Abstract
Ribonuclease P complexed with external guide sequence (EGS) bound to mRNA represents a unique nucleic acid-based gene interference approach for modulation of gene expression. Compared with other strategies, such as RNA interference, the EGS-based technology is unique because a custom-designed EGS molecule can hybridize with any mRNA and recruit intracellular ribonuclease P for specific degradation of the target mRNA. It has not been reported whether the EGS-based technology can modulate gene expression in mice. In this study, a functional EGS was constructed to target the mRNA encoding the protease (mPR) of murine cytomegalovirus (MCMV), which is essential for viral replication. Furthermore, a unique attenuated strain of Salmonella was generated for gene delivery of EGS in cultured cells and in mice. Efficient expression of EGS was observed in cultured cells treated with the generated Salmonella vector carrying constructs with the EGS expression cassette. Moreover, a significant reduction in mPR expression and viral growth was found in MCMV-infected cells treated with Salmonella carrying the construct with the functional EGS sequence. When MCMV-infected mice were orally treated with Salmonella carrying EGS expression cassettes, viral gene expression and growth in various organs of these animals were reduced and animal survival improved. Our study suggests that EGS RNAs, when expressed following Salmonella-mediated gene transfer, effectively inhibit viral gene expression and infection in mice. Furthermore, these results demonstrate the feasibility of developing Salmonella-mediated delivery of EGS as a unique approach for treatment that reduces viral diseases in vivo.
Keywords: antisense, gene therapy, herpesvirus
Nucleic acid-based gene interference strategies, such as antisense oligonucleotides, ribozymes, or DNAzymes and RNA interference (RNAi), represent powerful research tools and promising therapeutic agents for human diseases (1, 2). Ribonuclease P (RNase P), which has been found in all living organisms, catalyzes a hydrolysis reaction to remove the leader sequence of tRNA precursors by recognizing the common structure shared among all tRNAs (Fig. 1A) (3). Altman and colleagues proposed that RNase P can be recruited to cleave any mRNA using a custom-designed external guide sequence (EGS) that hybridizes with the target mRNA to form a structure resembling a tRNA substrate (Fig. 1 B–D) (4, 5). Compared with the RNAi approach, which induces the cellular RNA-induced silencing complex (RISC) RNase to cleave a target mRNA (1), the EGS-based technology is unique in inducing endogenous RNase P for targeted cleavage of particular RNAs. Moreover, the RNase P-mediated cleavage is highly specific and does not generate nonspecific “irrelevant cleavage” that is observed in RNase H-mediated cleavage induced by conventional antisense phosphothioate molecules (3, 6, 7). Thus, EGSs represent a new class of agents that may lead to highly effective and specific inhibition of gene expression (7–10). However, it has not been reported whether EGS-based technology is effective in reducing gene expression in mice.
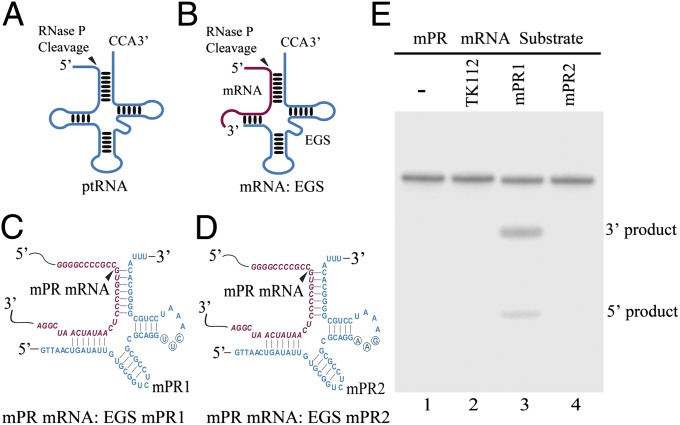
Fig. 1.
(A and B) Representation of a natural substrate (ptRNA) (A) and a hybridized complex of a target RNA (e.g., mRNA) and an EGS that resembles the structure of a tRNA (B). The truncated line corresponds to a region of the D loop of a tRNA (B). (C and D) Complexes of mPR mRNA sequence with EGS mPR1 (C) and mPR2 (D). The sequence of the mPR mRNA around the targeting site is shown in red and the EGS sequence is shown in blue. The sequences of mPR1 that were equivalent to the T-stem and loop, and variable region of a tRNA molecule were derived from tRNAser. The circled positions in the T-loop of EGS mPR1 represent the nucleotides that are mutated (5′-UUC-3′ → AAG) to generate mPR2. (E) Cleavage of mPR mRNA substrate mpr39 by RNase P in the presence of different EGSs. EGS RNAs (20 nM) were incubated with [32P]-labeled mpr39 RNA substrate (10 nM) and RNase P (5 units) (lanes 2–4). No EGS was added to the reaction mixture in lane 1.
Human cytomegalovirus (CMV) causes significant morbidity and mortality in immunocompromised or immunologically immature individuals, including neonates, AIDS patients, and transplant recipients (11). The emergence of drug-resistant strains of CMV has posed a need for the development of new treatment strategies. Murine cytomegalovirus (MCMV) infection of mice resembles in many ways its human counterpart with respect to pathogenesis, thus providing an animal model for studying CMV infection in vivo and for screening novel agents and developing new antiviral approaches (11). For example, macrophages represent the major reservoir for human and murine CMV as viruses can establish both primary and latent infections in these cells. Thus, blocking CMV infection and replication in macrophages is central in treating and preventing CMV-associated diseases.
