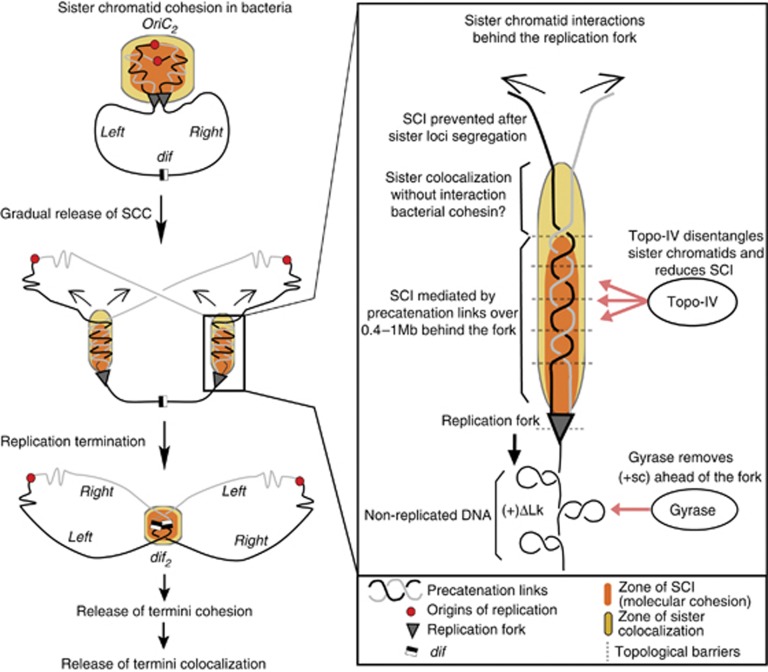
Model for establishment and release of sister chromatid cohesion in E. coli. In E. coli (middle panel) SSC is gradually released behind the replication fork in such a way that segregation of sister loci begins before replication terminates. On the present model, SCC is mediated by pre-catenation links that promote sister chromatids interactions (SCI, orange areas) and colocalisation (yellow zone). Interestingly, colocalisation does not necessarily confer the ability to interact physically. For Topo-IV, the main decatenase in E. coli, is proposed to release SSC by removing pre-catenation links created by replication. Concomitantly, DNA gyrase acts before the replication fork to relax positive supercoils (+sc). SCC is then established immediately after replication, and released after a time frame required for resolution of topological links. Topological barriers or chromosome segregation are able to limit the diffusion of precatenation The persistence of SCI for a 10–30 min period after replication suggests that a region about 400 kb to 1MB is involved in cohesive precatenation.
Roles for sister chromatid cohesion in bacteria
As observed in eukaryotes, SCI in E. coli is established during S phase. This process has functional implications. First, it should ensure the availability of the sister chromatid to repair newly synthesized damaged DNA duplexes near the replication fork (Cox et al, 2000). We observed a long SCI period for the dif proximal locus in the terminus of replication. In our study, this locus is the only one that would still present SCI for a long time after the end of S phase (Figure 6). The prolonged SCI at the dif locus could favor chromosome dimmers resolution by the XerCD/dif system (Deghorain et al, 2012). SC cohesion in eukaryotes plays another important role by securing chromosome segregation. The processes controlling chromosome segregation in bacteria are not fully understood. A number of segregation machineries have been shown to interact with the Ori region (for review see (Possoz et al, 2012)). However, the involvement of active mechanisms to promote the segregation of the chromosome bulk is not known. In this regard, SCC may help to control the timing of sister chromatids separation in the two cell halves.
