Abstract
The “apoptotic ring” is characterized by the phosphorylation of histone H2AX at serine 139 (γ-H2AX) by DNA-dependent protein kinase (DNA-PK). The γ-H2AX apoptotic ring differs from the nuclear foci patterns observed in response to DNA-damaging agents. It contains phosphorylated DNA damage response proteins including activated Chk2, activated ATM, and activated DNA-PK itself but lacks MDC1 and 53BP1, which are required to initiate DNA repair. Because DNA-PK can phosphorylate heat shock protein 90α (HSP90α) in biochemical assays, we investigated whether HSP90α is involved in the apoptotic ring. Here we show that HSP90α is phosphorylated by DNA-PK on threonines 5 and 7 early during apoptosis and that both phosphorylated HSP90α and DNA-PK colocalize in the apoptotic ring. We also show that DNA-PK is a client of HSP90α and that HSP90α is required for full DNA-PK activation, γ-H2AX formation, DNA fragmentation, and apoptotic body formation. In contrast, HSP90 inhibition by geldanamycin markedly enhances TRAIL-induced DNA-PK and H2AX activation. Together, our results reveal that HSP90α is a substrate and chaperone of DNA-PK in the apoptotic response. The response of phosphorylated HSP90α to TRAIL and its localization to the γ-H2AX ring represent epigenetic features of apoptosis that offer insights for studying and monitoring nuclear apoptosis.
Keywords: programmed cell death, caspase, camptothecin, staurosporine, FasL
Molecular chaperones enable the folding or unfolding and the assembly or disassembly of macromolecular structures. Heat shock protein 90 (HSP90) is an evolutionarily conserved molecular chaperone required for the stability and function of more than 200 proteins (1–4). HSP90 consists of three domains: an amino-terminal region that contains an ATP binding site and drug-binding site and co-chaperone–interacting motifs, a central domain with docking sites for client proteins and co-chaperones, and a C-terminal domain that contains the dimerization motif, a second drug-binding region, and interaction sites for additional co-chaperones (2, 5–7). The HSP90 family includes four members: HSP90α (HSP90AA1), HSP90β (HSP90AB1), glucose-regulated protein 94 (GRP94) (HSP90B1), and tumor necrosis factor receptor-associated protein 1 (TRAP1). HSP90α and β are highly homologous (86% sequence identity), but HSP90α contains two threonine residues (T5 and T7) that are not present in HSP90β. Both proteins are primarily cytoplasmic, but a small fraction (<10%) is also found in the nucleus (8, 9). In addition, HSP90α can be expressed at the cell surface and secreted into the extracellular space under metastatic conditions (2, 10–12). HSP90β is constitutively and ubiquitously expressed, whereas HSP90α is heat inducible and tissue specific (13). The HSP90 homolog GRP94 is found in the endoplasmic reticulum where it is essential for the maturation and secretion of insulin-like growth factors (14). TRAP1 is localized in mitochondria, where it functions as a protective factor against oxidative stress (15, 16). HSP90 proteins are involved in cellular homeostasis (17), transcriptional regulation (18), and chromatin remodeling (19). Because HSP90 is a molecular chaperone of numerous oncoproteins, it is considered to be a crucial facilitator of oncogene addiction (a unique characteristic of cancer cells that makes them dependent on one or more oncogenic proteins for their survival) (2) and is a validated anticancer drug target (2).
One promising approach for cancer therapy is based on biological agents that induce apoptosis by activating caspases. One such agent is recombinant human Apo2L/TNF-related apoptosis-inducing ligand (TRAIL), a proapoptotic receptor agonist that activates the two membrane death receptors (DR4 and DR5) selectively in cancer cells. It is derived from human endogenous TRAIL, a 281-amino acid polypeptide produced by immune cells and involved in the innate immune response, autoimmunity, and tumor immunosurveillance (20–23). Recombinant Apo2L/TRAIL is in clinical trial for non-small cell lung cancers (http://www.clinicaltrials.gov).
One of the early landmarks of apoptosis is the phosphorylation of histone H2AX at serine 139 (γ-H2AX) by activated DNA-dependent protein kinase (DNA-PK) (24–27). We previously reported that this epigenetic process begins at the periphery of the nucleus and forms a characteristic “apoptotic ring” in confocal microscopy imaging before diffusing to the whole chromatin and persisting at later stages of apoptosis even when the nucleus fragments into apoptotic bodies. The γ-H2AX apoptotic ring, although morphologically distinct from the DNA damage response (DDR) focal pattern observed after DNA damage (28), also contains phosphorylated DDR proteins, including activated checkpoint kinase 2 (Chk2) (phosphorylated on T68), activated ataxia telangiectasia mutated (ATM) (phosphorylated on S1981), and activated DNA-PK itself (phosphorylated on T2609) (24–26). However, it lacks MDC1 and 53BP1 because of the caspase-induced cleavage of MDC1, which inactivates the initiation of DNA repair (25).
In the present study, we investigated whether nuclear HSP90α is involved in the apoptotic ring. This hypothesis stemmed from two earlier studies unrelated to apoptosis that showed that DNA-PKcs is a client of HSP90 (http://www.picard.ch/downloads/Hsp90facts.pdf) (29) and that DNA-PK can phosphorylate HSP90α at its two N-terminal threonine residues (threonines 5 and 7; T5/7) in biochemical assays (30). Using a specific antibody against HSP90α phosphorylated on T5/7, we show that TRAIL and other apoptotic inducers cause massive, rapid, and persistent nuclear phosphorylation of HSP90α by DNA-PK. We also show that HSP90α acts as a chaperone of DNA-PK and as a coactivator necessary for efficient execution of nuclear apoptosis.
Results
HSP90α Phosphorylation Is an Extensive and Early Process During Apoptosis.
