Abstract
Using solution synchrotron X-ray scattering, we measure the variation of DNA-DNA d-spacings in bacteriophage λ with mono-, di- and poly-valent salt concentrations, for wild-type (48.5 kbp) and short-genome-mutant (37.8 kbp) strains. From the decrease in d-spacings with increasing salt, we deduce the relative contributions of DNA self-repulsion and bending to the energetics of pack-aged phage genomes. We quantify the DNA-DNA interaction energies within the intact phage by combining the measured d-spacings in the capsid with measurements of osmotic pressure in DNA assemblies under the same salt conditions in bulk solution. In the commonly used Tris-Mg buffer, the DNA-DNA interaction energies inside the phage capsids are shown to be about 1 kT/base pair, an order of magnitude larger than the bending energies.
The tight packaging of DNA in chromatin, sperm, and viral capsids is closely connected with gene regulation, fertility, and viral infectivity [1]. DNA occupies about 15% of the volume of chromatin in its most compacted state [2], up to 45% when condensed by protamine in sperm heads, and as much as 60% of the inner capsid volume of many viruses such as bacteriophage λ [3]. Phage capsids are permeable not only to water but also to small ions [4], so that the remaining volume fraction is occupied by aqueous solution that mediates the DNA self-repulsion and bending stress [5, 6]. Accordingly, the pressure inside phage capsids has been found to depend strongly on salt concentration, especially on the presence of polyvalent cations, varying from several tens of atmospheres down to a few atmospheres [7–9]. Generation of these high pressures is associated with the work performed by powerful DNA packaging motors [10, 11]. Such pressurized genome storage has been proposed as a viral tactic for infecting bacteria by initiating DNA ejection into the host cell [5, 9, 12]; completion of genome delivery depends on other mechanisms [13], notably active processes such as transcription [14]. Theory has suggested that, for any given set of salt concentrations, the key structural property of the packaged DNA is the average inter-axial spacing of neighboring strands [12, 15, 16], which determines the DNA-DNA self-repulsion.
More than 30 years ago, Earnshaw and Harrison [17] observed X-ray diffraction by condensed pellets of P22 and λ phages and measured DNA inter-axial distances of the local hexagonal packing of the genomes in wild-type and deletion mutants. They reported Bragg spacings on the order of 25 Å, which increased with the decrease in genome length in a way that suggested near-constancy of the volume occupied by the packaged DNA. On the basis of their observations they proposed several alternative scenarios for the organization of the DNA, e.g., coaxial spool vs. ball-of-string vs. chain-folded configurations. More recently, these issues have been addressed experimentally by cryo-electron microscopy [18–21] and theoretically by molecular dynamics simulations [22]. But controversy remains concerning the spatial organization of the confined DNA and the relative importance of DNA-DNA interactions vs. DNA bending elasticity in determining the operative pressures [12, 22].
In the present Letter we report DNA-DNA d-spacings for intact λ phages in solution, determined from synchrotron small-angle X-ray scattering (SAXS), as functions of the concentrations of mono-, di- and poly-valent cations. The salt concentrations and the genome length control the relative contributions to packaging stress from DNA self-repulsion and bending. Bulk-solution osmotic stress measurements [23] of d-spacing in ordered DNA arrays as a function of salt concentration are used to quantify the contributions of DNA interactions to capsid stress, which are shown to be large compared to those from DNA bending elasticity.
Following previous protocols [9], Luria-Bertani (LB) broth cultures of E. coli strain c600, in its early exponential growth phase (1011 cells/liter), are infected with each λ phage stock with a multiplicity of infection of 0.1. Cell density first increases and then drops due to bacterial lysis and phage release; chloroform is added before harvest to lyse the remaining cells, and cell debris is removed by gentle centrifugation. Phage particles are precipitated in 10% polyethylene-glycol (PEG) 8000 and purified by CsCl density-gradient sedimentation. Equilibrium dialysis was used to vary the ionic conditions of the phage particles. The CsCl salt was first exchanged into TM buffer (50 mM Tris pH 7.5, 10 mM MgCl2) by dialyzing 2 mL against 1000 mL volume at 4 °C with four buffer changes over one week. λ phage solution in this TM buffer was then dialyzed for two weeks against: 50 mM Tris pH 7.5 buffer in the absence of MgCl2 [stock solution (a)]; TM buffer with 0.3 mM Cobalt3+ hexammine [solution (b)]; and TM buffer with 0.3 mM Spermine4+ [solution (c)]. Successive salt concentration series were made up by adding salt to the appropriate stock solution: NaCl to stock (a) for the [Na+] series, MgCl2 to phage in solution (a) or in TM buffer for the [Mg2+] series, Cobalt3+ hexammine to stock (b) for the [Cobalt3+] series, and Spermine4+ to stock (c) for the [Spermine4+] series. We used TM buffer as the background buffer in polyvalent cation series to be consistent with the solution conditions used in the literature [7]. Initial dialysis against polyvalent cation solutions was necessary to sufficiently exchange the counterion atmosphere around the DNA phosphate groups (~3 mM equivalent concentration).