In developing EGSs for gene therapy applications in vivo, one of the most important issues is the delivery of these agents to specific types of cells and tissues. Attenuated strains of invasive bacteria such as Salmonella have been shown to function as a carrier system for delivery of nucleic acid-based vaccines and anti-tumor short hairpin RNAs (12–15). In these studies, attenuated Salmonella was introduced with plasmid constructs containing the transgenes under the control of a eukaryotic expression promoter. The bacteria were then used to target specific cells such as dendritic cells, macrophages, and epithelial cells, leading to efficient transgene expression (13). Compared with other vectors, Salmonella may represent unique delivery agents for gene therapy in vivo as they can be administrated orally, a noninvasive delivery route with significant advantage, and can target specific tissues and cells. For example, macrophages represent the major in vivo reservoir for Salmonella following their systemic dissemination and therefore are considered an optimal target for a Salmonella-based gene target therapy (13, 14).
In this study, we constructed a functional EGS to target the mRNA encoding the protease of murine cytomegalovirus (MCMV) (mPR), which is essential for viral DNA encapsidation and replication (11, 16). We also generated a unique attenuated strain of Salmonella that was used for gene delivery of EGS in cultured cells and in mice. Using MCMV infection of mice as the model, we provide direct evidence that EGS RNA is effective in inhibiting viral gene expression and infection in mice, leading to improved survival of animals. Our study investigates the activity of EGS in mice.
Results
In Vitro RNase P-Mediated Cleavage of Target mRNA Induced by Constructed EGSs.
We used an in vivo mapping approach with dimethyl sulfate (DMS) (10) to determine the accessible regions of the mPR mRNA. A position 160 nucleotides downstream from the mPR translational initiation codon was chosen as the cleavage site for RNase P. This site appeared to be one of the regions most accessible to DMS modification and is likely accessible to EGS binding. Two EGSs were constructed (Fig. 1 C and D): mPR1, which resembles a part of the tRNAser structure and contains a T loop and stem and a variable region (Fig. 1C). EGS mPR2 was derived from mPR1 by introducing base-substitution mutations in three positions of the T-loop (Fig. 1D). The nucleotides in these three positions are highly conserved among tRNA molecules and are important for folding of the tRNA molecules and their recognition by RNase P (3); thus, mutations in these positions inactivate EGS activity. To determine if EGS with an incorrect guide sequence could affect the level of the target mRNA, EGS TK112, which was derived from tRNAser and targeted the mRNA of thymidine kinase (TK) of herpes simplex virus 1 (HSV-1) (10), was also included in the study.
EGS RNAs were synthesized in vitro from the DNA sequences coding the EGSs and subsequently incubated with human RNase P and substrate mpr39, which contained an mPR mRNA sequence of 39 nucleotides. Apparent cleavage of mpr39 by RNase P was observed in the presence of mPR1 (Fig. 1E, lane 3) whereas cleavage was barely detectable in the presence of mPR2 or TK112 (lanes 2 and 4). The binding between the EGSs and mpr39 was assayed in the absence of human RNase P, and the binding affinity of mPR2 to mpr39 (Kd = 430 ± 70 nM) was similar to that of mPR1 (Kd = 475 ± 75 nM). Because mPR2 had an identical targeting sequence and exhibited similar binding affinity to the mPR mRNA sequence as mPR1 but was unable to induce RNase P-mediated cleavage—possibly due to the T-loop mutations disrupting the recognition of EGS–mPR mRNA complex by RNase P—this EGS can be used as a control for the antisense effect in our experiments (see below).
Salmonella-Mediated Delivery for EGS Expression in Cultured Cells.
We cloned DNA sequences encoding mPR1, mPR2, and TK112 into expression vector pU6, which contains the small nuclear U6 RNA promoter for expressing EGS and a green fluorescence protein (GFP) expression cassette. The pU6-EGS constructs were transformed into Salmonella strain SL201 for gene delivery studies. SL201 was derived from auxotrophic strain SL7207 (17) and, in addition, contained a deletion of the msbB gene. SL7207 has been shown to function efficiently as a gene delivery carrier for the expression of several transgenes in mammalian cells (14, 15). The msbB gene encodes an enzyme important for the biosynthesis of the lipopolysaccharide (LPS), which is a major virulence and proimflammatory factor expressed on the surface of Salmonella (18). Deletion of msbB is expected to further reduce the virulence/toxicity of Salmonella and facilitate intracellular lysis of bacteria and release of the transgene construct, leading to efficient expression of the delivered gene in target cells and in animals.
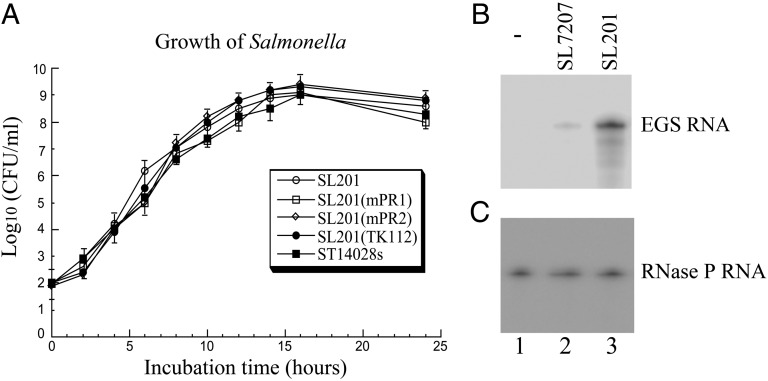
The presence of the EGS sequence cassette itself did not affect the viability of the bacterial carrier as we observed no difference in the growth kinetics of Salmonella carrying no constructs or various pU6-EGS constructs in LB broth (Fig. 2A). When murine J774 macrophages were infected with Salmonella carrying pU6-EGS constructs, more than 80% of cells were GFP positive at 24 h post infection, demonstrating efficient gene transfer mediated by Salmonella. Northern blot analysis confirmed the EGS expression in these cells, using mouse RNase P RNA as the internal control (Fig. 2 B and C). The levels of EGS RNAs in cells treated with SL201 carrying pU6-mPR1 were about fivefold higher than those with SL7207 carrying the same plasmid (lanes 2 and 3), suggesting that SL201 is a more effective delivery vector, possibly as a result of less toxicity and more efficient intracellular lysis of Salmonella due to the deletion of msbB, leading to more release of pU6-mPR1 and a higher level of gene expression.
