Abstract
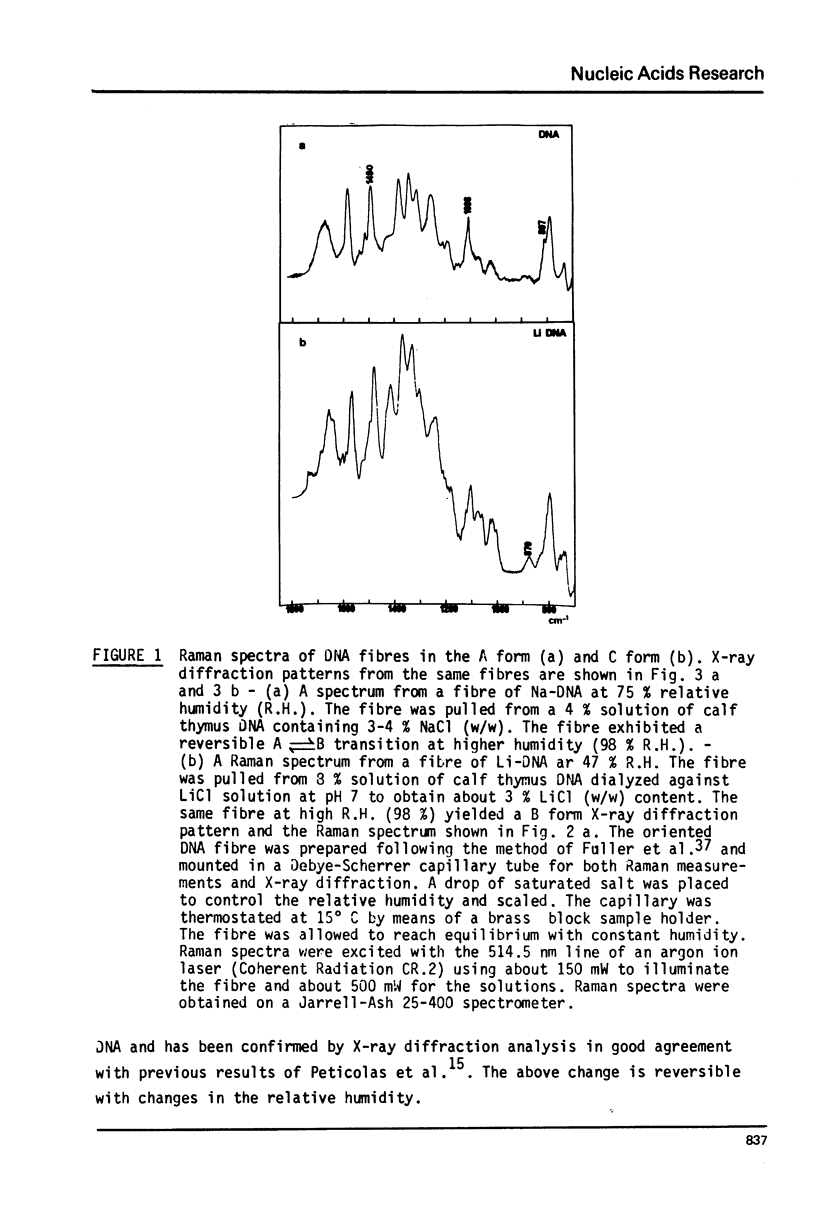
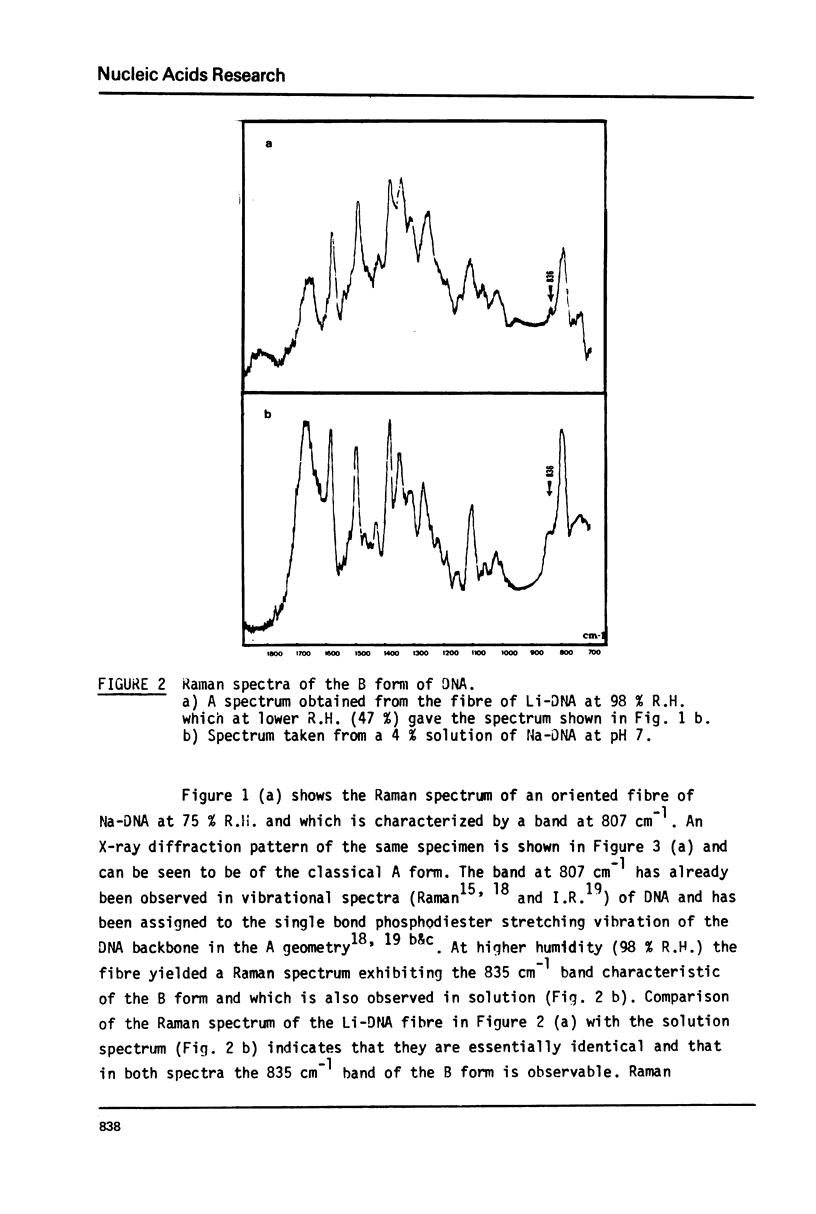
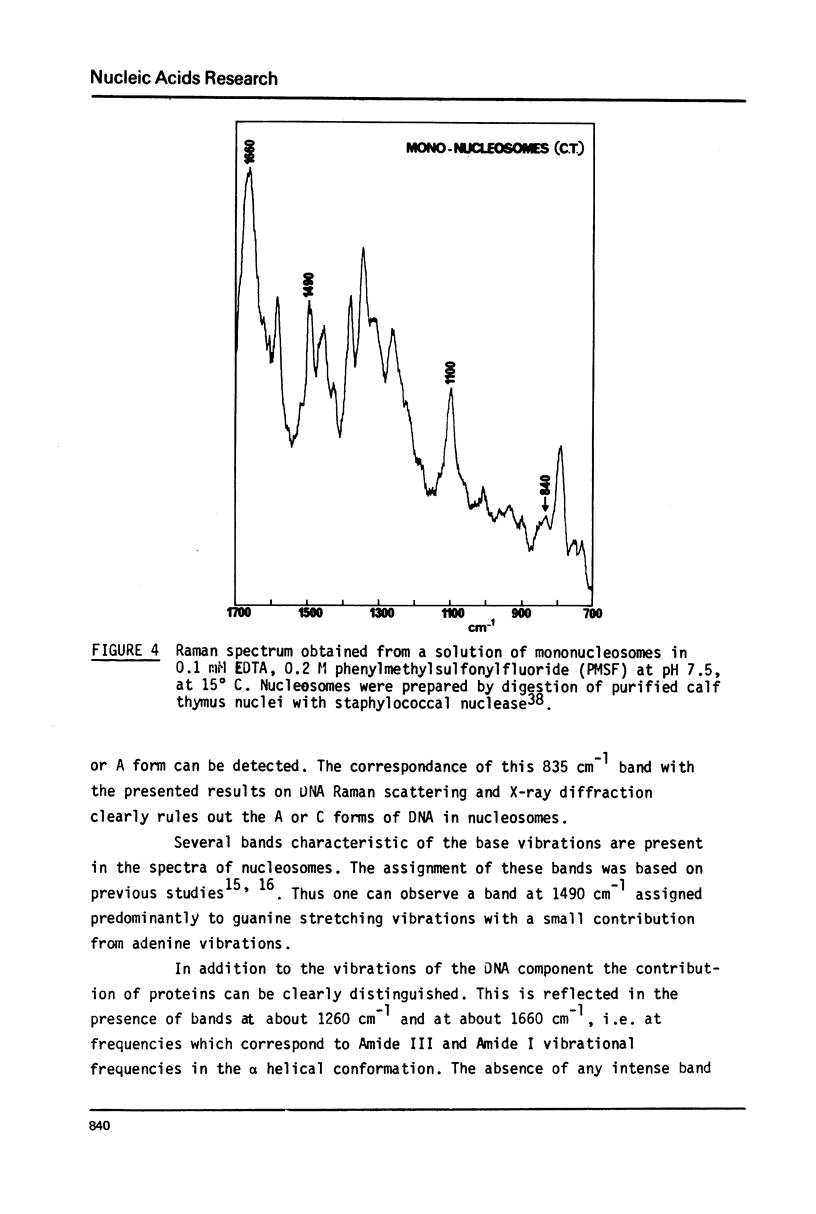
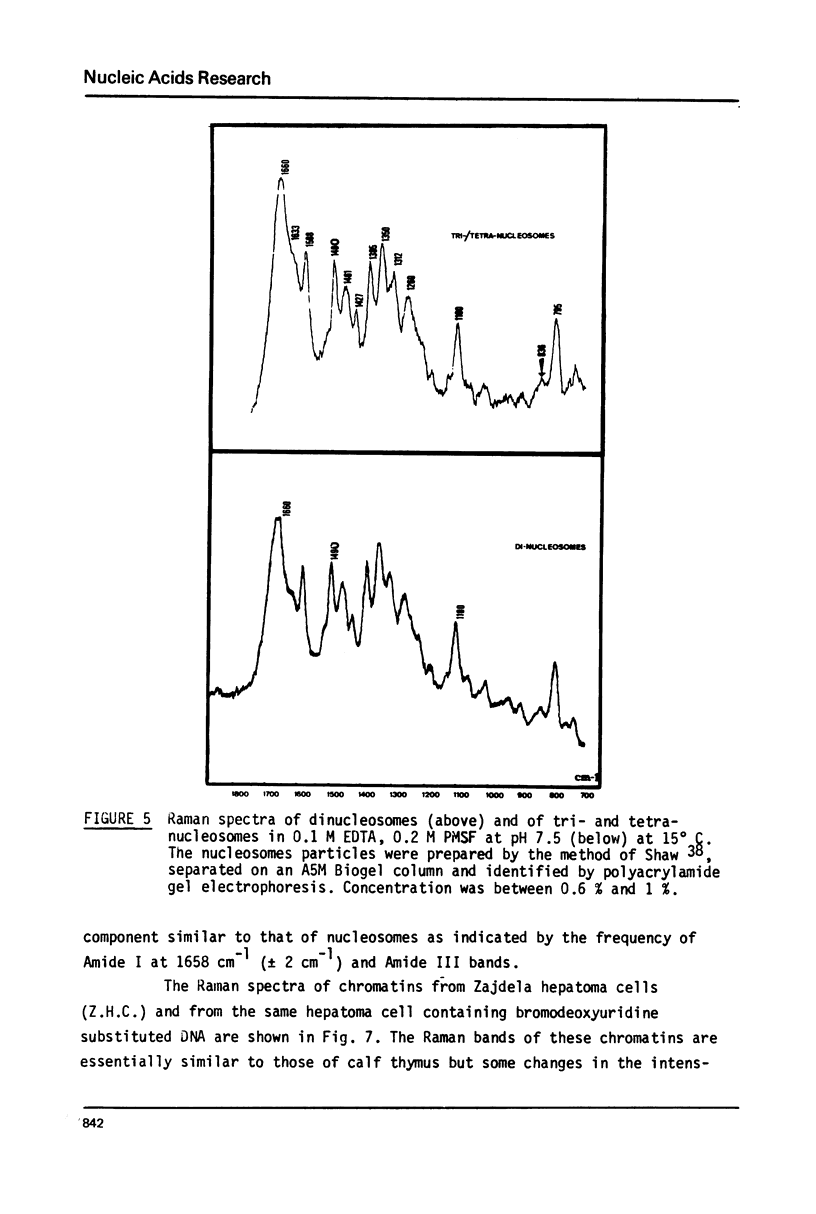
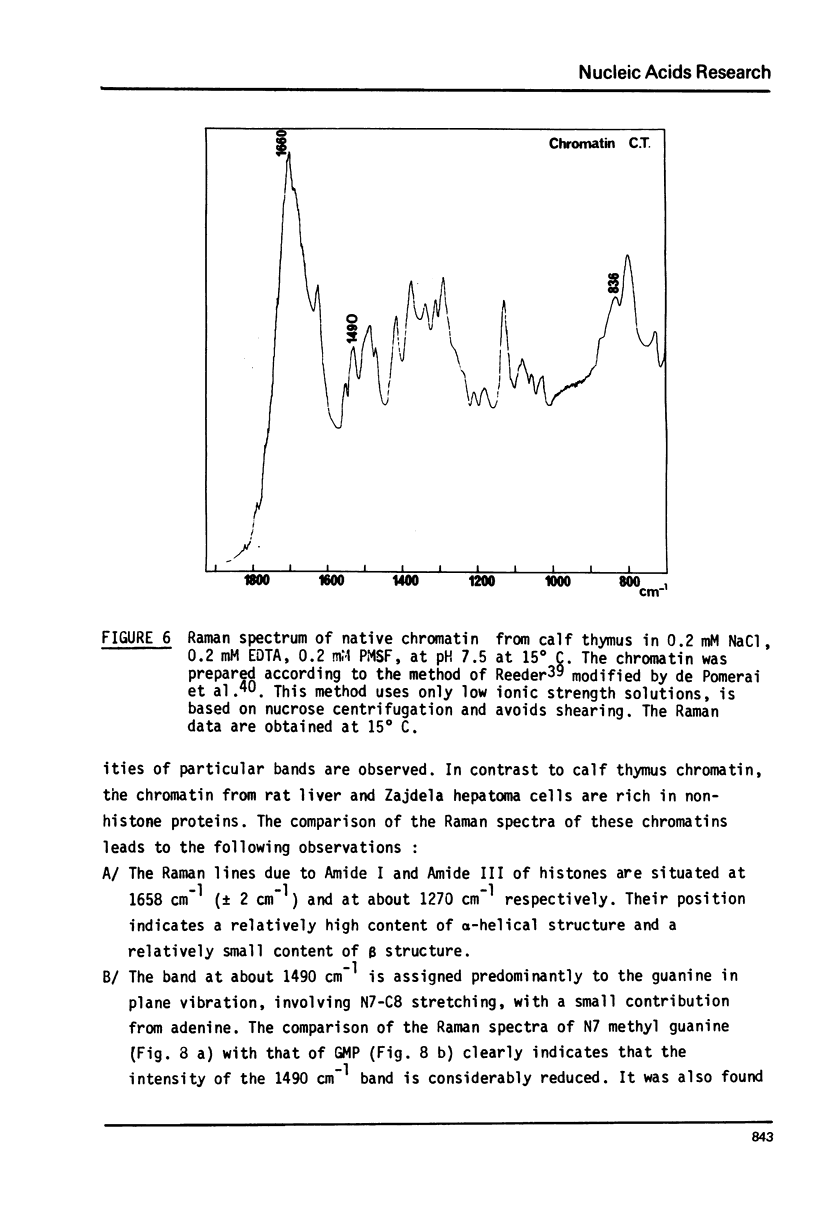
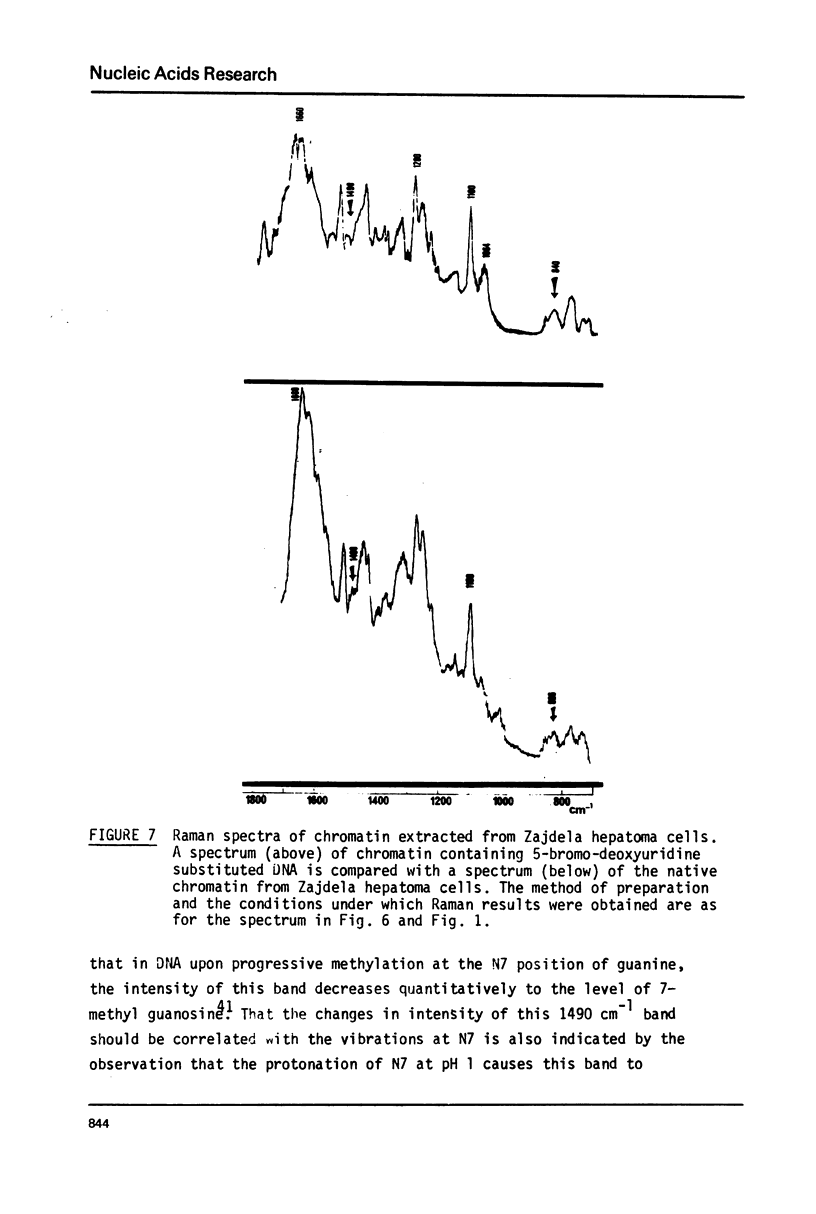
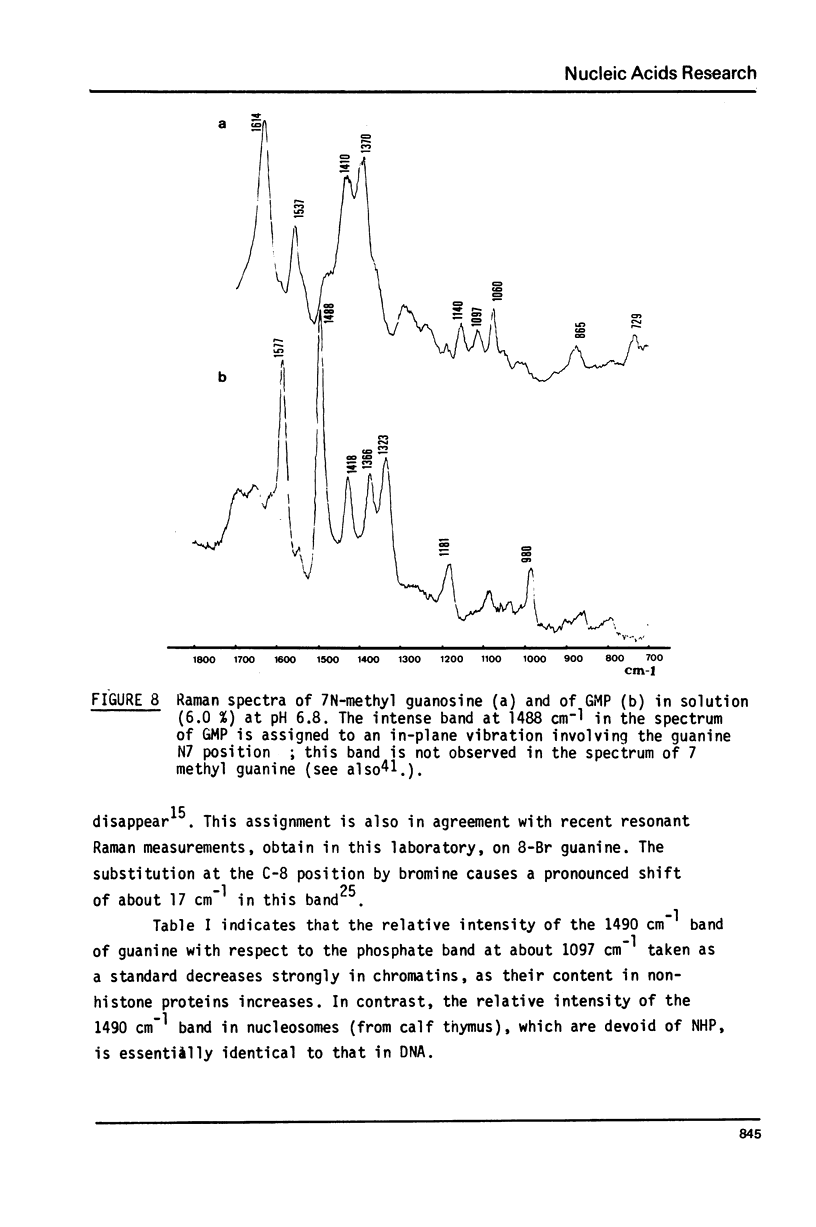
Studies of native chromatins and of isolated nucleosomes (from calf thymus) show that the DNA is in the B form or modified B form. This was determined by Raman spectroscopy of chromatins, of nucleosomes (from calf thymus) and of DNA fibres and directly correlated with X-ray diffraction studies. The Raman spectra of three forms of DNA (A, B and C) have been characterized in fibres both by X-ray diffraction and Raman spectroscopy on the same sample. In particular, the Raman spectrum of the C form of DNA is characterized by a band of about 870 cm(-1). For the first time, chromatins of