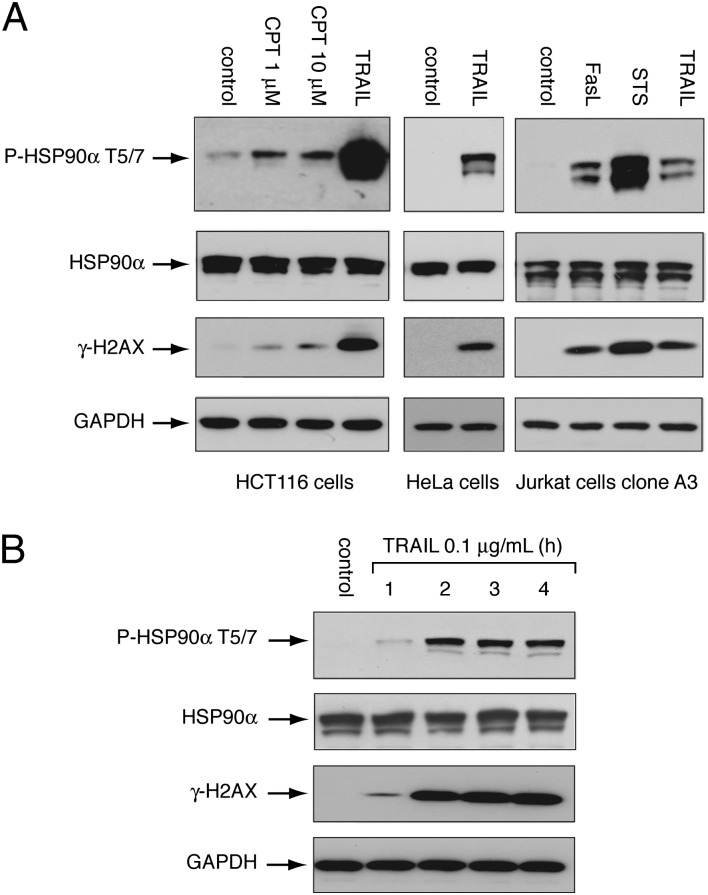
Fig. 1A shows that several diverse apoptotic agents [TRAIL, FasL (31), and staurosporine (32)] induce HSP90α phosphorylation at threonines 5 and 7 (P-HSP90α-T5/7) in various cell lines (colon carcinoma HCT116 cells, cervix carcinoma HeLa cells, and leukemic Jurkat cells). HSP90α phosphorylation was associated with the concurrent phosphorylation of H2AX (Fig. 1A). Two bands were observed for total HSP90α and P-HSP90α-T5/7; these bands correspond to the two different splicing isoforms of HSP90α (HSP90AA1-1 and HSP90AA1-2). We also found that DNA damage following camptothecin treatment, which is known to induce γ-H2AX (28), induced P-HSP90α-T5/7, albeit to a much lower level than seen in response to apoptosis-inducing agents (Fig. 1A). In the rest of the study, we focused on TRAIL-induced apoptosis because TRAIL induces a robust HSP90α-T5/7 phosphorylation and initiates its effect from the cell surface without targeting chromatin, thus eliminating the possibility that DNA-PK activation could be initiated by agents that directly induce DNA damage. Also, TRAIL has been the focus of our recent studies on chromatin modifications in the early stages of apoptosis under conditions similar to those presented here (24–26).
Fig. 1.
Extensive and rapid HSP90α phosphorylation at threonines 5 and 7 in apoptotic cells. (A) HCT116 and HeLa cells were treated with camptothecin (CPT, 1 μM, 10 μM, 3 h) or TRAIL (0.1 μg/mL, 3 h). Jurkat cells were treated with FasL (0.1 μg/mL, 6 h), TRAIL (0.1 μg/mL, 6 h), or staurosporine (STS, 0.1 μM, 6 h). P-HSP90α-T5/7, total HSP90α, and γ-H2AX were analyzed by Western blotting. GAPDH was used as a loading control. (B) TRAIL induces rapid HSP90α phosphorylation. HCT116 cells were treated with 0.1 μg/mL TRAIL for the indicated times. P-HSP90α-T5/7, total HSP90α, and γ-H2AX were analyzed by Western blotting. GAPDH was used as a loading control.
Because we previously showed that the apoptotic DDR (including phosphorylation of H2AX, DNA-PK, Chk2, and ATM) is initiated within 1 h after TRAIL treatment (24–26), we compared the kinetics of HSP90α and H2AX phosphorylation in response to TRAIL. Fig. 1B shows that TRAIL induces rapid phosphorylation of HSP90α, starting at 1 h after TRAIL treatment. This phosphorylation was maximal at ∼2 h, similar to H2AX phosphorylation (Fig. 1B).
Apoptotic HSP90α Phosphorylation Is an Early Characteristic of the Apoptotic Ring.
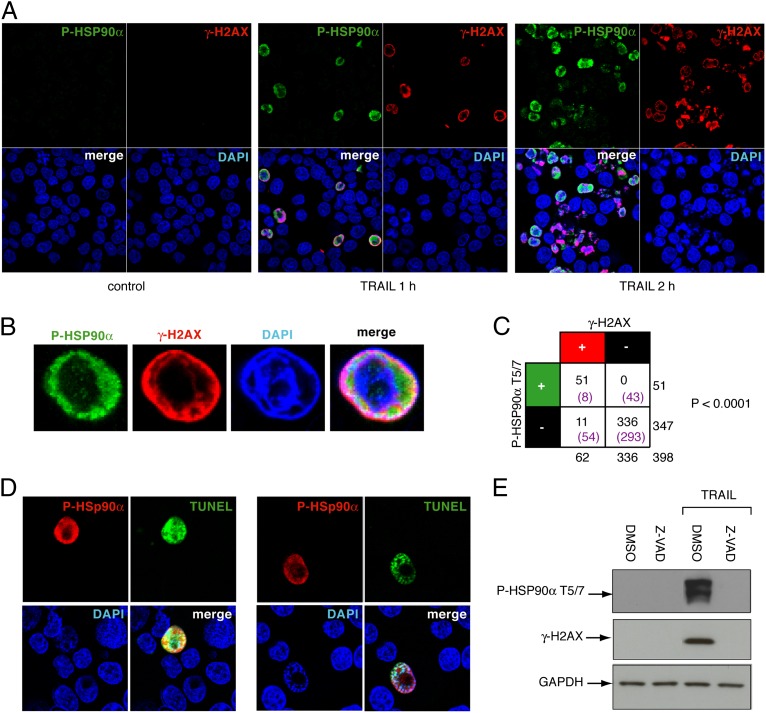
To characterize better the nuclear distribution of P-HSP90α-T5/7 and its relationship with the apoptotic γ-H2AX ring, we performed immunofluorescence microscopy experiments with γ-H2AX costaining. One hour after TRAIL treatment, only a fraction of the cells became positive for P-HSP90α-T5/7 (Fig. 2A). Such a stochastic response is consistent with our previous findings for the apoptotic γ-H2AX response (24–26). Moreover, the cells that showed the P-HSP90α-T5/7 response were the same cells that were undergoing the γ-H2AX response (Fig. 2 A–C). Overall, the cells that demonstrated P-HSP90α-T5/7 staining were positive for γ-H2AX staining also (Fig. 2C).
