Abstract
NAC1 is a transcriptional co-repressor protein that is essential to sustain cancer cell proliferation and migration. However, the underlying molecular mechanisms of NAC1 function in cancer cells remain unknown. In this study, we show that NAC1 functions as an actin monomer binding protein. The conserved BTB protein interaction domain in NAC1 is the minimal region for actin binding. Disrupting NAC1 complex function by dominant negative or siRNA strategies reduced cell retraction and abscission during late stage cytokinesis, causing multinucleation in cancer cells. In Nac1-deficient murine fibroblasts, restoring NAC1 expression was sufficient to partially avert multinucleation. We found that siRNA-mediated silencing of the actin binding protein profilin-1 in cancer cells caused a similar multinucleation phenotype and that NAC1 modulated the binding of actin to profillin-1. Taken together, our results indicate that the NAC1/actin/profilin-1 complex is crucial for cancer cell cytokinesis, with a variety of important biological and clinical implications.
Keywords: actin, cytokinesis, NAC1, profilin-1
Introduction
The Bric-a-Brac Tramtrack Broad complex (BTB) family of proteins shares a conserved BTB protein-protein interaction domain (1, 2) that is present in many DNA and actin binding proteins (3). Nucleus accumbens-associated 1 (NAC1) encoded by NACC1 is a member of the family, and the gene was first identified by the observation that the level of NAC1 transcript was elevated in the rat nucleus accumbens in response to cocaine administration (4). Subsequently, NAC1 has been shown to participate in diverse biological functions ranging from maintenance of stemness and proliferation of embryonic stem cells (5, 6) to the pathogenesis of human cancer (7–12). Like other members of the BTB family, the homodimeric or heterodimeric interaction mediated by its BTB domain is thought to be essential for the various functional activities of NAC1 (7).
The biological role of NAC1 in cancer has recently emerged, as NAC1 has been found to be overexpressed in several types of human cancer including ovarian high-grade serous carcinoma, one of the most lethal neoplastic diseases in women (7, 8, 12–15). One of the mechanisms underlying NAC1 overexpression in ovarian cancer is amplification of the NAC1-encoded gene, NACC1, and analysis of The Cancer Genome Atlas (TCGA) data set demonstrates NACC1 as one of the top potential “driver” genes, showing the highest correlation between DNA and RNA copy number in ovarian high-grade serous carcinomas (11). Moreover, NAC1 upregulation is associated with disease aggressiveness and contributes to the development of chemoresistance (9, 10, 16). NAC1 is essential for survival and growth of ovarian cancer cells that overexpress NAC1, because NAC1-silencing or inactivation through ectopic expression of a deletion mutant containing only the BTB domain significantly reduced proliferation of ovarian cancer cells (7–10). Molecular mechanisms underlying the essential role of NAC1 in cancer cell survival are thought to involve multiple pathways including activation of the Gadd45 cell survival pathway (8, 9), fatty acid metabolism (17), and the HMGB-1 (High-mobility group protein B1) mediated autophagic response (16).
In addition, the possibility that NAC1 is also involved in non-nuclear functions is indicated by the dynamic changes in subcellular localization of NAC1 at different phases of cell cycle progression (18). In non-mitotic cells, NAC1 accumulated in distinct nuclear punctate bodies, which were dissolved into a diffuse pattern of distribution in the cytoplasm during mitosis. NAC1 nuclear bodies immediately reappeared on completion of mitosis (18), suggesting that NAC1 plays a role specifically during cell division. In this study we sought to investigate the role of NAC1 in cell division and to identify molecular mechanisms by which it contributes to the survival of tumor cells. Our results demonstrate that NAC1 is an actin monomer binding protein that regulates actin assembly in cancer cells during cytokinesis, a finding that provides a new perspective on the regulation of this fundamental biological process in tumor cells.