Fig. 2.
(A) Analysis of growth in LB broth of Salmonella wild-type strain ST14028s and vector strain SL201 and its derivatives that carry constructs pU6-mPR1, pU6-mPR2, and pU6-TK1. (B and C) Northern analyses of the EGS RNA expression in J774 macrophages treated with strain SL201 carrying the empty vector pU6 (-, lane 1) and pU6-mPR1 (lane 3 in B and C) or with strain SL7207 carrying pU6-mPR1 (lane 2 in B and C). RNA samples (25 μg) were separated on 2% agarose gels that contained formaldehyde, transferred to nitrocellulose membranes, and hybridized to a [32P]-radiolabeled probe that contained the DNA sequence coding for mPR1 RNA (B) or mouse RNase P RNA (C).
Inhibition of MCMV Gene Expression and Replication by Salmonella-Mediated Delivery of EGS.
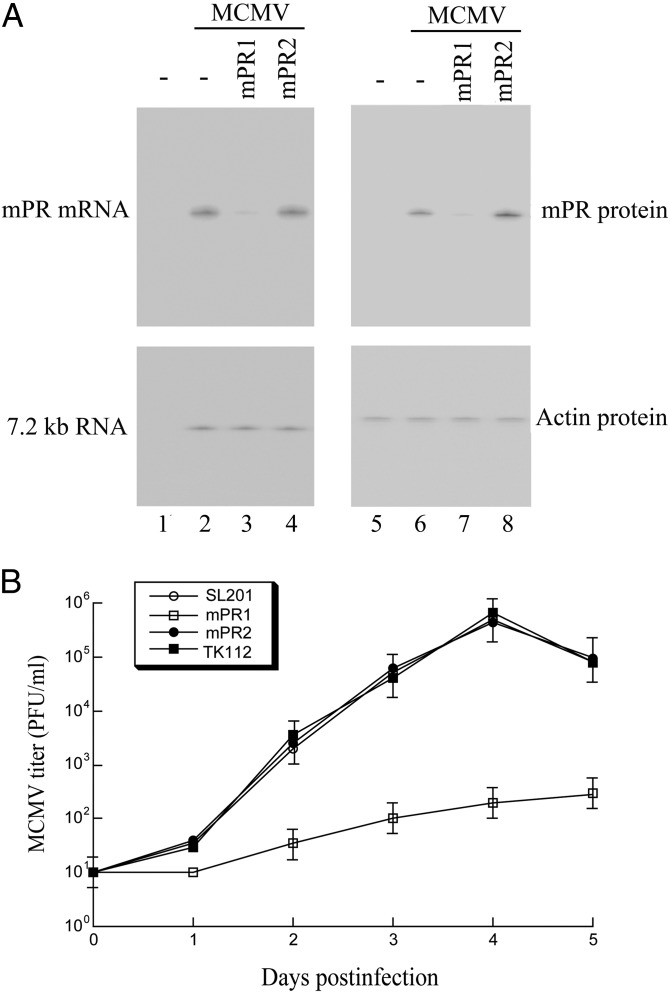
To determine the effect of Salmonella-mediated delivery of EGS on MCMV gene expression, we first treated J774 cells with SL201 carrying EGS constructs. The Salmonella-containing cells were then isolated by FACS analysis based on GFP expression and infected with MCMV. The expression of MCMV mPR mRNA was determined by Northern blot analysis. The level of the MCMV 7.2-kb-long viral transcript (7.2 kb RNA), the expression of which is not regulated by mPR under the assay conditions (11), was used as the internal control for the quantification of the expression of mPR mRNA (Fig. 3A, lanes 1–4). At 48 h post infection, a reduction of 85 ± 6% in the mPR mRNA level was observed in cells treated with SL201 carrying pU6-mPR1 whereas a reduction of less than 10% was observed in cells with SL201 containing pU6-mPR2 or pU6-TK112 (Fig. 3A, lanes 1–4 and Table 1). Consistent with observations in previous studies (7–10), no specific products of the cleavage of the target mRNAs by RNase P were detected in either Northern blot analyses or 5′ rapid amplification cDNA ends (RACE) PCR assays between the RNA samples from cultured cells treated with SL201 carrying pU6-mPR1 and those with SL201 carrying pU6-mPR2 or pU6-TK112. This result is presumably because these RNAs, which lack either a cap structure or a poly(A) sequence, are rapidly degraded by intracellular RNases.
Fig. 3.
(A) Expression levels of MCMV mRNAs (lanes 1–4) and proteins (lanes 5–8). J774 cells were first treated with Salmonella carrying pU6 (-, lanes 1–2 and 5–6), pU6-mPR1 (lanes 3 and 7), and pU6-mPR2 (lanes 4 and 8). The cells were then either mock-infected (lanes 1 and 5) or infected with MCMV (lanes 2–4 and 6–8) and were harvested at 12–72 h post infection. In Northern analysis (lanes 1–4), RNA samples (20 μg) were separated on agarose gels, transferred to nitrocellulose membranes, and hybridized to [32P]-radiolabeled probes that contained the sequence of the MCMV 7.2-kb transcript and mPR mRNA. For Western analyses (lanes 5–8), protein samples (30 μg) were separated in SDS-polyacrylamide gels, and the membranes were stained with the antibodies against mouse actin and MCMV mPR. (B) Growth of MCMV in J774 cells that were treated with Salmonella carrying constructs pU6 (SL201), pU6-mPR1 (mPR1), pU6-mPR2 (mPR2), and pU6-TK112 (TK112). Error bars indicate the SD. Error bars that are not evident indicate that the SD was less than or equal to the height of the symbols.