Fig. 2.
TRAIL induces rapid HSP90α phosphorylation and localization of HSP90α to the apoptotic ring. (A) P-HSP90α-T5/7 and γ-H2AX confocal immunofluorescence staining in HCT116 cells treated with TRAIL. P-HSP90α-T5/7 was labeled in green and γ-H2AX in red; nuclei were stained blue with DAPI. (B) Representative P-HSP90α-T5/7 and γ-H2AX confocal microscopy in a single TRAIL-treated HCT116 cell. P-HSP90α-T5/7 was labeled in green and γ-H2AX in red; the nucleus was stained blue with DAPI. (C) Table showing the highly significant relationship between P-HSP90α-T5/7 and γ-H2AX. Double-positive, single-positive, and double-negative cells were scored after 1 h TRAIL treatment. Expected numbers from the contingency table are in parentheses. Result of χ2 test is shown at right (P < 0.0001). (D) Representative TUNEL experiment showing P-HSP90α-T5/7 staining in apoptotic nuclei (TRAIL 0.1 μg/mL, 1 h). P-HSP90α-T5/7 was labeled in red, and apoptotic nuclei were labeled in green. (E) Caspase requirement for TRAIL-induced P-HSP90α-T5/7. HCT116 cells were treated with Z-VAD-fmk at 100 μM for 1 h before exposure to TRAIL at 0.1 μg/mL for 3 h. P-HSP90α-T5/7 and γ-H2AX were analyzed by Western blotting. GAPDH was used as a loading control.
Analysis of the P-HSP90α-T5/7 response at the level of individual cells showed that in untreated cells P-HSP90α was almost undetectable except for centrosomal staining (more than 70% of cells examined presented a centrosomal staining; Fig. S1). Upon TRAIL treatment, as cells initiated their apoptotic γ-H2AX ring response, P-HSP90α staining was colocalized with the apoptotic γ-H2AX ring and was exclusively nuclear (Fig. 2B). TUNEL assays showed that the cells that were positive for γ-H2AX and P-HSP90α-T5/7 rings also were positive for TUNEL staining (Fig. 2D), demonstrating the presence of DNA breaks in cells positive for P-HSP90α. As cells progressed toward a more extensive apoptotic response, both the P-HSP90α-T5/7 and γ-H2AX staining became diffuse throughout the nucleus and affected a greater fraction of the cells. However, both the P-HSP90α and γ-H2AX staining remained colocalized (Fig. 2A).
Finally, pretreatment by the broad-spectrum caspase inhibitor z-Val-Ala-DL-Asp-fluoromethylketone (Z-VAD-fmk) abolished TRAIL-induced phosphorylation of both HSP90α and H2AX (Fig. 2E), demonstrating that P-HSP90α and γ-H2AX are induced by the apoptosis response initiated by TRAIL.
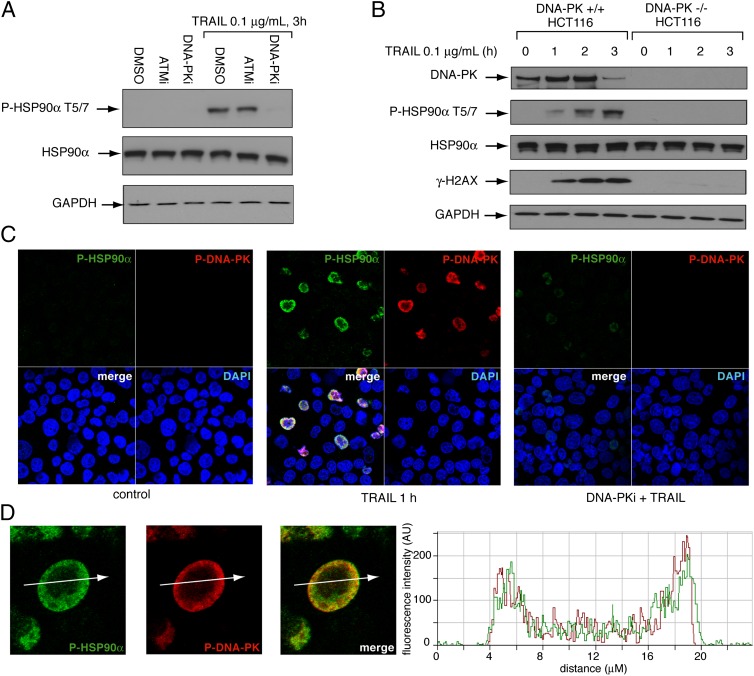
DNA-PK Is the Primary Kinase for HSP90α Phosphorylation, and Activated DNA-PK Colocalizes with Phosphorylated HSP90α-T5/7.
Because both ATM and DNA-PK are activated in response to TRAIL (26), we tested whether ATM or DNA-PK is implicated in HSP90α-T5/7 phosphorylation. Pretreatment with the ATM inhibitor (ATMi) KU-55933 (33) under conditions that blocked ATM autophosphorylation on S1981 (24) (Fig. S2) had no effect on HSP90α phosphorylation (Fig. 3A). In contrast, pretreatment with Nu7441, a specific DNA-PK inhibitor (DNA-PKi) (34, 35), completely suppressed HSP90α phosphorylation induced by TRAIL (Fig. 3A). Similar results were obtained by immunofluorescence microscopy. Nu7441 effectively suppressed both HSP90α and DNA-PK phosphorylation (Fig. 3C). Moreover TRAIL-stimulated HSP90α-T5/7 phosphorylation was totally abrogated in DNA-PK–knockout cells (Fig. 3B). Together, these results demonstrate that DNA-PK phosphorylates HSP90α at T5/7 in apoptotic cells. Single-cell analyses also revealed the colocalization of P-DNA-PK and P-HSP90α-T5/7 after TRAIL treatment (Fig. 3D).
