Abstract
The crystal structures of two nucleosides, 5-carbamoylmethyluridine (1) and 5-carboxymethyluridine (2), were determined from three-dimensional x-ray diffraction data, and refined to R = 0.036 and R = 0.047, respectively. Compound 1 is in the C3'-endo conformation with chi +5.2 degrees (anti), psiinfinity = +63.4 degrees and psialpha = +180.0 degrees (tt); 2 is in the C2'endo conformation with chi +49.4 degrees (anti), psiinfinity -60.5 degrees and psialpha +60.0 degrees (gg). For each derivative, the plane of the side chain substituent is skewed with respect to the plane of the nucleobase; for 1, the carboxamide group is on the same side of the uracil plane vis a vis the ribose ring; for 2, the carboxyl group is on the opposite side of this plane. No base pairing is observed for either structure. Incorporation of structure 1 into a 3'-stacked tRNA anticodon appears to place 08 within hydrogen bonding distance of the 02' hydroxyl of ribose 33, which may limit the ability of such a molecule of tRNA to "wobble".
Full text
PDF


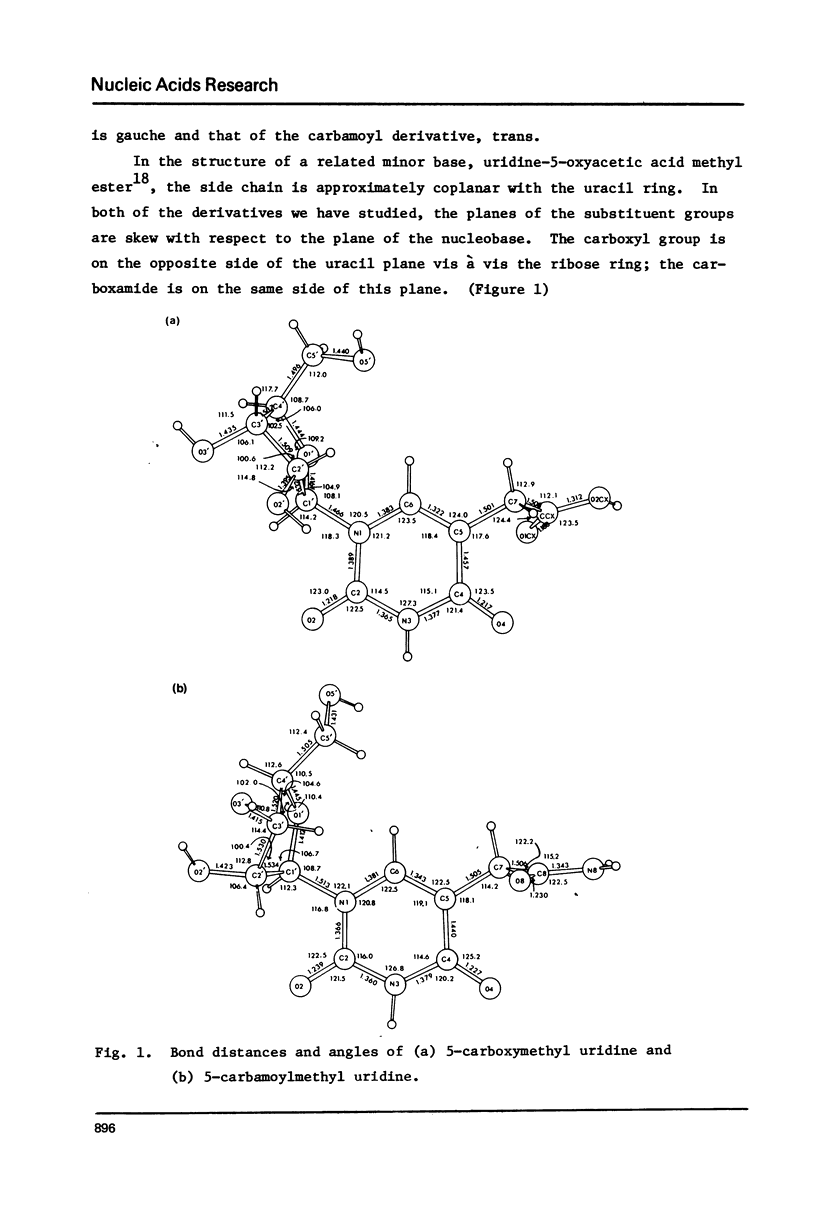
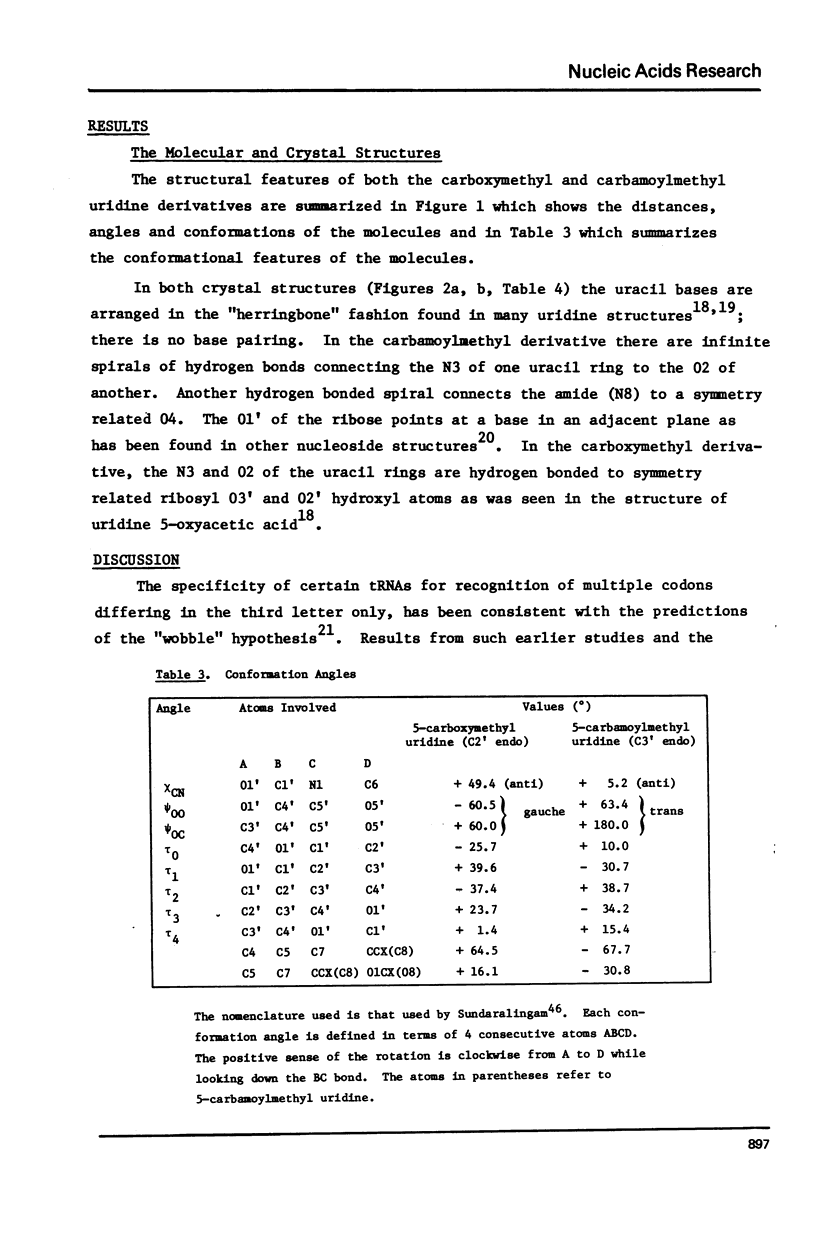
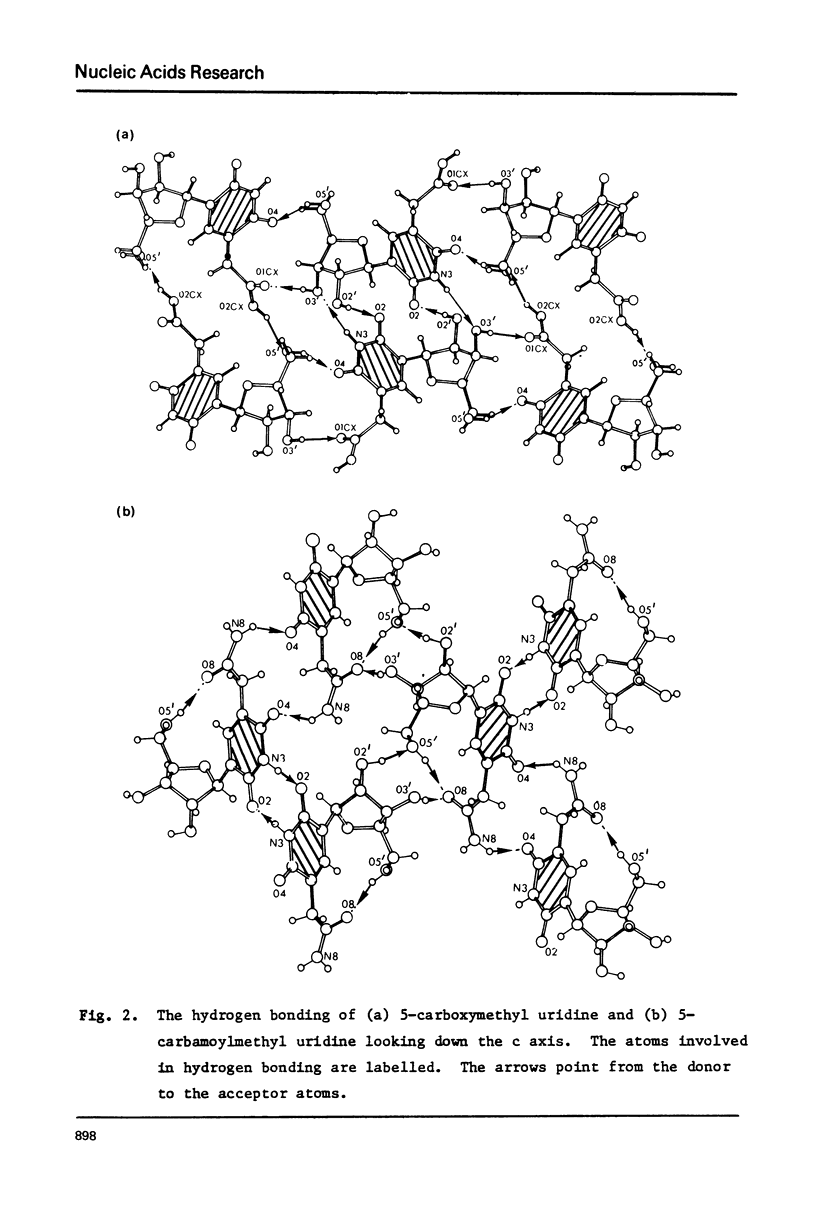





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel'rod V. D., Kryukov V. M., Isaenko S. N., Bayev A. A. Nucleotide sequence in tRNA Val-2a from baker's yeast. FEBS Lett. 1974 Sep 1;45(1):333–336. doi: 10.1016/0014-5793(74)80874-4. [DOI] [PubMed] [Google Scholar]
- Baczynskyj L., Biemann K., Hall R. H. Sulfur-containing nucleoside from yeast transfer ribonucleic acid: 2-thio-5(or 6)-uridine acetic acid methyl ester. Science. 1968 Mar 29;159(3822):1481–1483. doi: 10.1126/science.159.3822.1481. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Seidman J. G., Guthrie C., McClain W. H. Transfer RNA biosynthesis: the nucleotide sequence of a precursor to serine and proline transfer RNAs. Proc Natl Acad Sci U S A. 1974 Feb;71(2):413–416. doi: 10.1073/pnas.71.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg C. E., Thomas J. M., Sundaralingam M., Rao S. T. Stereochemistry of nucleic acids and their constituents. X. Solid-state base-stacking patterns in nucleic acid constituents and polynucleotides. Biopolymers. 1971;10(1):175–219. doi: 10.1002/bip.360100113. [DOI] [PubMed] [Google Scholar]
- Cameron V., Uhlenbeck O. C. Removal of Y-37 from tRNA phe yeast alters oligomer binding to two loops. Biochem Biophys Res Commun. 1973 Feb 5;50(3):635–640. doi: 10.1016/0006-291x(73)91291-6. [DOI] [PubMed] [Google Scholar]
- Carbon J., Squires C., Hill C. W. Glycine transfer RNA of Escherichia coli. II. Impaired GGA-recognition in strains containing a genetically altered transfer RNA; reversal by a secondary suppressor mutation. J Mol Biol. 1970 Sep 28;52(3):571–584. doi: 10.1016/0022-2836(70)90420-1. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Lane B. G. 5-carboxymethyluridine, a novel nucleoside derived from yeast and wheat embryo transfer ribonucleates. Biochemistry. 1968 Oct;7(10):3441–3453. doi: 10.1021/bi00850a020. [DOI] [PubMed] [Google Scholar]
- Gray M. W. Structural analysis of O2'-methyl-5-carbamoylmethyluridine, a newly discovered constituent of yeast transfer RNA. Biochemistry. 1976 Jul 13;15(14):3046–3051. doi: 10.1021/bi00659a017. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Söll D. G., Crothers D. M. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J Mol Biol. 1976 May 25;103(3):499–519. doi: 10.1016/0022-2836(76)90214-x. [DOI] [PubMed] [Google Scholar]