different origins with increasing content of non-histone proteins have been investigated by Raman spectroscopy. The site of interaction of the non-histone proteins appears to involve the N7 position of guanine while the histone core does not interact at this site. It is proposed that the mechanism of specific recognition in chromatin involves the large groove.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler K., Beyreuther K., Fanning E., Geisler N., Gronenborn B., Klemm A., Müller-Hill B., Pfahl M., Schmitz A. How lac repressor binds to DNA. Nature. 1972 Jun 9;237(5354):322–327. doi: 10.1038/237322a0. [DOI] [PubMed] [Google Scholar]
- Bram S. The secondary structure of DNA in solution and in nucleohistone. J Mol Biol. 1971 May 28;58(1):277–288. doi: 10.1016/0022-2836(71)90246-4. [DOI] [PubMed] [Google Scholar]
- Brown E. B., Peticolas W. L. Conformational geometry and vibrational frequencies of nucleic acid chains. Biopolymers. 1975 Jun;14(6):1259–1271. doi: 10.1002/bip.1975.360140614. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr, Kraut J. A proposed model for interaction of polypeptides with RNA. Proc Natl Acad Sci U S A. 1974 Feb;71(2):283–287. doi: 10.1073/pnas.71.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champeil P., Tran T. P., Brahms J. A new approach to the characterization of the B and A forms of DNA by I.R. spectroscopy. Biochem Biophys Res Commun. 1973 Dec 10;55(3):881–887. doi: 10.1016/0006-291x(73)91226-6. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Adler A. J., Fasman G. D. Conformational prediction and circular dichroism studies on the lac repressor. J Mol Biol. 1975 Jul 25;96(1):29–45. doi: 10.1016/0022-2836(75)90180-1. [DOI] [PubMed] [Google Scholar]