Fig. 3.
DNA-PK is the kinase phosphorylating HSP90α in apoptotic cells. (A) Effects of specific ATMi and DNA-PKi on the phosphorylation of HSP90α after TRAIL treatment. HCT116 cells were treated with the ATMi or DNA-PKi (10 μM, 1 h) before the addition of TRAIL (0.1 μg/mL, 3 h). P-HSP90α-T5/7 and total HSP90α were analyzed by Western blotting. GAPDH was used as a loading control. (B) DNA-PK–knockout cells fail to phosphorylate HSP90α and H2AX in response to TRAIL. DNA-PK+/+ and DNA-PK−/− HCT116 cells were treated as indicated. DNA-PK, P-HSP90α-T5/7, total HSP90α, and γ-H2AX were analyzed by Western blotting. GAPDH was used as a loading control. (C) P-HSP90α-T5/7 and P-DNA-PK-T2609 confocal immunofluorescence staining in HCT116 cells treated with DNA-PKi (10 μM, 1 h) before the addition of TRAIL (0.1 μg/mL, 1 h). P-HSP90α-T5/7 was labeled in green and P-DNA-PK-T2609 in red; nuclei were stained blue with DAPI. (D) Relative distribution of P-HSP90α-T5/7 and P-DNA-PK-T2609 in a single cell. (Left) Confocal microscopy images. (Right) Intensity tracing.
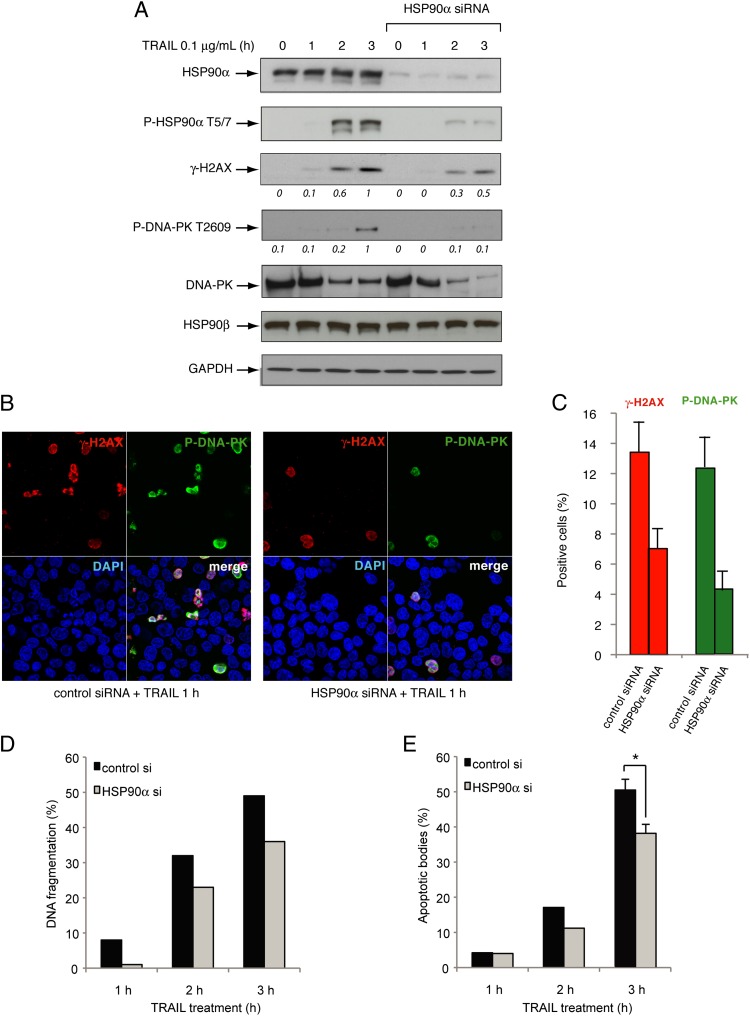
HSP90α Down-Regulation Reduces DNA-PK Activation, DNA Fragmentation, and Apoptotic Body Formation.
HSP90α down-regulation by siRNA transfection reduced TRAIL-induced activation/phosphorylation of DNA-PK and γ-H2AX induction (Fig. 4 A–C), as demonstrated by Western blotting (Fig. 4A) and immunofluorescence microscopy (Fig. 4 B and C). We also observed that the reduction of DNA-PK protein levels, which has been related to caspase-mediated cleavage of DNA-PK (36), decreased to a greater extent upon TRAIL treatment in cells whose HSP90α expression was reduced by siRNA than in control cells (Fig. 4A). These results indicate that HSP90α acts as a cofactor for DNA-PK stability and activity during apoptosis. Moreover, we observed that down-regulation of HSP90α decreased TRAIL-induced DNA fragmentation measured by filter elution assay (Fig. 4D) and also reduced TRAIL-induced apoptotic body formation measured as sub-G1 fraction by FACS analysis (Fig. 4E). However, caspase-8 activation, which is at the very beginning of the TRAIL activation pathway (37), was not affected by HSP90α siRNA (Fig. S3), demonstrating that HSP90α siRNA does not inhibit TRAIL activity directly at the receptor level. Together, these results indicate the importance of HSP90α for the execution of nuclear apoptosis. Importantly, silencing of HSP90α did not affect HSP90β expression (Fig. 4A), emphasizing the specificity of the role played by HSP90α in these processes.
Fig. 4.