Topo-IV modulated sister-chromatid cohesion
We showed that transient inactivation of Topo-IV dramatically increased the level of SCI in three of the four loci tested. Topo-IV was the main E. coli enzyme involved in removing catenanes links between two circular DNA molecules (Zechiedrich et al, 1997). This indicated that the entanglement of replicated sister sequences facilitated their interaction, likely by delaying their segregation (Figure 6). The work presented on Figure 5 allows estimating that the absence of Topo-IV increases sister chromatids cohesion by two fold. It is consistent with the observation that Topo-IV activity modulated the lifespan of sister sequence colocalization around the chromosome in E. coli (Figure 4) and (Wang et al, 2008) and C. crescentus (Wang and Shapiro, 2004). Based on these observations, the pairing of sister chromatids could result from the formation of precatenanes. The superhelical tension accumulated in the front of the fork as positive supercoils would be converted into precatenanes behind the fork, upon free rotation of the replication fork (Postow et al, 2001).We propose that precatenanes formed behind the replication fork enhance sister chromatid cohesion (see pink zone on Figure 6). The reduction of SCI observed upon mild overexpression of Topo-IV suggests that in wt cells precatenanes form some of the cohesion links between sister chromatids. We cannot rule out the hypothesis that cohesion is established through a different mechanism (a yet uncharacterized cohesin like activity) and that precatenation would reinforce this mechanism. Such interplay between cohesins and DNA tangles has been recently observed during SCC in yeast (Farcas et al, 2012). Precatenanes have been observed in vivo on replicating plasmids (Peter et al, 1998; Schvartzman et al, 2011), but have not been directly observed on chromosomes. Peter and colleagues demonstrated that the positive supercoils ((+) ΔLk) generated by plasmid replication diffuses throughout a topological domain so that it is distributed both ahead, (+) ΔLk, and behind the replication fork, precatenanes (Peter et al, 1998). Therefore, topological barriers that limit the diffusion of precatenanes are required to explain the different extent of SCI observed for the different loci tested. For example the high transcription rate of ribosomal DNA could serve as topological barriers and concentrate the precatenation links around the snaps regions near the origin (Joshi et al, 2011). Alternatively, the segregation of the chromosome per se could force the precatenanes to accumulate in the regions in contact with the replication forks (Figure 6). Various speed or tension imposed by the segregation of an upstream region would therefore modulate the extent of cohesion of the downstream regions.
Modulation of precatenation during the cell cycle
The partition of the work between DNA gyrase and Topo-IV during replication remains unclear. It is assumed that the gyrase is in charge of the (+) ΔLk ahead of the replication fork. In the absence of gyrase, Topo-IV provides sufficient (+) ΔLk unwinding to support replication of a small part of the chromosome at one-third of the wt rate (Khodursky et al, 2000). In the absence of Topo-IV, replication proceeds until the end of the chromosome at the wt rate (Wang et al, 2008). Topo-IV activity appeared to reach its peak after the S phase (Espeli et al, 2003). Therefore, Topo-IV is predicted to play a limited role in the dissipation of the (+) ΔLk waves ahead of the replication forks. Our system allowed for the estimation of the extent of the cohesion step, from 10 to 30 min, following replication. At the estimated replication rate of 42 kb/min/fork, the replication forks move 400 kb away from any replicated loci within 10 min, suggesting that sister chromatids are linked over 400–800 kb behind the replication fork. This result raised a question about the regulation of Topo-IV activity. Is the Topo-IV amount limiting compared to the introduction of precatenanes links? We estimate that Topo-IV activity, ∼3000 Lk/s/cell (Espeli et al, 2003) is in large excess compared to the precatenane formation rate, ∼50 Lk/s/fork (Peter et al, 1998). It is likely that a control of Topo-IV activity is required to maintain a sufficient level of precatenation to support sister chromatid cohesion. This regulation could rely on the differential subcellular localization of ParE and ParC subunits, as observed in E. coli (Espeli et al, 2003) and C. crescentus (Wang and Shapiro, 2004).