SAXS experiments were carried out at the X21 station at the National Synchrotron Light Source (NSLS) at the Brookhaven National Lab. The x-ray beam was set at 10.0 keV and 0.3 × 0.3 mm. Solutions of 40 µl were injected into an in-vacuum flow-through quartz capillary (1 mm diameter, 10 µm wall thickness) allowing windowless SAXS operation to reduce background scattering and avoid variations between capillaries. Sample to detector (1024 × 1024 pixels of 160 µm size) distance was 1.35 meters. Eight exposures of 30 seconds each showed identical scattering patterns that were averaged to improve the counting statistics.
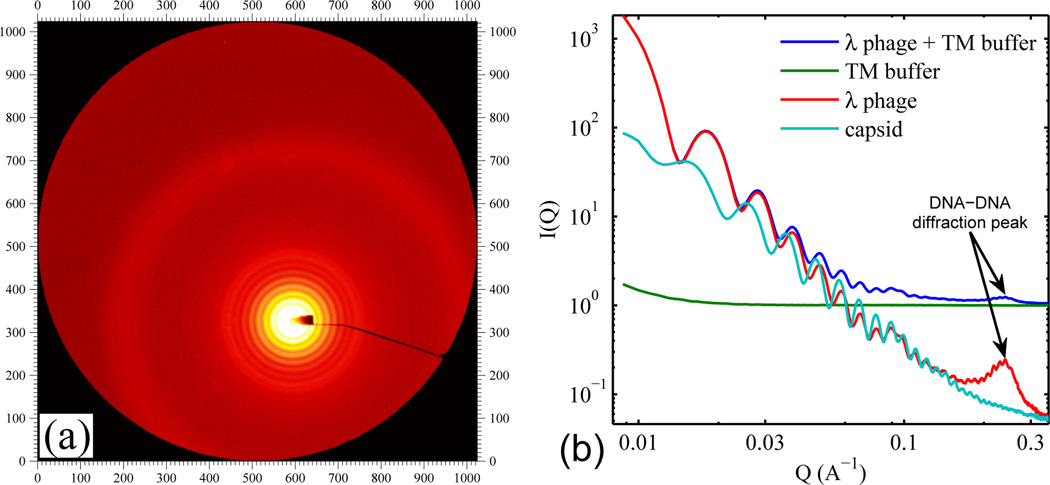
Fig. 1a shows raw SAXS data from a 2 × 1013 virus/mL solution of purified wild-type λ phage. A striking feature is the set of distinct concentric rings around the beam stop (see off-center yellow core). The radially integrated intensity (Fig. 1b) shows that these concentric rings give rise to pronounced oscillations at low Q values, which originate from the regular icosahedral shape of λ phage and indicate the high purity of the sample. Once the background scattering from the buffer is measured (see green curve) and subtracted (see lower two curves), the DNA-DNA diffraction peak at higher Q (around 0.25 Å−1) stands out clearly. This peak is absent in the SAXS profile of empty λ capsid (which is also shown in Fig. 1b – see aqua curve at bottom), confirming that locally-hexagonally-packed DNA is the origin of the red peak.
FIG. 1.
(a) Raw SAXS 2D image of the wild-type λ phage (48.5 kbp genomic DNA length) in TM buffer. (b) Radially integrated scattering intensity I(Q) where , 2θ is the scattering angle, and λ here is the x-ray wavelength. The blue and green I(Q) curves are the raw profiles from the wild-type λ phage solution and from the buffer. The red (“λ phage”) and aqua (empty “capsid”) curves have the buffer I(Q) subtracted. Note that the scattering from phage is an order of magnitude smaller than the background at high Q.