HSP90α down-regulation reduces the apoptotic DNA-PK activation, γ-H2AX formation, DNA fragmentation, and apoptotic body formation. (A) HCT116 cells were treated with TRAIL 72 h after transfection with siRNA against HSP90α or negative control siRNA. HSP90α, P-HSP90α-T5/7, γ-H2AX, P-DNA-PK-T2609, total DNA-PK, and HSP90β were analyzed by Western blotting. GAPDH was used as a loading control. The numeric values indicated in italics under the blots were obtained by densitometry analysis (ImageQuant software) and represent the ratios of γ-H2AX or P-DNA-PK to GAPDH (taking the 3-h control siRNA as 1) (B) γ-H2AX and P-DNA-PK-T2609 confocal immunofluorescence staining in HCT116 cells treated with TRAIL (0.1 μg/mL, 1 h) 72 h after transfection with siRNA against HSP90α or negative control siRNA. P-DNA-PK was labeled in green and γ-H2AX in red; nuclei were stained blue with DAPI. (C) Quantification of the experiment presented in B. The percentages of γ-H2AX– and P-DNA-PK–positive cells are represented by red and green columns, respectively. Error bars indicate 95% confidence intervals. One thousand cells were counted. (D) Effect of HSP90α on DNA fragmentation measured by filter elution assay. HCT116 cells transfected with siRNA against HSP90α or negative control siRNA were treated with TRAIL (0.1 μg/mL) for the indicated times. The y-axis represents the percentage of DNA fragmentation. (E) Effect of HSP90α on apoptotic body formation measured as sub-G1 by FACS analysis. HCT116 cells transfected with siRNA against HSP90α or negative control siRNA were treated with TRAIL (0.1 μg/mL) for the indicated times. The y-axis represents the percentage of cells with sub-G1 DNA. *P < 0.5, unpaired t test.
Discussion
Our study shows that HSP90α is phosphorylated extensively by DNA-PK at threonines 5 and 7 in the nucleus of cells that engage in programmed cell death. This phosphorylation is not limited to TRAIL-induced apoptosis but also is induced by Fas ligand and the ubiquitous apoptosis inducer staurosporine (31, 32). P-HSP90α-T5/7 localizes in the apoptotic ring together with activated/phosphorylated DNA-PK and γ-H2AX. We also demonstrate that HSP90α acts as a chaperone for DNA-PK and plays a necessary role in the execution of nuclear apoptosis.
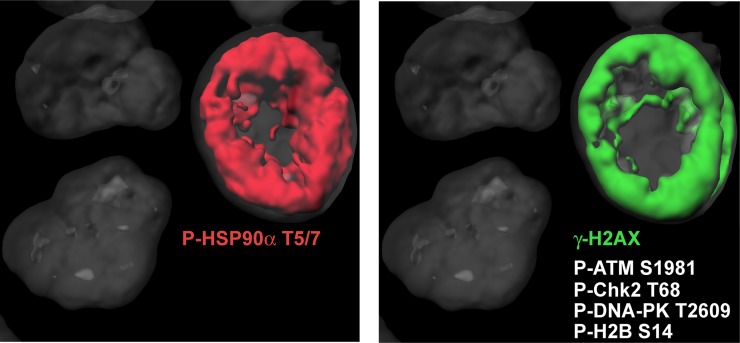
Apoptosis was defined initially by its landmark nuclear characteristics, including pyknotic nuclei and internucleosomal DNA fragmentation (38). High-molecular-weight genomic fragments were found to occur early in apoptosis in response to the nuclease caspase-activated DNase that initiates the breakdown of the cell’s genetic material (39). Epigenetic modifications also take place during the early phases of apoptosis. Recently we reported that phosphorylation of H2AX forms diffusely at the nuclear periphery of cells that apparently are still normal by light microscopy but are committed to apoptosis (24–26). This γ-H2AX nuclear staining, which we refer to as “the apoptotic γ-H2AX ring,” constitutes an epigenetic landmark of early apoptosis. It spreads to the whole nucleus and persists throughout the apoptotic process as nuclei condense with diffuse γ-H2AX staining before their fragmentation into apoptotic bodies. Histone H2B also is phosphorylated on serine 14 during apoptosis and localizes to the γ-H2AX ring (24), and histone H2AX phosphorylation on tyrosine 142 has been reported during apoptosis (40, 41). The current study reveals that P-HSP90α-T5/7 is an abundant component of the apoptotic ring and is found in cells undergoing DNA fragmentation. Thus, P-HSP90α-T5/7 now can be added to the list of phosphorylated and activated DDR proteins that are involved in apoptosis, along with Chk2, H2AX, ATM, and DNA-PK (Fig. 5 and Movie S1).
Fig. 5.
Members of the apoptotic ring. 3D picture of HCT116 cells treated with TRAIL (0.1 μg/mL, 1 h). P-HSP90α was labeled in red, γ-H2AX in green, and the nucleus in gray. P-HSP90α-T5/7, γ-H2AX, P-ATM, P-Chk2, P-DNA-PK, and P-H2B have been identified in the apoptotic ring. See Movie S1.
While our studies were in progress, an independent study reported HSP90α-T5/7 phosphorylation by DNA-PK in response to DNA damage (42). These findings, together with ours described herein, highlight the ubiquitous nature of HSP90α phosphorylation by DNA-PK in response to DNA breaks. However, although both phenomena are mediated by DNA-PK, apoptotic HSP90α-T5/7 phosphorylation differs from DDR-induced HSP90α-T5/7 phosphorylation in several ways. First, the intensity of apoptotic P-HSP90α-T5/7 is markedly greater than that generated by DNA damage inducers such as the topoisomerase I inhibitor camptothecin, a well-characterized inducer of γ-H2AX and DDR responses (Fig. 1) (43). Second, apoptotic P-HSP90α-T5/7 is distributed broadly and rapidly in the apoptotic ring, contrary to the much weaker, focally distributed and delayed phosphorylation of HSP90α in γ-H2AX DDR foci (42). Third, the apoptotic ring, unlike DDR foci, is defective for DNA repair because it lacks many DDR effector proteins (such as 53BP1). This is due to the fact that caspase-mediated cleavage of MDC1, which normally binds to γ-H2AX and initiates the DDR, dissociates the forkhead-associated and breast cancer C-terminal domains of MDC1 and inactivates DNA repair in the apoptotic ring (24–26).
Our study provides evidence for the functional importance of HSP90α as a chaperone for DNA-PK in apoptosis. HSP90α not only promotes formation of the apoptotic ring but also promotes apoptotic DNA fragmentation and the generation of apoptotic bodies (Fig. 4). HSP90α (but not HSP90β; see below) enables the full activation/phosphorylation of DNA-PK, as demonstrated by our experiments showing that down-regulation of HSP90α decreases TRAIL-induced phosphorylation of DNA-PK and consequently the phosphorylation of the DNA-PK substrates H2AX and HSP90α itself.