Table 1.
Levels of inhibition of the expression of viral genes in J774 cells
| EGS (%) |
|||||
| Viral gene class | SL201 | TK112 | mPR1 | mPR2 | |
| mie1 mRNA | α | 0 | 0 | 1 | 1 |
| m155 mRNA | γ | 0 | 1 | 0 | 1 |
| mPR mRNA | γ | 0 | 2 | 85 ± 6 | 7 |
| M83 protein | γ | 0 | 1 | 1 | 0 |
| M112 protein | β, γ | 0 | 1 | 0 | 0 |
| mPR protein | γ | 0 | 0 | 86 ± 8 | 7 |
The J774 cells were treated with Salmonella SL201 carrying constructs pU6-mPR1 (mPR1), pU6-mPR2 (mPR2), and pU6-TK112 (TK112) and were compared with the levels of inhibition in cells that were treated with SL201 carrying empty vector pU6 (SL201). The values shown are the arithmetic means of three independent experiments performed in triplicate; the values of SD that were less than 5% are not shown.
Using the level of actin protein as the internal control, Western analysis detected a reduction of about 86% in the mPR protein level in cells treated with SL201 carrying pU6-mPR1 (Fig. 3A, lanes 5–8). A low level of inhibition (∼7%) was found in cells treated with SL201 carrying pU6-mPR2 (Fig. 3A and Table 1), presumably due to an antisense effect because mPR2 exhibited a similar binding affinity to the target sequence as mPR1 but was unable to induce RNase P-mediated cleavage. Salmonella-mediated gene delivery of anti-mPR EGS also effectively inhibited MCMV growth. At 4 d post infection, a reduction of about 3,000-fold in viral yield was observed in cells treated with Salmonella carrying pU6-mPR1, whereas no significant reduction was found in cells treated with SL201 containing pU6-mPR2 or pU6-TK112 (Fig. 3B).
It is expected that inhibition of the expression of mPR, which is essential for viral DNA encapsidation, would not affect the expression of other viral genes, including immediate-early (α), early (β), and late (γ) genes (11). We examined the mRNA levels of mie1 (an α-gene) and m155 (a γ-gene) and the protein levels of M112 (a βγ-gene) and M83 (a γ-gene) and observed no significant difference in the expression of these genes among Salmonella-treated cells (Table 1). These results suggest that the Salmonella-mediated delivery of mPR1 specifically inhibits the expression of its target and does not affect overall viral gene expression.
Inhibition of MCMV Infection and Virulence by Salmonella-Mediated Delivery of EGS in Mice.
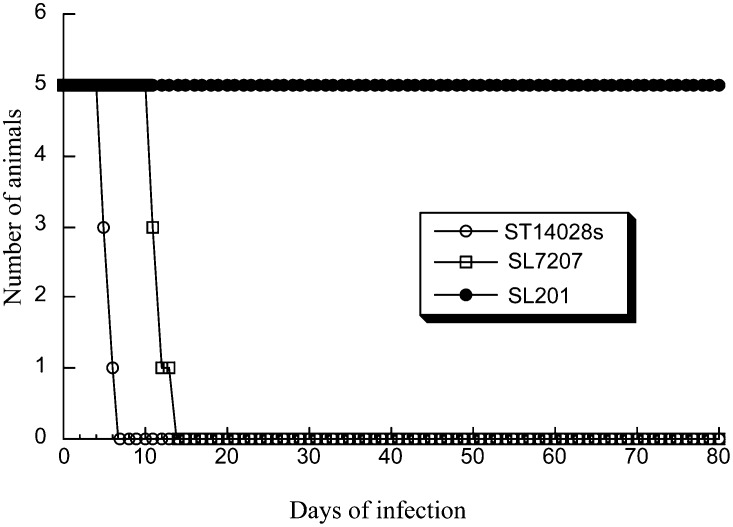
Immunodeficient SCID mice are extremely susceptible to MCMV infection and represent an excellent animal model for evaluating therapeutic approaches in blocking CMV infection and preventing viral-associated diseases in vivo (11). Viral replication in the spleen and liver plays an important role in MCMV pathogenesis during primary infection (11). To study Salmonella-mediated delivery of EGS in vivo, we intragastrically inoculated SCID mice with EGS-containing SL201. Gene delivery mediated by SL201 was efficient and durable as a substantial amount of EGS and GFP-positive cells was detected in the livers and spleens of mice (Fig. 4A). Furthermore, SL201 exhibited much less virulence in vivo than the parental strain SL7207 and the wild-type strain ST14028s (Fig. 5). All mice infected with SL201 (1 × 109 cfu/mouse) remained alive at 80 d post inoculation. In contrast, mice inoculated with a much lower dose of ST14028s (1 × 103 cfu/mouse) and SL7207 (5 × 105 cfu/mouse) died within 7 and 15 d post inoculation, respectively (Fig. 5).