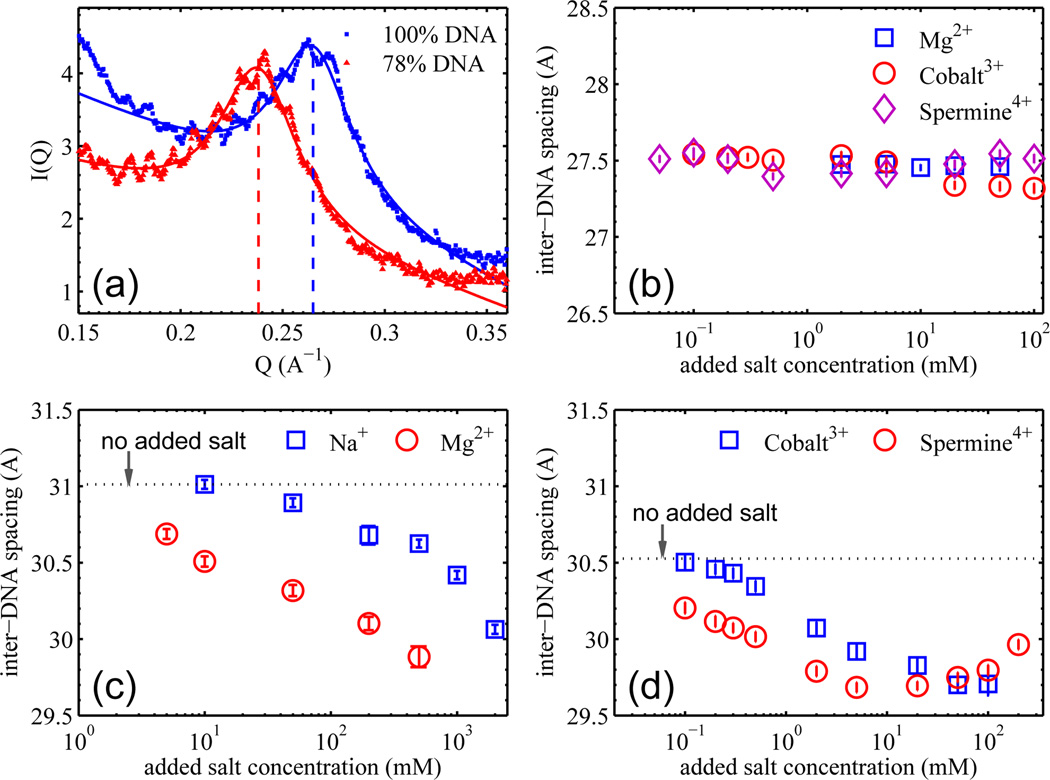
The DNA diffraction peaks from two λ strains (with 100% and 78% of the wild-type λ-DNA length) are shown expanded in Fig. 2a. The well-defined peak positions directly determine the d-spacings to an accuracy of 0.1 Å. The ripples on top of these diffraction peaks correspond to a spatial periodicity of 55 nm – the size of the viral capsid, suggestive of coherent diffraction of viral DNA as a whole. As seen in Fig. 2a, varying the DNA length (from 78 to 100%) significantly shifts the position of the DNA diffraction peak (from 0.238 to 0.264 Å−1). Each peak can be fitted with a Lorentz function plus a linear background (Fig. 2a). While addition of a damped sine wave fits the ripples reasonably well, it does not change the pertinent DNA diffraction peak parameters (e.g., peak position and width). Once the peak position QBragg is determined, the d-spacing follows from , corresponding to the local hexagonal packing. Fitting the peaks in Fig. 2a gives inter-axial separations of 30.5 and 27.5 Å for λ phage strains with 78% and 100% of the wild-type DNA length, in agreement with the values reported by Earnshaw and Harrison (30.3 and 27.5 Å respectively) [17]. From these d-spacings and the known DNA lengths, the volume occupied by DNA can be computed and is found to be essentially identical (to within a few percent) not only for the 78% and 100% DNA length, but also for 85% (data not shown). This can be qualitatively understood in terms of DNA taking up nearly all the available capsid volume as its strong self-repulsion overwhelms the bending stiffness.
FIG. 2.