Although inhibition of HSP90 has been reported to reduce DNA-PK activation in response to ionizing radiation (44), a number of studies (45–47), including ours (Fig. S4), have found that HSP90 inhibitors enhance TRAIL-induced apoptosis. HSP90α stabilizes DNA-PK and limits its degradation by caspases (36), and TRAIL treatment enhances the HSP90α/DNA-PK interaction (Fig. 4 and Fig. S5). HSP90 inhibitors are well known to favor client protein release from HSP90 and in some cases to cause transient activation of the client in the process (48, 49). We speculate that HSP90 inhibitor could act synergistically with TRAIL to enhance the efficiency of DNA-PK release from HSP90 and hence promote more robust DNA-PK activation.
In contrast to pharmacologic inhibition of HSP90, genetic depletion of HSP90α decreases both TRAIL-induced DNA-PK activation and TRAIL-induced apoptosis (Fig. 4). Besides emphasizing that drug inhibition and genetic depletion seldom lead to equivalent outcomes, our data should be considered in light of the fact that HSP90 inhibitors do not target HSP90α specifically but also target other HSP90 isoforms, including HSP90β (2). We found that molecular silencing of HSP90β, which does not contain the N-terminal threonines 5 and 7 phosphorylated by DNA-PK in response to TRAIL, does not reduce TRAIL-induced γ-H2AX but increases P-HSP90α (Fig. S6). Thus, HSP90 inhibitors used in this context may have unpredicted off-target effects that are not seen with specific depletion of HSP90α.
Thus, HSP90α can be viewed as a facilitator of apoptosis by enabling robust activation of DNA-PK and allowing the completion of the apoptotic program leading to DNA fragmentation and formation of apoptotic bodies. The physiologic importance of such complete nuclear apoptosis should not be overlooked, because this process might be critical to avoid the release of toxic and potentially immunogenic cellular components, including nucleic acids and nuclear proteins, from apoptotic cells and tissues (39, 50, 51).
The recognition of the P-HSP90α-T5/7 apoptotic ring, its coincidence with the γ-H2AX apoptotic ring, and the fact that these epigenetic markers can be measured readily in single cells throughout the apoptotic process up to and including the formation of apoptotic bodies identify the P-HSP90α-T5/7 apoptotic modification as a potentially powerful biomarker for translational research and provide insight into the molecular components that orchestrate nuclear apoptosis.
Materials and Methods
Chemicals.
Recombinant human soluble TRAIL was obtained from Alexis (Axxora). Anti-Fas was from Upstate. Camptothecin and staurosporine were from Sigma-Aldrich. The DNA-PKi NU7441 and the ATMi KU-55933 were from Tocris Bioscience. The broad-spectrum caspase inhibitor Z-VAD-fmk was from Bachem. The HSP90 inhibitors geldanamycin and its derivative 17-allylamino-17-demethoxygeldanamycin were from Sigma-Aldrich and National Cancer Institute, respectively.
Cell Lines.
Human colon carcinoma HCT116, cervix carcinoma HeLa, and leukemic Jurkat cell lines were obtained from American Type Culture Collection. HCT116 DNA-PK−/− cells were obtained from Rob Howes (Horizon Discovery, Cambridge, U.K.).
Western Blotting and Antibodies.
Cells were washed twice in PBS and lysed at 4 °C in buffer containing 1% SDS and 10 mM Tris⋅HCl (pH 7.4) supplemented with protease inhibitors (Roche Applied Science) and phosphatase inhibitors (Sigma-Aldrich). Viscosity of the samples was reduced by brief sonication. Equal amounts of proteins were boiled for 5 min in Tris⋅glycine-SDS sample buffer (Invitrogen) or were heated at 70 °C for 10 min in lithium dodecyl sulfate sample buffer (Invitrogen), separated by Tris⋅glycine or Tris⋅acetate polyacrylamide gels (Invitrogen), and electroblotted onto nitrocellulose membranes (Bio-Rad). The membranes were saturated with milk, incubated overnight at 4 °C with primary antibodies, washed, and then incubated for 45 min with the secondary antibodies peroxidase-conjugated goat anti-mouse IgG, peroxidase-conjugated goat anti-rabbit IgG, or peroxidase-conjugated goat anti-rat IgG (Santa Cruz Biotechnology). Signals were revealed by autoradiography using the Enhanced Chemiluminescence detection kit (Pierce).
Primary antibodies used were anti–P-ATM-S1981 (4526; Cell Signaling Technology), anti–DNA-PK (NA57; Calbiochem, EMD Biosciences), anti–P-DNA-PK-T2609 (ab18356; Abcam), anti-GAPDH (2118; Cell Signaling Technology), anti–γ-H2AX (05-636; Upstate), anti-HSP90α (ADI-SPA-840;, Enzo Life Sciences), anti–P-HSP90α-T5/7 (3488; Cell Signaling Technology), anti-HSP90β (PA3-012; Thermo Fisher Scientific), and anti–γ-tubulin (T5326; Sigma-Aldrich).
siRNA.
siRNA targeting HSP90α was obtained from Dharmacon (ON-TARGETplus SMARTpool, catalog no. L-005186–00-0005; ON-TARGETplus siRNA, catalog no. J-005186-08-0050). siRNA targeting HSP90β was obtained from Dharmacon (ON-TARGETplus SMARTpool, catalog number L-005187-00-0005). Negative control siRNA was obtained from Dharmacon (ON-TARGETplus control pool, catalog no. D-001810-10-05). Cells were seeded in six-well plates at a density of 100,000 cells per well (200,000 cells per well for HSP90β siRNA) 16 h before transfection. For each sample, 500 pmoles of siRNA were mixed with 250 μL (Invitrogen) (mix A). Five microliters of lipofectamine 2000 (Invitrogen) were mixed with 250 μL Opti-MEM and incubated for 5 min at room temperature (mix B). After mixes A and B were combined and further incubated for 20 min at room temperature, the siRNA/Lipofectamine complexes were added to 2 mL of culture medium containing the cells. After 5 h, the medium was replaced with regular medium, and cells were incubated for a further 72 h (48 h for HSP90β siRNA).
Immunofluorescence Microscopy.