Materials and Methods
Traction force microscopy
The method for embedding 0.1 μm RFP (red fluorescent protein) methoxy fluorescent beads in the polymerized acrylamide substrate (0.1% methylene-bis-acrylamide and 5% acrylamide) was detailed in a previous report (19) and carried out on a glass bottom cell culture dish. A collagen layer was overlaid before SKOV3 N130 tTA cells were seeded in the dish at a suitable density for time-lapse imaging. An image frame was collected once every 10 minutes to minimize the deleterious effects related to the excitation beam, and to construct time-lapse videos for frame-by-frame measurements of the displacement of the beads. Photomicrographs were taken at 40x magnification to ensure sufficient resolution for resolving miniscule bead displacement using the ‘Tracking’ function in the Nikon Elements software. A total of 80 beads were tracked over the 3 conditions. The comparison of traction generated by the cells was accomplished by measuring maximal displacement of beads from origin along the x and y axis over time and plotting the data as a distribution.
Measurement of the contractile ring diameter
N130-induced and non-induced cells were fixed and stained for actin and DAPI. Dividing cells were randomly selected (n=20 for each condition) and the diameter of their contractile ring was directly measured using the ImageJ software (National Institutes of Health, Bethesda, Maryland).
Plotting of cell displacement trajectories
SKOV3 and HeLa cells seeded in collagen-coated plates were imaged at low magnification every 10 minutes overnight. Using the image recognition software (Metamorph/Metavue) to accomplish tracking of the displacement of individual cells, the x and y coordinates of each cell on each frame were recorded. Stage drift of the moving stage microscope was accounted for in the calculations. Trajectories of individual cells after lentiviral shRNA treatments were plotted after Day 3 and Day 6.
Establishment of primary cell cultures
To establish a primary culture of mouse lung fibroblast in the NAC1 knockout genotype, lung tissue specimens were minced, digested using collagenase (10 μg/ml) at 37°C on a rotisserie rotator, and sieved to isolate single cells. Cells were cultured in RPMI 1640 supplemented with 10% (v/v) FBS and penicillin/streptomycin (Invitrogen).
To establish a primary culture of mouse ovarian surface epithelial cells (mOSE), we harvested the ovaries from female mice of about 10 weeks of age and the mOSE cells were dissociated from the ovarian surface after incubating with 0.25% trypsin-EDTA at 37°C for 30 minutes followed by gentle agitation. The mOSE cells were washed and plated in Dulbecco’s modified Eagle’s medium-F12 media (10% Fetal bovine serum, 5% insulin-transferrin-selenium-A and 5% penicillin streptomycin) in a culture dish. Fresh media was replaced every 2–3 days and the cells were harvested at subconfluence for experimental purposes.
Generation of binding isotherm of NAC1/N130/Profilin-1 and actin
The construction of the binding isotherm was carried out (20) with actin at fixed low concentrations in supernatant, and using increasing amounts of NAC1 immobilized on anti-V5 beads to overcome the limited solubility of NAC1 in aqueous condition. The amount of actin in solution was quantified by high resolution scanning and processing by band densitometric analysis using the BioRad ChemiDoc XRS QuantityOne software in comparison to a standard curve based on actin input and silver staining.
Cell lines and culture conditions
The cell lines used in this study, including SKOV3, HeLa, MCF7, TOV21G and HEK293T, were purchased from ATCC (Rockville, MD, USA), authenticated with short tandem repeat (STR) based genotyping by the Fragment Analysis Facility at the Johns Hopkins University, and matched to their profiles at the ATCC STR profile database for human cell lines. Transfection was performed using the Lipofectamine-2000 reagent (Invitrogen). Nocodazole (100 ng/ml) was used to synchronize the cells at the mitotic phase. Nuclear and cytoplasmic purification of the lysates was carried out as described using the hypotonic swelling method; the purity of the nuclei were verified by microscopy (21).
Recombinant protein production
The NAC1-V5 construct was cloned into the pQE bacterial expression vector for recombinant protein production and purification using Ni-NTA beads (Qiagen). The insoluble protein was refolded using the Pierce protein refolding kit buffer 3 (Pierce) and immobilized on anti-V5 agarose beads (Sigma). Purified recombinant actin and profilin-1 were purchased from Cytoskeleton Inc. (Denver, CO).