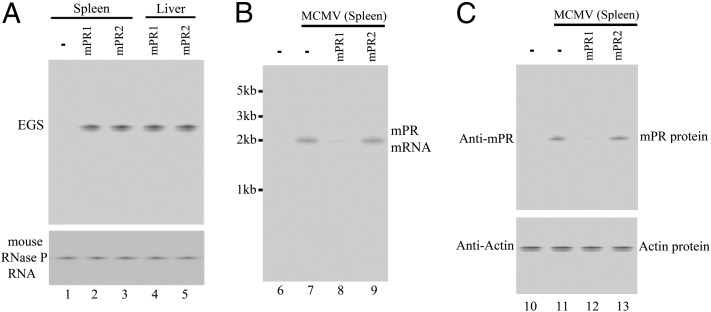
Fig. 4.
Expression of EGS RNA (A), viral mRNAs (B), and proteins in mice (C). Spleens and livers were isolated from SCID mice that were intragastrically inoculated with SL201 carrying different constructs and either mock-infected (lanes 1–5, 6, and 10) or infected with MCMV (lanes 7–9 and 11–13) and harvested at 14 d post infection. Northern and Western analyses were carried out using RNA (A and B) or protein samples (C) isolated from different organs of mice that were treated with SL201 carrying pU6 (-, lanes 1, 6–7, and 10–11), pU6-mPR1 (lanes 2, 4, 8, and 12), or pU6-mPR2 (lanes 3, 5, 9, and 13). The levels of the mouse RNase P RNA and actin protein were used as the internal controls.
Fig. 5.
Toxicity and virulence of different Salmonella strains in SCID mice. SCID mice (five animals per group) were infected intragastrically with the wild-type strain ST14028 (1 × 103 cfu), and vector strains SL7207 (5 × 105 cfu) or SL201 (1 × 109 cfu) carrying pU6-mPR1, and the survival of the animals was recorded.
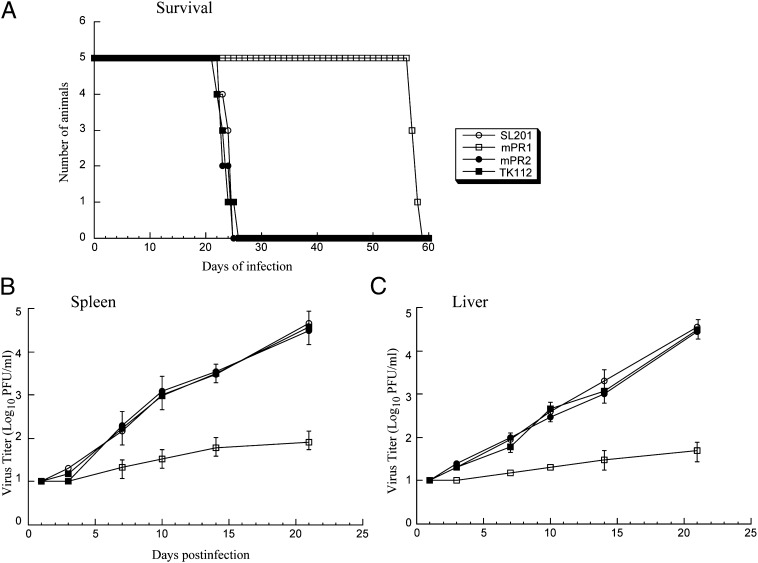
To study the antiviral effect of Salmonella-mediated oral delivery of EGS in vivo, SCID mice were intraperitoneally infected with MCMV, followed by oral inoculation of Salmonella carrying EGS constructs 36 h later. To further allow sustained expression of EGSs, we repeated oral inoculation of Salmonella every 5 d. Treatment of SL201 carrying pU6-mPR2 or pU6-TK112 had no effect on animal survival compared with untreated animals as all mice died within 26 d post infection (Fig. 6A). In contrast, in MCMV-infected mice treated with SL201 expressing mPR1, life span improved significantly as no animals died before 55 d post infection (Fig. 6A).
Fig. 6.
(A) Mortality of the SCID mice infected with MCMV, followed by oral inoculation of Salmonella SL201 (1 × 108 cfu/animal) carrying the empty vector pU6 alone (SL201) and the vector carrying different EGS expression cassettes (mPR1, mPR2, and TK112). We intraperitoneally infected CB17 SCID mice (five animals per group) with 1 × 104 pfu MCMV, 36 h before Salmonella inoculation and repeated Salmonella inoculation every 5 d. (B and C) Growth of MCMV in the spleens (B) and livers (C) of the infected SCID mice. The viral titers represent the average obtained from triplicate experiments. Error bars indicate the SD.
Salmonella-mediated oral delivery of EGS appeared to block MCMV infection in the treated mice. At 14 d post infection, substantial expression of viral mPR mRNA as well as mPR protein was readily detectable in livers and spleens of mice receiving SL201 carrying pU6-mPR2 and pU6-TK112, whereas little expression of mPR was detected in mice treated with SL201 carrying pU6-mPR1 (Fig. 4 B and C). At 21 d post infection, the viral titers in the spleens and livers of animals treated with pU6-mPR1–containing SL201 were lower than those in animals treated with SL201 carrying control constructs by 500- and 700-fold, respectively (Fig. 6 B and C).