Cells were washed with PBS, fixed with 4% (wt/wt) formaldehyde in PBS for 20 min, washed with PBS, postfixed, permeabilized with cold (−20 °C) 70% (wt/wt) ethanol for 20 min, washed with PBS, blocked with 8% (wt/wt) BSA in PBS for 1 h, washed with PBS, incubated with the first antibody (P-DNA-PK-T2609, 1:250; γ-H2AX, 1:500; P-HSP90α-T5/7, 1:100; γ-tubulin, 1:500) in 1% BSA in PBS for 2 h, washed with PBS, incubated with secondary antibody conjugated with Alexa Fluor 488 or 568 for 1 h, washed with PBS, and mounted by using Vectashield mounting medium with DAPI to counterstain the DNA (Vector Laboratories). Confocal images were acquired sequentially with Zeiss AIM software on a Zeiss LSM 510 NLO Confocal system (Carl Zeiss Inc.). 3D pictures and the movie were realized with the Bitplane Imaris software v. 6-2.1. The line intensity profiles were realized with the Zeiss AIM software v3.2.
TUNEL Assay Coupled with γ-H2AX Staining.
The TUNEL assay was performed with the In Situ Cell Death Detection kit (Roche). Briefly, cells were fixed with 4% (wt/wt) formaldehyde in PBS for 60 min, washed with PBS, permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate for 2 min on ice, washed with PBS, incubated with TUNEL reaction mixture for 60 min at 37 °C, and washed with PBS. Then the γ-H2AX staining was performed as described above.
DNA Fragmentation by Filter Elution Assay.
The DNA fragmentation by filter elution assay has been described previously (52). Briefly, cells were incubated with [2-14C] thymidine (0.02 μCi/mL) for 2 d and chased overnight in radioisotope-free medium. After transfection and drug treatment, cells were loaded onto a protein-adsorbing filter (Metricel Membrane Filter, 0.8-μm pore size, 25-mm diameter; Pall Corporation), washed with PBS, and lysed in 0.2% sodium sarkosyl, 2 M NaCl, 0.04 M EDTA, pH 10. Then the filters were washed with 0.02 M EDTA, pH 10. DNA was depurinated by incubation of filters in 1 M HCl at 65 °C and then was released from the filters with 0.4 M NaOH at room temperature. Radioactivity was counted by liquid scintillation spectrometry in each fraction (wash, lysis, EDTA wash, and filter). DNA fragmentation was measured as the fraction of disintegrations/min (dpm) in the wash plus lysis fraction plus EDTA wash relative to the total intracellular dpm.
Sub-G1 Analysis.
Cells were washed with PBS, fixed, and permeabilized with cold (− 20 °C) 70% (wt/wt) ethanol overnight. The next day, cell pellets were washed again with PBS, resuspended in PBS buffer containing 0.2% Nonidet P-40 and 0.5 mg/mL RNase A, incubated at room temperature for 15 min, and put on ice 10 min before the addition of 50 μg/mL of propidium iodide. DNA content was determined with a FACScan flow cytometer (Becton Dickinson) and quantified with CellQuest software (Becton Dickinson).
Caspase Activity Assay.
Cells were washed twice in PBS, lysed in 150 mM NaCl, 50 mM Tris⋅HCl (pH 8.0), 0.1% SDS, 1% Nonidet P-40, and 0.5% sodium deoxycholate for 30 min at 4 °C and were centrifuged (10,000 × g at 4 °C). Fifteen micrograms of proteins from the resulting supernatant were incubated in 100 mM Hepes (pH 7.0), 1 mM EDTA, 0.1% CHAPS, 10% glycerol, and 20 mM DTT in the presence of 100 μM of the fluorogenic peptide substrate Z-Ile-Glu-Thr-Asp-AFC, caspase-8 (Calbiochem). 7-Amino-4-trifluoromethylcoumarin (AFC) released from the substrate was excited at 400 nm to measure emission at 505 nm. Fluorescence was monitored continuously at 37 °C for 30 min in a dual luminescence fluorimeter (SpectraMax Gemini XS; Molecular Devices). Caspase activities were determined as initial velocities expressed as relative intensity per minute per milligram protein.
Supplementary Material
Acknowledgments
These studies were supported by the Center for Cancer Research Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203617109/-/DCSupplemental.
See Commentary on page 12844.
References
- 1.Neckers L. Heat shock protein 90: The cancer chaperone. J Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 2.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 4.Zhao R, et al. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnelly A, Blagg BS. Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr Med Chem. 2008;15:2702–2717. doi: 10.2174/092986708786242895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prodromou C, Pearl LH. Structure and functional relationships of Hsp90. Curr Cancer Drug Targets. 2003;3:301–323. doi: 10.2174/1568009033481877. [DOI] [PubMed] [Google Scholar]
- 8.Akner G, Mossberg K, Sundqvist KG, Gustafsson JA, Wikström AC. Evidence for reversible, non-microtubule and non-microfilament-dependent nuclear translocation of hsp90 after heat shock in human fibroblasts. Eur J Cell Biol. 1992;58:356–364. [PubMed] [Google Scholar]
- 9.Perdew GH, Hord N, Hollenback CE, Welsh MJ. Localization and characterization of the 86- and 84-kDa heat shock proteins in Hepa 1c1c7 cells. Exp Cell Res. 1993;209:350–356. doi: 10.1006/excr.1993.1320. [DOI] [PubMed] [Google Scholar]
- 10.Becker B, et al. Induction of Hsp90 protein expression in malignant melanomas and melanoma metastases. Exp Dermatol. 2004;13:27–32. doi: 10.1111/j.0906-6705.2004.00114.x. [DOI] [PubMed] [Google Scholar]
- 11.Eustace BK, et al. Functional proteomic screens reveal an essential extracellular role for hsp90 alpha in cancer cell invasiveness. Nat Cell Biol. 2004;6:507–514. doi: 10.1038/ncb1131. [DOI] [PubMed] [Google Scholar]
- 12.Sidera K, Patsavoudi E. Extracellular HSP90: Conquering the cell surface. Cell Cycle. 2008;7:1564–1568. doi: 10.4161/cc.7.11.6054. [DOI] [PubMed] [Google Scholar]
- 13.Sreedhar AS, Kalmár E, Csermely P, Shen YF. Hsp90 isoforms: Functions, expression and clinical importance. FEBS Lett. 2004;562:11–15. doi: 10.1016/s0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- 14.Wanderling S, et al. GRP94 is essential for mesoderm induction and muscle development because it regulates insulin-like growth factor secretion. Mol Biol Cell. 2007;18:3764–3775. doi: 10.1091/mbc.E07-03-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felts SJ, et al. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J Biol Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- 16.Hua G, Zhang Q, Fan Z. Heat shock protein 75 (TRAP1) antagonizes reactive oxygen species generation and protects cells from granzyme M-mediated apoptosis. J Biol Chem. 2007;282:20553–20560. doi: 10.1074/jbc.M703196200. [DOI] [PubMed] [Google Scholar]