Materials and methods
General procedures and constructs
The wild type strain was E. coli K12 MG1655 (ΔlacMluI). The LacloxP cassette was constructed by the integration of a double stranded oligonucleotide 5′-CGTAATAACTTCGTATAATGTATGCTATACGAAGTTATGGATCCCCGGGTACCGAGCTCATAACTTCGTATAATGTATGCTATACGAAGTTATCCTA-3′ into the ClaI restriction site of the lacZ gene. The LacloxP cassette was integrated in the chromosome, using a vector derived from pKD4 called pGBKD3-Laclox and pGBKD3-Lacloxrif. These vectors contained the LacloxP or the Lacloxrif cassettes adjacent to the chloramphenicol resistance gene. Laclox::Cm and the parS::Cm were inserted into the intergenic regions of non-essential genes (Espeli et al, 2008) using the standard ‘lambda red’ technique (Datsenko and Wanner, 2000). Expression of the Cre recombinase was driven by a pSC101 carrying the arabinose-inducible Cre gene derived from pFX465, kindly given by FX Barre. The Δ30ParB-GFP fusion protein was expressed from plasmid pALA2705. The mukB::Kn, DnaC2::Tet, parC1215TS::Tet, gyrB203ts::Tet, matP::Cm and ssb-ypet::Cm strains were constructed using standard techniques.
Cre induction assay
Bacterial strains containing the LacloxP insertions of interest were grown in appropriate medium and temperature to OD600=0.2. The production of Cre was induced by adding 0.1% arabinose, and induction was stopped by a 104 fold dilution with fresh culture medium or by freezing the cells in liquid nitrogen. Cell samples were immediately plated on X-gal containing plates to measure the ratio (Lac+/total colonies). All experiments were performed in minimal medium A supplemented with 0.2% glycerol and 0.12% casamino acids, except for the synchronization experiment with the dnaC2 strain where glycerol was substituted with 0.2% succinate. Kanamycin (50 μg/ml), streptomycin (100 μg/ml), tetracycline (15 μg/ml) and chloramphenicol (30 μg/ml) were added when required. PCR was performed with the ExTaq (TaKaRa) Taq DNA polymerase, on genomic DNA extracted from liquid nitrogen frozen cells using the Macherey Nagel genomic extraction kit.
Cell cycle features and microscopy
The cell cycle features were experimentally characterized using generation time, SSB-YPet foci pattern and rifampicin/cephalexin flow cytometry (Mercier et al, 2008). The following equation from Helmstetter gives the average number of loci/genes per cell in the population (Helmstetter, 1996): average number of loci=2[C(1−x)+D]/τ where x is the fraction of the C period at which the locus replicates, and τ is the generation time. Therefore, the number of generation from locus replication to division, R=ln(average number of loci)/ln(2). Similarly, the number of generations from focus separation to cell division, S=ln(average number of foci)/ln(2) (Nielsen et al, 2007). The colocalization periods (CP) is given by CP=(R−S) × τ. Cells carrying a parS insertion and plasmid pALA2705 were grown to OD600=0.2, concentrated 50-fold by centrifugation and placed on an agarose pad on a slide. Microscopy analysis was performed as described before (Espeli et al, 2008). For each experiment, 200–400 cells were counted.
Supplementary Material
Acknowledgments
We want to thank Michele Valens for her great experimental assistance, François Xavier Barre for the gift of plasmid pFX465, Hsiu-Fang Fan and Ludovic le Chat for critical reading of the manuscript. The work performed in FB lab was funded by the program ANR 08-Blan-0119 2009–2012. The work performed in OE lab was funded by the program ANR JCJC 2010-2014 SISTERS.