Discussion
For EGSs to be successful as a therapeutic tool, one of the most important issues is targeted delivery of these agents to specific types of cells and tissues. This study demonstrates the activity of EGS RNAs in mice. In this study, we have designed an EGS RNA targeting the MCMV mPR mRNA. Furthermore, we have generated an attenuated strain of Salmonella, SL201, which exhibited high gene transfer activity and low cytotoxicity/pathogenicity in vivo. A reduction of about 85% in the expression levels of protease mRNA and protein and a reduction of about 3,000-fold in viral production were observed in MCMV-infected cells that were treated with SL201 carrying pU6-mPR1. In contrast, a reduction of less than 10% in the levels of mPR expression and viral production was observed in cells that were treated with SL201 carrying pU6-mPR2 or pU6-TK112 that expressed control EGSs. When MCMV-infected SCID mice were orally inoculated with SL201 carrying different EGS sequences, the expression of EGS RNAs was detected in several tissues, including spleen and liver. All infected animals that received SL201 only or SL201 carrying the pU6-mPR2 or pU6-TK112 constructs died within 26 d post infection whereas those treated with SL201 carrying pU6-mPR1 remained alive until 55 d post infection. Furthermore, viral titers in the spleen and liver of the infected animals treated with SL201 carrying pU6-mPR1 were lower than those in animals treated with SL201 only or SL201 with pU6-mPR2 or pU6-TK112. Control EGS TK112 targets an unrelated mRNA whereas mPR2 contains the identical guide sequence to mPR1 but is inactive in directing RNase P-mediated cleavage. These results suggest that the observed reduction in MCMV gene expression and growth in the cells and mice that were treated with Salmonella carrying pU6-mPR1 may be due to the specific targeted RNase P-mediated cleavage induced by the EGS as opposed to the antisense effect of the guide sequence or other nonspecific effects such as potential immune responses induced by SL201.
Several lines of evidence in our study indicate that EGS RNAs expressed following the Salmonella-mediated gene delivery are active and specific in mice. First, the viability and gene transfer ability of the Salmonella vectors were not significantly affected by the presence of EGS sequences (Fig. 2). Following Salmonella-mediated gene delivery, EGS RNAs were readily detected in cultured cells and in the livers and spleens of the animals. Furthermore, animals treated with SL201 carrying EGS constructs via oral inoculation of over 1 × 109 cfu exhibited no adverse signs for at least 80 d (Fig. 5), suggesting that oral inoculation of SL201 and the expression of EGS RNAs exhibited little pathogenicity or cytotoxicity in vivo. Second, the EGS appeared to be active in directing RNase P-mediated cleavage in vivo. Reduced mPR expression, decreased viral titers, and increased survival were observed in mice that were inoculated with SL201 carrying pU6-mPR1 but not with control constructs pU6-mPR2 or pU6-TK112. Third, the antiviral effect associated with the expression of functional EGS mPR1 appeared to be due to the reduction of mPR expression as a result of RNase P-mediated cleavage of mPR mRNA directed by mPR1. Only the levels of the target mPR but not other viral genes examined (e.g., mie1, M83, M112, and m155) were significantly reduced in cells treated with SL201 carrying pU6-mPR1 (Table 1). These results suggest that, following Salmonella-mediated delivery, EGSs effectively and specifically direct RNase P-mediated cleavage of its target mRNA, resulting in a decreased expression of mAP and reduction of viral growth, leading to inhibition of viral infection and increasing survival of infected animals.
To provide direct evidence of EGS RNA-directed RNase P cleavage of the target mRNA in vivo, it is important to analyze the cleavage products. However, no specific cleavage products were detected in either Northern blot analyses or 5′ RACE PCR assays between the RNA samples from cultured cells or spleen tissues of mice treated with SL201 carrying pU6-mPR1 and those with SL201 carrying pU6-mPR2 or pU6-TK112, presumably because these RNAs, which lack either a cap structure or a poly(A) sequence, are rapidly degraded by intracellular RNases. Compared with other studies using uninfected cells and tissues, the RNase P-specific cleavage products may be even more difficult to be detected in the presence of CMV infection in our study. In our study, intracellular RNAs may be more unstable in CMV-infected cells because CMV, like all other herpesviruses, is believed to encode a highly active nonspecific RNase that serves to shut off host functions by degrading host mRNAs to facilitate viral infection (11). We do not provide absolute proof of guide RNA-directed cleavage by RNase P. To show RNAi for in vivo destruction of targeted mRNAs, one must demonstrate the specificity of the reaction by RACE PCR products and prove that the site of cleavage is mediated by Argonaute 2 (1, 2). Further experiments to identify and characterize the cleavage products and study the mechanism of how an EGS directs RNase P to cleave its target mRNA in vivo will address this important issue.
Our results also suggest that Salmonella-based vectors are effective in delivering EGS for gene-targeting applications in vitro and in vivo. As a gene delivery tool, Salmonella-based vectors exhibit several unique and attractive features. First, Salmonella-based vectors are low cost, easy to prepare, store, and transport. Second, one of the most interesting aspects associated with the Salmonella vector is the oral route of administration of these bacteria (19, 20). Third, attenuated mutants with more than one independent deletion can be generated and are safe even in immunocompromised hosts (21). Fourth, whereas some safety considerations for the use of Gram-negative bacteria refer to the toxic effect of LPS, this concern has mostly been alleviated by oral delivery and by the fact that Salmonella strains have been widely used as vaccines in humans (19, 20). Fifth, oncogenesis promotion of the bacterial infection has not been shown and integration of Salmonella-delivered DNA in the host-cell genome is not common. Thus, Salmonella represents an attractive and promising gene delivery tool for gene therapy for human diseases, including those caused by human CMV.
Different bacterial components such as unmethylated CpG motifs and LPS can elicit various immune responses, including activation of TLR9 and TLR4 (22, 23), some of which are beneficial to the host and others of which are detrimental. Bacterial vectors with reduced cytotoxicity can be generated by introducing mutations to inactivate specific bacterial components (19). Alternatively, bacteria carrying transgenes that modulate specific responses can be constructed (23). Indeed, our newly constructed mutant SL201, which was derived from SL7207 and, in addition, contained a deletion of msbB, exhibited little if any virulence while being highly efficient for gene delivery. These results demonstrate the feasibility of developing vector strains exhibiting low pathogenecity/toxicity and high-gene-delivery efficiency in vivo.