- 17.Dezwaan DC, Freeman BC. HSP90: The Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- 18.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 19.Tariq M, Nussbaumer U, Chen Y, Beisel C, Paro R. Trithorax requires Hsp90 for maintenance of active chromatin at sites of gene expression. Proc Natl Acad Sci USA. 2009;106:1157–1162. doi: 10.1073/pnas.0809669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taieb J, et al. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med. 2006;12:214–219. doi: 10.1038/nm1356. [DOI] [PubMed] [Google Scholar]
- 21.Wang S. The promise of cancer therapeutics targeting the TNF-related apoptosis-inducing ligand and TRAIL receptor pathway. Oncogene. 2008;27:6207–6215. doi: 10.1038/onc.2008.298. [DOI] [PubMed] [Google Scholar]
- 22.Yagita H, Takeda K, Hayakawa Y, Smyth MJ, Okumura K. TRAIL and its receptors as targets for cancer therapy. Cancer Sci. 2004;95:777–783. doi: 10.1111/j.1349-7006.2004.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JJ, Knudsen S, Mazin W, Dahlgaard J, Zhang B. A 71-gene signature of TRAIL sensitivity in cancer cells. Mol Cancer Ther. 2012;11:34–44. doi: 10.1158/1535-7163.MCT-11-0620. [DOI] [PubMed] [Google Scholar]
- 24.Solier S, Pommier Y. The apoptotic ring: A novel entity with phosphorylated histones H2AX and H2B and activated DNA damage response kinases. Cell Cycle. 2009;8:1853–1859. doi: 10.4161/cc.8.12.8865. [DOI] [PubMed] [Google Scholar]
- 25.Solier S, Pommier Y. MDC1 cleavage by caspase-3: A novel mechanism for inactivating the DNA damage response during apoptosis. Cancer Res. 2011;71:906–913. doi: 10.1158/0008-5472.CAN-10-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solier S, Sordet O, Kohn KW, Pommier Y. Death receptor-induced activation of the Chk2- and histone H2AX-associated DNA damage response pathways. Mol Cell Biol. 2009;29:68–82. doi: 10.1128/MCB.00581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukherjee B, et al. DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair (Amst) 2006;5:575–590. doi: 10.1016/j.dnarep.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Bonner WM, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falsone SF, Gesslbauer B, Tirk F, Piccinini AM, Kungl AJ. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 2005;579:6350–6354. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 30.Lees-Miller SP, Anderson CW. The human double-stranded DNA-activated protein kinase phosphorylates the 90-kDa heat-shock protein, hsp90 alpha at two NH2-terminal threonine residues. J Biol Chem. 1989;264:17275–17280. [PubMed] [Google Scholar]
- 31.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertrand R, Solary E, O’Connor P, Kohn KW, Pommier Y. Induction of a common pathway of apoptosis by staurosporine. Exp Cell Res. 1994;211:314–321. doi: 10.1006/excr.1994.1093. [DOI] [PubMed] [Google Scholar]
- 33.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 34.Leahy JJ, et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 36.Bharti A, et al. Inactivation of DNA-dependent protein kinase by protein kinase Cdelta: Implications for apoptosis. Mol Cell Biol. 1998;18:6719–6728. doi: 10.1128/mcb.18.11.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley SK, Ashkenazi A. Targeting death receptors in cancer with Apo2L/TRAIL. Curr Opin Pharmacol. 2004;4:333–339. doi: 10.1016/j.coph.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Lecoeur H. Nuclear apoptosis detection by flow cytometry: Influence of endogenous endonucleases. Exp Cell Res. 2002;277:1–14. doi: 10.1006/excr.2002.5537. [DOI] [PubMed] [Google Scholar]
- 39.Samejima K, Earnshaw WC. Trashing the genome: The role of nucleases during apoptosis. Nat Rev Mol Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 40.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quanz M, et al. Heat shock protein 90α (Hsp90α) is phosphorylated in response to DNA damage and accumulates in repair foci. J Biol Chem. 2012;287:8803–8815. doi: 10.1074/jbc.M111.320887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furuta T, et al. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem. 2003;278:20303–20312. doi: 10.1074/jbc.M300198200. [DOI] [PubMed] [Google Scholar]
- 44.Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66:9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- 45.Ma Y, Lakshmikanthan V, Lewis RW, Kumar MV. Sensitization of TRAIL-resistant cells by inhibition of heat shock protein 90 with low-dose geldanamycin. Mol Cancer Ther. 2006;5:170–178. doi: 10.1158/1535-7163.MCT-05-0129. [DOI] [PubMed] [Google Scholar]
- 46.Vasilevskaya IA, O’Dwyer PJ. 17-Allylamino-17-demethoxygeldanamycin overcomes TRAIL resistance in colon cancer cell lines. Biochem Pharmacol. 2005;70:580–589. doi: 10.1016/j.bcp.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 47.Kim YJ, Lee SA, Myung SC, Kim W, Lee CS. Radicicol, an inhibitor of Hsp90, enhances TRAIL-induced apoptosis in human epithelial ovarian carcinoma cells by promoting activation of apoptosis-related proteins. Mol Cell Biochem. 2012;359:33–43. doi: 10.1007/s11010-011-0997-9. [DOI] [PubMed] [Google Scholar]
- 48.Donzé O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koga F, et al. Hsp90 inhibition transiently activates Src kinase and promotes Src-dependent Akt and Erk activation. Proc Natl Acad Sci USA. 2006;103:11318–11322. doi: 10.1073/pnas.0604705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napirei M, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 51.Yasutomo K, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 52.Solary E, Bertrand R, Kohn KW, Pommier Y. Differential induction of apoptosis in undifferentiated and differentiated HL-60 cells by DNA topoisomerase I and II inhibitors. Blood. 1993;81:1359–1368. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.