Author contributions: CL, EG, OE performed the experiments; CL, EG, FB, and OE designed the experiments; CL, EG, FB and OE wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adachi S, Fukushima T, Hiraga S (2008) Dynamic events of sister chromosomes in the cell cycle of Escherichia coli. Genes Cells 13: 181–197 [DOI] [PubMed] [Google Scholar]
- Bates D, Kleckner N (2005) Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ (2000) The importance of repairing stalled replication forks. Nature 404: 37–41 [DOI] [PubMed] [Google Scholar]
- Cui Y, Petrushenko ZM, Rybenkov VV (2008) MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol 15: 411–418 [DOI] [PubMed] [Google Scholar]
- Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C (2007) MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol 65: 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deghorain M, Pages C, Meile JC, Stouf M, Capiaux H, Mercier R, Lesterlin C, Hallet B, Cornet F (2012) A defined terminal region of the E. coli chromosome shows late segregation and high FtsK activity. PLoS One 6: e22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F (2012) A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J 31: 3198–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Levine C, Hassing H, Marians KJ (2003) Temporal regulation of topoisomerase IV activity in E. coli. Mol Cell 11: 189–201 [DOI] [PubMed] [Google Scholar]
- Espeli O, Mercier R, Boccard F (2008) DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68: 1418–1427 [DOI] [PubMed] [Google Scholar]
- Fan HF (2012) Real-time single-molecule tethered particle motion experiments reveal the kinetics and mechanisms of Cre-mediated site-specific recombination. Nucleic Acids Res (advance online publication, 29 March 2012; doi:10.1093/nar/gks274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Uluocak P, Helmhart W, Nasmyth K (2012) Cohesin’s concatenation of sister DNAs maintains their intertwining. Mol Cell 44: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompone G, Ehrlich SD, Michel B (2003) Replication restart in gyrB Escherichia coli mutants. Mol Microbiol 48: 845–854 [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J (2009) Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137: 685–696 [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Hamilton DL, Abremski K (1984) Site-specific recombination by the bacteriophage P1 lox-Cre system. Cre-mediated synapsis of two lox sites. J Mol Biol 178: 481–486 [DOI] [PubMed] [Google Scholar]
- Hayama R, Marians KJ (2010) Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc Natl Acad Sci USA 107: 18826–18831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter CE (1996) Timing of Synthetic Activities in the Cell Cycle Vol. 2: Washington, DC: ASM Press, [Google Scholar]
- Hoess RH, Abremski K (1985) Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol 181: 351–362 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L (1999) The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA 96: 10661–10666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi MC, Bourniquel A, Fisher J, Ho BT, Magnan D, Kleckner N, Bates D (2011) Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc Natl Acad Sci USA 108: 2765–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR (2000) Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc Natl Acad Sci USA 97: 9419–9424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23: 787–799 [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F (2008) Asymmetry of chromosome Replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet 4: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (1998) Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135: 475–485 [DOI] [PubMed] [Google Scholar]
- Mossessova E, Levine C, Peng H, Nurse P, Bahng S, Marians KJ (2000) Mutational analysis of Escherichia coli topoisomerase IV. I. Selection of dominant-negative parE alleles. J Biol Chem 275: 4099–4103 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Youngren B, Hansen FG, Austin S (2007) Dynamics of Escherichia coli chromosome segregation during multifork replication. J Bacteriol 189: 8660–8666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR (1998) The structure of supercoiled intermediates in DNA replication. Cell 94: 819–827 [DOI] [PubMed] [Google Scholar]
- Petrushenko ZM, Lai CH, Rai R, Rybenkov VV (2006) DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J Biol Chem 281: 4606–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possoz C, Junier I, Espeli O (2012) Bacterial chromosome segregation. Front Biosci 17: 1020–1034 [DOI] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR (2001) Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci USA 98: 8219–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ (2008) Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133: 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JB, Martinez-Robles ML, Hernandez P, Krimer DB (2011) Plasmid DNA replication and topology as visualized by two-dimensional agarose gel electrophoresis. Plasmid 63: 1–10 [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW (1996) GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol 6: 1599–1608 [DOI] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ (2009) Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137: 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunako Y, Onogi T, Hiraga S (2001) Sister chromosome cohesion of Escherichia coli. Mol Microbiol 42: 1233–1241 [DOI] [PubMed] [Google Scholar]
- Wang SC, Shapiro L (2004) The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc Natl Acad Sci USA 101: 9251–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Reyes-Lamothe R, Sherratt DJ (2008) Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev 22: 2426–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers HL, Bernander R (1998) Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J Bacteriol 180: 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Niki H (2004) migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J 23: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechiedrich EL, Khodursky AB, Cozzarelli NR (1997) Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev 11: 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.