- Hickey E. D., Weber L. A., Baglioni C., Kim C. H., Sarma R. H. A relation between inhibition of protein synthesis and conformation of 5'-phosphorylated 7-methylguanosine derivatives. J Mol Biol. 1977 Jan 15;109(2):173–183. doi: 10.1016/s0022-2836(77)80027-2. [DOI] [PubMed] [Google Scholar]
- Ishikura H., Yamada Y., Nishimura S. Structure of serine tRNA from Escherichia coli. I. Purification of serine tRNA's with different codon responses. Biochim Biophys Acta. 1971 Jan 28;228(2):471–481. doi: 10.1016/0005-2787(71)90052-9. [DOI] [PubMed] [Google Scholar]
- KALLEN R. G., SIMON M., MARMUR J. The new occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA:5-hydroxymethyl uracil. J Mol Biol. 1962 Aug;5:248–250. doi: 10.1016/s0022-2836(62)80087-4. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Sussman J. L. pi turn is a conformational pattern in RNA loops and bends. Nature. 1976 Apr 15;260(5552):645–646. doi: 10.1038/260645a0. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Irie T., Yoshida M., Takeishi K., Ukita T. The primary structure of yeast glutamic acid tRNA specific to the GAA codon. Biochim Biophys Acta. 1974 Oct 11;366(2):168–181. doi: 10.1016/0005-2787(74)90331-1. [DOI] [PubMed] [Google Scholar]
- Kyogoku Y., Lord R. C., Rich A. The effect of substituents on the hydrogen bonding of adenine and uracil derivatives. Proc Natl Acad Sci U S A. 1967 Feb;57(2):250–257. doi: 10.1073/pnas.57.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Atomic co-ordinates for yeast phenylalanine tRNA. Nucleic Acids Res. 1975 Sep;2(9):1629–1637. doi: 10.1093/nar/2.9.