Results
NAC1 inactivation or silencing causes a multinucleation phenotype in cancer cells
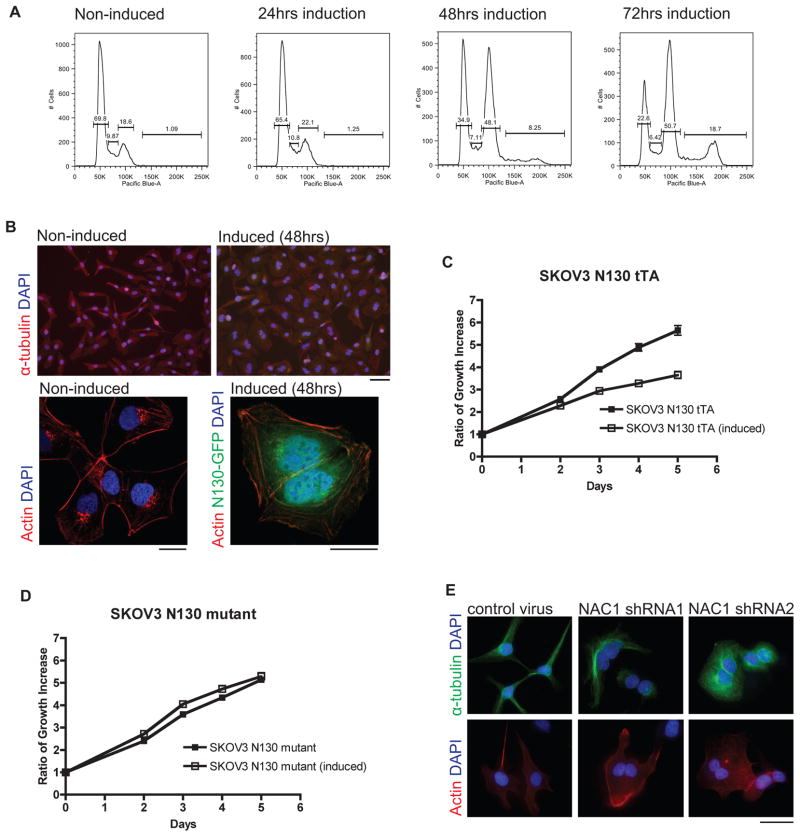
In order to document the biological roles of NAC1 during cancer cell division, we used two independent and complementary approaches. First, we ectopically expressed a truncated NAC1 construct, N130, in SKOV3 human ovarian cancer cells and HeLa cells to competitively inhibit the functional activity of wild type NAC1. The N130 construct encodes exclusively the BTB domain, which is the minimal motif required for the homo-dimerization of NAC1; expression of N130 has been shown to competitively disrupt NAC1 homodimerization (7). Following induction of N130 expression in SKOV3, an accumulation of polyploid cells at 4n was observed at 48 hours. After 72 hours, another distinct population of polyploid cells at 8n was detected (Fig. 1A). By direct visualization using immunofluorescence staining, we found that the increase in DNA content corresponded to the increase in the number of multinucleated cells (Fig. 1B). The induction of the BTB domain in HeLa cells caused a similar multinucleation phenotype (Supplementary Fig. S1A and S1B). The multinucleation induced by N130 induction was also accompanied by a significant reduction in cellular proliferation in SKOV3 cells (Fig. 1C).
Figure 1. NAC1 inactivation or knockdown in ovarian cancer cells causes a multinucleation phenotype.
A. Flow cytometric analysis shows that induced expression of a NAC1 deletion mutant (N130) containing the BTB domain after 48 hours in SKOV3 cells causes an accumulation of polyploid cells at 4n. After 72 hours, another distinct population of polyploid cells at 8n can be observed. B. Immunofluorescence microscopy demonstrates polyploid cells with two nuclei, which accounts for the doubling of DNA content in SKOV3 cells at 48 hours. Specimens were counterstained with DAPI for the nucleus. Top set: Scale bar, 50 μM. Bottom set: Images obtained using confocal microscope and 40x objective. Scale bar, 25 μM. C. Induction of N130 (BTB domain only) results in growth suppression. D. Induction of N130mut fails to reduce cellular proliferation. E. Silencing of NAC1 by lentiviral shRNA in SKOV3 cells leads to a multinucleated phenotype. Scale bar, 50 μM.