(a) The DNA-DNA diffraction peaks from the λ cI60 and b221 strains with DNA lengths of 100% and 78% of the wild-type, respectively. Solid lines are the fits to the peak using a Lorentz function with a linear background. (b) DNA-DNA d-spacing as a function of added multivalent cation concentration for λ cI60 phage; the background buffer is TM buffer for all three series. There is no [Na+] series because the cI60 phage did not survive the dialysis against 50 mM Tris only, probably due to the elevated DNA pressure breaking the capsid. (c) DNA-DNA d-spacing as a function of added mono- and di-valent cation concentration for λ b221 phage; the background buffer is 50 mM pH 7.5 Tris only. The dotted line indicates the d-spacing with no added salt (i.e., 50 mM Tris only). (d) DNA-DNA d-spacing as a function of added tri- and tetra-valent cation concentration for λ b221 phage; the background buffer is TM buffer. The dotted line indicates the d-spacing with no added salt (i.e., TM buffer only). The Y axis range in panels (b), (c), & (d) is set to 2 Å for easy comparison.
But the repulsions are not completely dominant, as can be shown by measuring the change in d-spacing with varying salt conditions. More explicitly, because salt decreases DNA-DNA repulsion [23], adding salt should lead to smaller d-spacings and lowering of bending energy. For example, if there were no cost associated with bending the DNA, the molecule would occupy the entire capsid volume in order to maximize d-spacing and minimize self-repulsion. Conversely, if there were no self-repulsion, the stiff DNA would be crammed near the inner capsid wall, which maximizes its radius of curvature and minimizes bending stress. In reality, a tug of war between the competing tendencies results in a compromise. Varying the magnitude of either contribution shifts the balance between them. Conveniently, this can be done by changing concentrations of cations, which screen the repulsions (and, in the case of the polyvalent ions, can mediate an effective attraction – see below) between neighboring DNA segments. The balance between self-interaction and bending can also be varied by changing the length of packaged DNA via mutant strains as discussed earlier.
Fig. 2b shows the dependence of DNA-DNA d-spacings on ionic conditions of the full length DNA of 48.5 kbp. As the di-, tri- and tetra-valent counterions are added, the DNA-DNA separation shows less than 1% variation, remaining essentially constant. Its 27.5 Å d-spacing implies a surface-surface separation between DNA duplexes of just 7.5 Å. This minimal effect of adding salts is not surprising: at such close surface separation, DNA-DNA repulsion remains dominant under all ionic conditions [23], and bending stress is unable to push DNA strands closer. On the other hand, the 37.8 kbp DNA in b221 strain (the shortest DNA length strain available) has a 30.5 Å d-spacing in the TM buffer, and the DNA-DNA repulsion is significantly weaker. Accordingly, additions of Na+ and Mg2+ (Fig. 2c), and of Cobalt3+ hexammine and Spermine4+ (Fig. 2d) all result in significant decreases of DNA-DNA separation. The Na+ and Mg2+ data (Fig. 2c) are particularly instructive. Because the DNA-DNA interaction is always repulsive and favors maximal DNA spacing, the observed changes in d-spacings attest to the contribution of bending elasticity to DNA packaging. The DNA-DNA surface separation decreases from 11 to 9.7 Å as the molecule crowds more on itself at large radius of curvature to lower the curvature energy. Similarly, addition of multivalent cations to the λ b221 strain (Fig. 2d) also results in progressively smaller DNA-DNA separations, while at ten times lower concentrations than for the mono- and di-valent cations. This is consistent with the greater screening power of multivalent ions, which can also even induce effective attractions between neighboring DNA strands at mM concentrations. Notably, the DNA-DNA separation increases again at >20 mM [Spermine4+] and >100 mM [Cobalt3+] (see Fig. 2d), consistent with the well-known “swelling” behavior of DNA at sufficiently high concentrations of multivalent cations [24, 25].