Our study uses Salmonella-mediated gene delivery of EGS for gene-targeting applications in vivo. Human CMV, a member of the human herpesvirus family that includes seven other different viruses such as HSV and Epstein–Barr virus, can engage in lytic replication as well as establish latent infections (11). To further evaluate the anti-CMV activity of EGS, the EGS can be delivered into the macrophages and their progenitor cells (e.g., monocytes), which are the major reservoir for CMV and where the virus can establish both lytic replication and latent infection (11). These experiments will demonstrate whether EGS is effective in blocking CMV infection and preventing viral-associated diseases in vivo. These studies, as well as those studies on how to construct highly active EGSs and unique Salmonella strains, should facilitate the development of the EGS-based technology as a promising gene-targeting approach for in vivo applications.
Materials and Methods
EGS and mPR mRNA Substrate.
The DNA sequence coding for EGS mPR1 was generated by PCR using construct pTK112 (10) as the template with 5′ primer oligomPR41 (5′-GGAATTCTAATACGACTCACTATAGGTTAACTGATATTGTGCGGTCTCC-3′) and 3′ primer oligomPR42 (5′-AAGCTTTAAATGTGCCCCGCAGGATTTGAACCTGCGCGCG-3′). The DNA sequence coding for EGS mPR2 was derived from that for mPR1 and contained point mutations (5′-TTC-3′ → AAG) at the three highly conserved positions in the T-loop of the EGS (Fig. 1). The DNA sequence that encodes substrate mpr39 was constructed by annealing oligonucleotides AF25 (5′-GGAATTCTAATACGACTCACTATAG-3′) and smPR (5′-CGGGATCCGATTGATATTGAGGGGCACGGCGGGCCCCCCTATAGTGAGTCGTATTA-3′). RNA substrate mpr39 and EGS RNAs were synthesized in vitro from the constructed DNA templates using T7 RNA polymerase. Purification of RNase P and the cleavage and binding assays were carried out as described previously (SI Materials and Methods) (10).
Expression of EGS RNAs by Salmonella-Mediated Delivery in Cultured Cells.
Salmonella strain SL201 was derived from the auxotrophic Salmonella typhimurium aroA strain SL7207 [a gift from Bruce A. D. Stocker (Stanford University, Stanford, CA)] (17) by deleting the coding sequence of msbB (SI Materials and Methods). We generated Salmonella carrying different EGS constructs by transforming SL201 with plasmids pU6, pU6-mPR1, pU6-mPR2, or pU6-TK112. Construct pU6 contained the GFP expression cassette and the small U6 RNA promoter used for the expression of EGS RNAs in mammalian cells.
In gene transfer experiments to express EGS RNAs, we infected mouse J774 cells [(1 × 106 cells/mL) pretreated with IFN-γ (150 U/mL) (R&D Systems Inc.) for at least 12 h] with Salmonella at a multiplicity of infection (MOI) of 10–20 bacteria/cell. To allow phagocytosis to occur, we incubated the cultures at 37 °C for 30 min. We then added to the cultures fresh medium containing gentamicin (20 μg/mL) and incubated them for the indicated time periods before harvesting cells. The expression of EGS RNAs was assayed using Northern analyses (10) (SI Materials and Methods).
Viral Infection and Assays for Viral Gene Expression and Growth.
Viruses, cells, and antibodies in the study are described in SI Materials and Methods. T-25 flasks of mouse J774 cells (∼106 cells) were first incubated with Salmonella at a MOI of 10–20 bacteria/cell at 37 °C for 30 min. The medium was aspirated and then replaced with fresh medium containing gentamicin (20 μg/mL) and incubated for 8 h to allow the expression of the EGSs. The Salmonella-containing cells were then subjected to FACS using a FACSVantage SE sorter (BD Biosciences), and a population of GFP-positive cells (usually 1–5 × 105 cells with a positive fluorescence of >99%) was isolated. We then cultured the isolated cells for 4 h, followed by mock or MCMV infection (a MOI of 0.5–1) for another 8–72 h (10). The levels of inhibition of viral growth in cells were determined as described previously (24) (SI Materials and Methods). The expression of specific mRNAs and proteins were assayed in Northern and Western analyses, respectively (10) (SI Materials and Methods).
Studies in Animals.
The protocol for all animal experiments was approved by the Animal Care and Use Committee of the University of California-Berkeley (Protocol #R240 and #R276). Four- to 6-wk-old CB17 SCID mice (Jackson Laboratory) were infected intraperitoneally with 1 × 104 pfu of MCMV and, at 36 h post infection, were inoculated with Salmonella intragastrically. For intragastric inoculation of mice, we first anesthetized the animals with isoflurane and then intragastrically inoculated them with 0.1–0.2 mL PBS containing 1 × 108 cfu Salmonella, using a feeding needle (25). We repeated the oral inoculation procedure every 5 d. To determine the delivery efficiency, we examined the GFP signal of the transfected cells (with fluorescence microscopy) and studied the expression of EGS RNAs (with Northern analyses) in mouse tissues.
To study viral growth and gene expression, groups of mice (at least five animals per group) were killed at 1, 3, 7, 10, 14, and 21 d post inoculation. The spleens and livers were harvested and sonicated as a 10% (wt/vol) suspension in a 1:1 mixture of DMEM and 10% skim milk. Viral titers of the samples were determined using plaque assays (SI Materials and Methods) (24). The expression of EGS and viral mRNAs was determined using Northern analyses, and the expression of viral proteins was assayed using Western analysis (SI Materials and Methods). The virulence and toxicity of Salmonella vectors was studied by intragastric inoculation of SCID mice (five animals per group) with Salmonella strain ST14028s (1 × 103 cfu), SL7207 (5 × 105 cfu), and SL201 (1 × 109 cfu) carrying pU6-mPR1. We determined the survival rates by monitoring the mortality of the animals for at least 80 d post infection.