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. Aminoacyl-tRNA binding at the recognition site is the first step of the elongation cycle of protein synthesis. Proc Natl Acad Sci U S A. 1977 May;74(5):1903–1907. doi: 10.1073/pnas.74.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K., Torii K., Iitaka Y., Tsuboi M., Nishimura S. Crystal and molecular structure of the methyl ester of uridin-5-oxyacetic acid: a minor constituent of Escherichia coli tRNAs. FEBS Lett. 1974 Nov 15;48(2):279–282. doi: 10.1016/0014-5793(74)80486-2. [DOI] [PubMed] [Google Scholar]
- Pongs O., Reinwald E. Function of Y in codon-anticodon interaction of tRNA Phe . Biochem Biophys Res Commun. 1973 Jan 23;50(2):357–363. doi: 10.1016/0006-291x(73)90848-6. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Seeman N. C., Wang A. H., Suddath F. L., Rich A. Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res. 1975 Dec;2(12):2329–2341. doi: 10.1093/nar/2.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerath K., Chia L. S., Gupta R. C., Randerath E. Structural analysis of nonradioactive RNA by postlabeling: the primary structure of baker's yeast tRNA Leu/CUA. Biochem Biophys Res Commun. 1975 Mar 3;63(1):157–163. doi: 10.1016/s0006-291x(75)80024-6. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Carbon J. Molecular mechanism for missense suppression in E. coli. Nature. 1974 Aug 2;250(465):412–414. doi: 10.1038/250412a0. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Carbon J. Nucleotide sequence studies of normal and genetically altered glycine transfer ribonucleic acids from Escherichia coli. J Biol Chem. 1975 Jul 25;250(14):5530–5541. [PubMed] [Google Scholar]
- Squires C., Carbon J. Normal and mutant glycine transfer RNAs. Nat New Biol. 1971 Oct 27;233(43):274–277. doi: 10.1038/newbio233274a0. [DOI] [PubMed] [Google Scholar]