To control for potential non-specific effects related to N130 expression, we ectopically expressed N130mut in SKOV3 cells. N130mut is a mutant form of N130 containing multiple introduced point mutations that prevent it from forming dimers with full length NAC1 (9). The results demonstrated that, as compared to the wild type N130, the expression of N130mut neither caused a significant reduction in cellular proliferation (Fig. 1D) nor induced multinucleation. We also performed RNAi to determine the specificity of N130-induced phenotypes. NAC1 expression was silenced using lentiviral shRNAs (Supplementary Fig. S2A), and like the effects of N130 induction, silencing of NAC1 in SKOV3 and HeLa cells caused multinucleation in cancer cells (SKOV3: Fig. 1E; HeLa: Supplementary Fig. S2B, S2C). We also found that shRNAs targeting NAC1 suppressed cellular proliferation in SKOV3, HeLa, and MCF7 cancer cells (Supplementary Fig. S2D), all of which express abundant NAC1 (Supplementary Fig. S2E), whereas NAC1 shRNA expression in the TOV21G tumor cell line, which has undetectable NAC1 expression (10), had a less conspicuous effect.
The multinucleation phenotype of cancer cells is caused by abscission failure during cytokinesis
Next, we applied time-lapse microscopy to observe the formation of multinucleation in NAC1-inactivated (N130-induced) cancer cells. As shown in the Supplemental videos, NAC1-inactivated cells (with N130-induction, Video S1) and control cells (without N130-induction, Video S2) underwent normal mitosis and displayed equatorial furrowing. However, in the later phases of cell division, NAC1-inactivated cells were characterized by incomplete constriction and abscission of the contractile ring, and as a result, many of the dividing cells became multinucleated (Video S1). This finding was recapitulated in time-lapse videos of dividing SKOV3 and HeLa cells after treatment with lentiviral NAC1 shRNAs (HeLa: Supplementary Fig. S3; SKOV3: data not shown). Whereas cells treated with control shRNA (Supplementary Fig. S3A) completed cytokinesis and formed two distinct daughter cells, NAC1 shRNA treated cells failed to complete the final stage of cytokinesis and became multinucleated (Supplementary Fig. S3B).
Failure of abscission during cytokinesis of cancer cells is a consequence of a loss in traction of dividing cells, impeded constriction of their contractile rings, and reduced migration potential
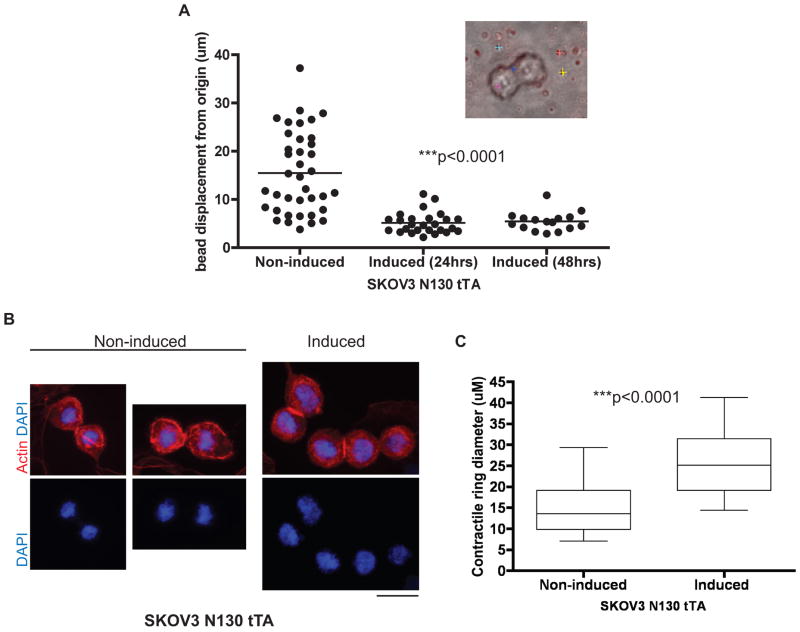
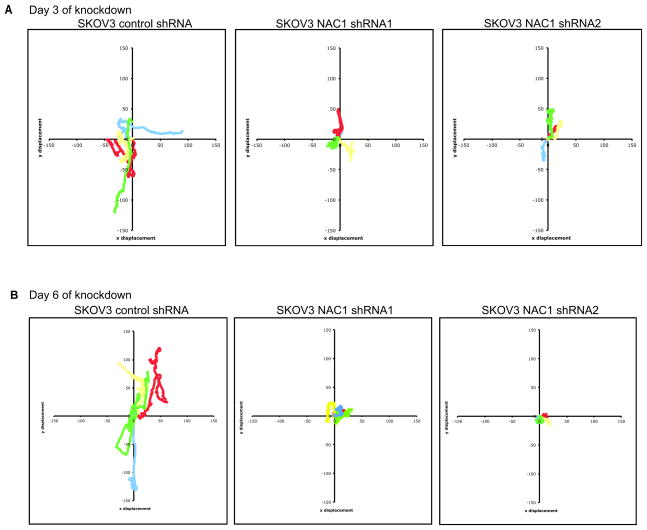
We hypothesize that the defect in late cytokinesis was a result of insufficient traction of the dividing cells to facilitate cellular division. To determine the traction generated by dividing cells, we used traction force microscopy to correlate the traction generated by dividing cells with the displacement of fluorescent beads embedded directly beneath the cells in a polyacrylamide substrate. Analysis of the displacement of each bead from its original location during the course of cell division revealed a significant decrease in the maximal displacement of beads by SKOV3 cells after NAC1 inactivation (Fig. 2A), suggesting that NAC1 proteins were important