- Church G. M., Sussman J. L., Kim S. H. Secondary structural complementarity between DNA and proteins. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1458–1462. doi: 10.1073/pnas.74.4.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H., Klug A. Kinky helix. Nature. 1975 Jun 12;255(5509):530–533. doi: 10.1038/255530a0. [DOI] [PubMed] [Google Scholar]
- De Santis P., Forni E., Rizzo R. Conformational analysis of DNA-basic polypeptide complexes: possible models of nucleoprotamines and nucleohistones. Biopolymers. 1974;13(2):313–326. doi: 10.1002/bip.1974.360130207. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Fuller W., Hutchinson F., Spencer M., Wilkins M. H. Molecular and crystal structures of double-helical RNA. I. An x-ray diffraction study of fragmented yeast RNA and a preliminary double-helical RNA model. J Mol Biol. 1967 Aug 14;27(3):507–524. doi: 10.1016/0022-2836(67)90055-1. [DOI] [PubMed] [Google Scholar]
- Gursky G. V., Tumanyan V. G., Zasedatelev A. S., Zhuze A. L., Grokhovsky S. L., Gottikh B. P. A model for the binding of lac repressor to the lac operator. Mol Biol Rep. 1976 Apr;2(5):427–434. doi: 10.1007/BF00366265. [DOI] [PubMed] [Google Scholar]
- Herskovits T. T., Brahms J. Structural investigations on DNA-protamine complexes. Biopolymers. 1976 Apr;15(4):687–706. doi: 10.1002/bip.1976.360150408. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Liquier J., Taboury J., Taillandier E., Brahms J. Infrared linear dichroism investigations of deoxyribonucleic complexes with histones H2B and H3. Biochemistry. 1977 Jul 12;16(14):3262–3266. doi: 10.1021/bi00633a034. [DOI] [PubMed] [Google Scholar]
- Mansy S., Engstrom S. K., Peticolas W. L. Laser Raman identification of an interaction site on DNA for arginine containing histones in chromatin. Biochem Biophys Res Commun. 1976 Feb 23;68(4):1242–1247. doi: 10.1016/0006-291x(76)90330-2. [DOI] [PubMed] [Google Scholar]