- Stout C. D., Mizuno H., Rubin J., Brennan T., Rao S. T., Sundaralingam M. Atomic coordinates and molecular conformation of yeast phenylalanyl tRNA. An independent investigation. Nucleic Acids Res. 1976 Apr;3(4):1111–1123. doi: 10.1093/nar/3.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Kim S. H. Idealized atomic coordinates of yeast phenylalanine transfer RNA. Biochem Biophys Res Commun. 1976 Jan 12;68(1):89–96. doi: 10.1016/0006-291x(76)90014-0. [DOI] [PubMed] [Google Scholar]
- Takemoto T., Takeishi K., Nishimura S., Ukita T. Transfer of valine into rabbit haemoglobin from various isoaccepting species of valyl-tRNA differing in codon recognition. Eur J Biochem. 1973 Oct 18;38(3):489–496. doi: 10.1111/j.1432-1033.1973.tb03084.x. [DOI] [PubMed] [Google Scholar]
- Tumaitis T. D., Lane B. G. Differential labelling of the carboxymethyl and methyl substituents of 5-carboxymethyluridine methyl ester, a trace nucleoside constituent of yeast transfer RNA. Biochim Biophys Acta. 1970 Dec 14;224(2):391–403. doi: 10.1016/0005-2787(70)90572-1. [DOI] [PubMed] [Google Scholar]
- Voet D., Rich A. The crystal structures of purines, pyrimidines and their intermolecular complexes. Prog Nucleic Acid Res Mol Biol. 1970;10:183–265. doi: 10.1016/s0079-6603(08)60565-6. [DOI] [PubMed] [Google Scholar]
- WYATT G. R., COHEN S. S. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953 Dec;55(5):774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Takeishi K., Ukita T. Anticodon structure of GAA-specific glutamic acid tRNA from yeast. Biochem Biophys Res Commun. 1970 Jun 5;39(5):852–857. doi: 10.1016/0006-291x(70)90401-8. [DOI] [PubMed] [Google Scholar]