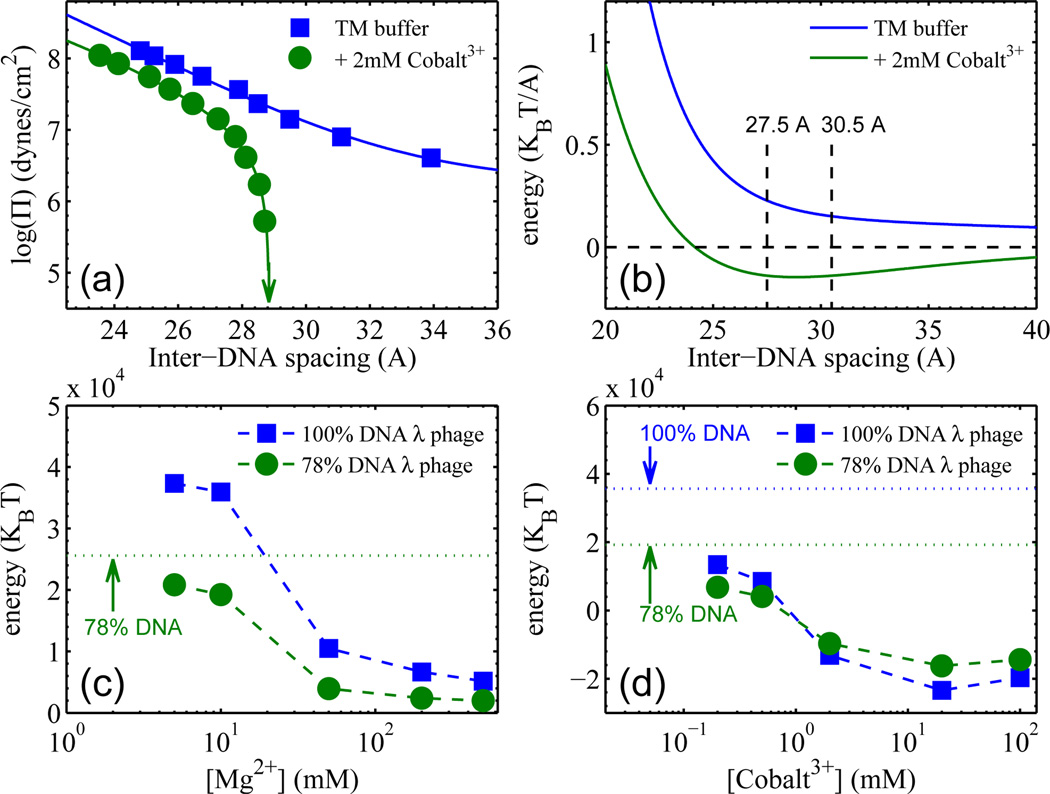
The above measurements of d-spacings vs. salt concentration make it possible to quantify DNA-DNA interaction energies in the virus from pressure vs. spacing measurements of DNA arrays in bulk solution. We systematically measured DNA d-spacings in solution as a function of imposed osmotic pressure, for each concentration of the cations used here in the SAXS measurements on intact phages. Results of this kind are shown in Fig. 3a, for the cases of TM buffer without and with 2 mM Co3+. By integrating the pressure curves (which do not involve any net bending of DNA strands) one obtains the DNA-DNA interaction energy per unit length [23], as shown in Fig. 3b for the case of TM buffer without and with 2 mM Co3+. Similarly, energy vs. separation curves can be obtained for each ionic concentration. It is then straightforward, using the measured DNA separations in the phage as a function of the same salt concentrations, to determine the DNA interaction energies (Fig. 3c and d) within the viral capsid; we simply multiply the appropriate interaction energy per unit length by the total DNA length.
FIG. 3.
(a) Measured DNA-DNA pressure vs d-spacing curves for bulk DNA solutions in TM buffer, without and with 2 mM Cobalt3+. The arrow indicates the d-spacing under zero osmotic pressure. (b) Interaction energy per unit length, vs d-spacing, obtained by integration of pressures from (a). (c) & (d) Total interaction energies of DNA inside phage capsids, for two DNA lengths, determined from results like those in (b). Background buffer is 50mM Tris pH 7.5 for (c), and TM buffer for (d). The energies for zero [Mg2+] and for zero [Cobalt3+], determined experimentally, are indicated by dotted horizontal lines in (c) and (d), respectively. Note again that there is no “zero [Mg2+]” point for the 100% DNA strain (cI60).