- Mansy S., Peticolas W. L. Detection of the sites of alkylation in DNA and polynucleotides by laser Raman spectroscopy. Biochemistry. 1976 Jun 15;15(12):2650–2655. doi: 10.1021/bi00657a026. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. The structure of the chromatin core particle in solution. Nucleic Acids Res. 1977 Sep;4(9):3199–3214. doi: 10.1093/nar/4.9.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilet J., Brahms J. Dependence of B-A conformational change in DNA on base composition. Nat New Biol. 1972 Mar 29;236(65):99–100. doi: 10.1038/newbio236099a0. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Transcription of chromatin by bacterial RNA polymerase. J Mol Biol. 1973 Oct 25;80(2):229–241. doi: 10.1016/0022-2836(73)90169-1. [DOI] [PubMed] [Google Scholar]
- Rill R., Van Holde K. E. Properties of nuclease-resistant fragments of calf thymus chromatin. J Biol Chem. 1973 Feb 10;248(3):1080–1083. [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw B. R., Corden J. L., Sahasrabuddhe C. G., Van Holde K. E. Chromatographic separation of chromatin subunits. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1193–1198. doi: 10.1016/s0006-291x(74)80410-9. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Thomas G. J., Jr, Prescott B., Olins D. E. Secondary structure of histones and DNA in chromatin. Science. 1977 Jul 22;197(4301):385–388. doi: 10.1126/science.560060. [DOI] [PubMed] [Google Scholar]
- de Pomerai D. I., Chesterton C. J., Butterworth P. H. The effect of heparin on the structure and template properties of chromatin. FEBS Lett. 1974 Jun 1;42(2):149–153. doi: 10.1016/0014-5793(74)80773-8. [DOI] [PubMed] [Google Scholar]