Supplementary Material
Acknowledgments
We thank Gerry Abenes, Yong Bai, Paul Rider, and Vincent Sheu for critical comments and technical assistance. X.J. is a recipient of a China Graduate Student Scholarship from the Chinese Ministry of Education. G.-P.V. and Y.-C.C. were partially supported by a predoctoral block grant from the University of California at Berkeley. This research has been supported by grants from the National Institutes of Health (AI041927, AI091536, and DE014842).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201620109/-/DCSupplemental.
References
- 1.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol. 2003;21:1457–1465. doi: 10.1038/nbt915. [DOI] [PubMed] [Google Scholar]
- 3.Gopalan V, Altman S. 2006. Ribonuclease P: Structure and catalysis. The RNA World, eds Gesteland R, Cech T, Atkins J. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY), Vol. 277, Chapter 6.1. Available at http://rna.cshl.edu.
- 4.Forster AC, Altman S. External guide sequences for an RNA enzyme. Science. 1990;249:783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- 5.Yuan Y, Hwang ES, Altman S. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci USA. 1992;89:8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma M, et al. Intracellular mRNA cleavage induced through activation of RNase P by nuclease-resistant external guide sequences. Nat Biotechnol. 2000;18(1):58–61. doi: 10.1038/71924. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, et al. Effective inhibition of Rta expression and lytic replication of Kaposi’s sarcoma-associated herpesvirus by human RNase P. Proc Natl Acad Sci USA. 2004;101:9073–9078. doi: 10.1073/pnas.0403164101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hnatyszyn H, Spruill G, Young A, Seivright R, Kraus G. Long-term RNase P-mediated inhibition of HIV-1 replication and pathogenesis. Gene Ther. 2001;8:1863–1871. doi: 10.1038/sj.gt.3301606. [DOI] [PubMed] [Google Scholar]
- 9.Plehn-Dujowich D, Altman S. Effective inhibition of influenza virus production in cultured cells by external guide sequences and ribonuclease P. Proc Natl Acad Sci USA. 1998;95:7327–7332. doi: 10.1073/pnas.95.13.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawa D, Wang J, Yuan Y, Liu F. Inhibition of viral gene expression by human ribonuclease P. RNA. 1998;4:1397–1406. doi: 10.1017/s1355838298980918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, et al., editors. Fields Virology. Philadelphia: Lippincott-William & Wilkins; 2007. pp. 2701–2772. [Google Scholar]
- 12.Darji A, et al. Oral somatic transgene vaccination using attenuated S. typhimurium. Cell. 1997;91:765–775. doi: 10.1016/s0092-8674(00)80465-1. [DOI] [PubMed] [Google Scholar]
- 13.Grillot-Courvalin C, Goussard S, Courvalin P. Bacteria as gene delivery vectors for mammalian cells. Curr Opin Biotechnol. 1999;10:477–481. doi: 10.1016/s0958-1669(99)00013-0. [DOI] [PubMed] [Google Scholar]
- 14.Paglia P, Terrazzini N, Schulze K, Guzmán CA, Colombo MP. In vivo correction of genetic defects of monocyte/macrophages using attenuated Salmonella as oral vectors for targeted gene delivery. Gene Ther. 2000;7:1725–1730. doi: 10.1038/sj.gt.3301290. [DOI] [PubMed] [Google Scholar]
- 15.Yang N, Zhu X, Chen L, Li S, Ren D. Oral administration of attenuated S. typhimurium carrying shRNA-expressing vectors as a cancer therapeutic. Cancer Biol Ther. 2008;7(1):145–151. doi: 10.4161/cbt.7.1.5195. [DOI] [PubMed] [Google Scholar]
- 16.Welch AR, Woods AS, McNally LM, Cotter RJ, Gibson W. A herpesvirus maturational proteinase, assemblin: Identification of its gene, putative active site domain, and cleavage site. Proc Natl Acad Sci USA. 1991;88:10792–10796. doi: 10.1073/pnas.88.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 18.Khan SA, et al. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998;29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 19.Clairmont C, et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis. 2000;181:1996–2002. doi: 10.1086/315497. [DOI] [PubMed] [Google Scholar]
- 20.Levine MM, et al. Safety, infectivity, immunogenicity, and in vivo stability of two attenuated auxotrophic mutant strains of Salmonella typhi, 541Ty and 543Ty, as live oral vaccines in humans. J Clin Invest. 1987;79:888–902. doi: 10.1172/JCI112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanCott JL, et al. Regulation of host immune responses by modification of Salmonella virulence genes. Nat Med. 1998;4:1247–1252. doi: 10.1038/3227. [DOI] [PubMed] [Google Scholar]
- 22.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 23.Krieg AM. Therapeutic potential of Toll-like receptor 9 activation. Nat Rev Drug Discov. 2006;5:471–484. doi: 10.1038/nrd2059. [DOI] [PubMed] [Google Scholar]
- 24.Abenes G, et al. Murine cytomegalovirus with a transposon insertional mutation at open reading frame m155 is deficient in growth and virulence in mice. J Virol. 2004;78:6891–6899. doi: 10.1128/JVI.78.13.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu S, Killoran PB, Fang FC, Riley LW. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun. 2002;70:451–461. doi: 10.1128/IAI.70.2.451-461.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.