The total interaction energy of packaged DNA is substantial, amounting to as much as 38000 kT for wild-type λ phage with 48,500 bp DNA in the commonly used TM buffer, as shown by the left-most square in Fig. 3c – note that TM buffer contains 10 mM Mg2+. The corresponding 0.8 kT per base pair repulsive energy is an order of magnitude larger than the bending energy per base pair associated with the average radius of curvature, say 10–15 nm (Ebending ~0.06 kT/bp). The magnitude of DNA-DNA interaction can be equivalently discussed in terms of DNA osmotic pressures. In TM buffer, the DNA-DNA interactions alone correspond to pressures of 38 and 11 atm for the cI60 and b221 strains, correlating well with the measured DNA ejection inhibition pressures of 32 and 12 atm [8], respectively. Note that reducing the DNA length by 22% dramatically reduces the DNA interaction pressure by over a factor of three. As expected, Figs. 3c&d show that adding di- and tri-valent cations effectively reduces the repulsive interaction energy of DNA. This is consistent with the observation that the internal force against DNA packaging is lowered by added salts due to ionic screening [26]. At mM [Co3+] the DNA interaction energy changes sign (see Fig. 3d) because of the counterion-induced attraction between neighboring DNA portions. Qualitatively similar behaviors are observed with Spermine4+ (data not shown), consistent with a series of earlier measurements reporting ejection fractions in the presence of fixed external osmotic pressure as a function of added cation concentration [7].
By combining SAXS probes of the structure of DNA in intact viral capsids with osmotic stress measurements of DNA in bulk solution, each as a function of salt concentrations, we have quantified DNA self-repulsion as the dominant contribution to capsid stress and have established experimentally the contribution of DNA bending elasticity to DNA packaging. We are currently working on SAXS probes of the DNA remaining in phage capsids upon partial ejection in the presence of external osmotic pressure, again as a function of mono-, di-, tri- and tetravalent salt concentrations, where we can systematically catch and pause the process of DNA ejection. The viral capsid here serves as a convenient device for physical interrogation of DNA mechanics in nanoscale confinement.
Acknowledgments
We thank Drs. Lin Yang, Sankar Adhya, Rotem Edgar for helpful advice and discussions. This research was supported by the Intramural Research Program of the NIH, Eunice Kennedy Shriver National Institute of Child Health and Human Development, and by NSF grant CHE07-14411.
References
- 1.Garcia HG, et al. Biopolymers. 2007;85:115. doi: 10.1002/bip.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson PJJ, Fairall L, Huynh VAT, Rhodes D. Proc. Natl. Acad. Sci. USA. 2006;103:6506. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurmemmedov E, Castelnovo M, Catalano CE, Evilevitch A. Q. Rev. Biophys. 2007;40:327. doi: 10.1017/S0033583508004666. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JE, Chiu W. Curr. Opin. Struc. Biol. 2007;17:237. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Gelbart W, Knobler C. Science. 2009;323:1682. doi: 10.1126/science.1170645. [DOI] [PubMed] [Google Scholar]
- 6.Gelbart WM, Knobler CM. Physics Today. 2008;61:42. [Google Scholar]
- 7.Evilevitch A, Fang LT, Yoffe AM, Castelnovo M, Rau DC, Parsegian VA, Gelbart WM, Knobler CM. Biophys. J. 2008;94:1110. doi: 10.1529/biophysj.107.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grayson P, Evilevitch A, Inamdar MM, Purohit PK, Gelbart WM, Knobler CM, Phillips R. Virology. 2006;348:430. doi: 10.1016/j.virol.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Proc. Natl. Acad. Sci. USA. 2003;100:9292. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. Nature. 2001;413:748. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 11.Fuller DN, et al. J. Mol. Biol. 2007;373:1113. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Biophys. J. 2003;84:1616. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Huici V, Salas M, Hermoso JM. Mol. Microbiol. 2004;52:529. doi: 10.1111/j.1365-2958.2004.03993.x. [DOI] [PubMed] [Google Scholar]
- 14.Molineux IJ. Mol. Microbiol. 2001;40:1. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 15.Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. Proc. Natl. Acad. Sci. USA. 2001;98:13671. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Biophys. J. 2005;88:851. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earnshaw WC, Harrison SC. Nature. 1977;268:598. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Nature. 2006;439:612. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Cell. 1997;91:271. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 20.Leforestier A, Livolant F. Proc. Natl. Acad. Sci. USA. 2009;106:9157. doi: 10.1073/pnas.0901240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comolli LR, et al. Virology. 2008;371:267. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Petrov AS, Harvey SC. Structure. 2007;15:21. doi: 10.1016/j.str.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Rau DC, Parsegian VA. Biophys. J. 1992;61:246. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raspaud E, Durand D, Livolant F. Biophys. J. 2005;88:392. doi: 10.1529/biophysj.104.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Rau DC. Biophys. J. 2005;89:1932. doi: 10.1529/biophysj.105.065060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Proc. Natl. Acad. Sci. USA. 2007;104:11245. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]