for the generation of traction in the dividing cancer cells. Using immunofluorescence microscopy to visualize actin structures, we compared the size of contractile rings between dividing N130-induced and non-induced cells. Although N130-induced cells correctly positioned the cleavage furrows and contractile rings, these cells displayed a larger average diameter of contractile rings than their non-induced counterparts, suggesting that the constriction of contractile rings was compromised in N130-induced cells (image: Fig. 2B, quantification: Fig. 2C). To determine if there was a loss in migration potential of the cells after silencing of NAC1 (Day 3 and Day 6), live-cell imaging was used to follow the cell displacement trajectories during an overnight observation. After Day 3 and Day 6 under control shRNA and NAC1 shRNA treatment, typical trajectories (Red, yellow, green, blue) of SKOV3 and HeLa individual cells were plotted by their x and y coordinates (Fig. 3 and Supplementary Fig. S4) and revealed that the silencing of NAC1 caused SKOV3 and HeLa cells to undergo restricted migration and greatly reduced displacement from their original location. As early as three days after NAC1 silencing, both SKOV3 and HeLa cells demonstrated restricted migration and reduced displacement from origin (Fig. 3A and Supplementary Fig. S4A). The silencing of NAC1 was sustained for as long as 6 days after shRNA treatment (Supplementary Fig. S4C). We demonstrated that the reduction in migration and displacement of NAC1 silenced cells was also observed at Day 6 of shRNA treatment in SKOV3 and HeLa cells (Fig. 3B and Supplementary Fig. S4B).
Figure 2. Biophysical effects of NAC1 inactivation or silencing include loss of cellular traction during division, impeded constriction of contractile ring, and reduced displacement from origin over time.
A. Traction force microscopy shows a significant reduction in cellular traction during cytokinesis when N130 (BTB domain only) was induced. A total of 80 beads were tracked in the experiment. ***p< 0.0001 using one-way ANOVA test. Inset: A representative cell undergoing mitosis on collagen-coated bis-polyacrylamide substrate with red fluorescent beads embedded underneath and tracked beads denoted by colored cross hairs. B. Immunofluorescence staining of actin structures in non-induced vs. induced SKOV3 N130 tTA cells enabling evaluation of the contractile ring diameter. Scale bar, 50 μM. C. Induction of N130 (BTB domain only) results in a reduction of average contractile ring diameter as compared to the non-induced SKOV3 cells during cell division. The contractile ring diameters of dividing cells between the two conditions (n=20 each) were measured. Error bars denote standard deviation. ***p<0.0001 using unpaired t-test.
Figure 3. Reduced migration and displacement of SKOV3 cells after NAC1 silencing.
Typical trajectories of individual SKOV3 cells after control lentiviral shRNA and NAC1 shRNA treatment after A. Day 3 and B. Day 6 denoted by red, yellow, green and blue tracks. Each color denotes the trajectory of a single cell.
Expression of NAC1 partially rescues normal cytokinesis in Nac1-null cells
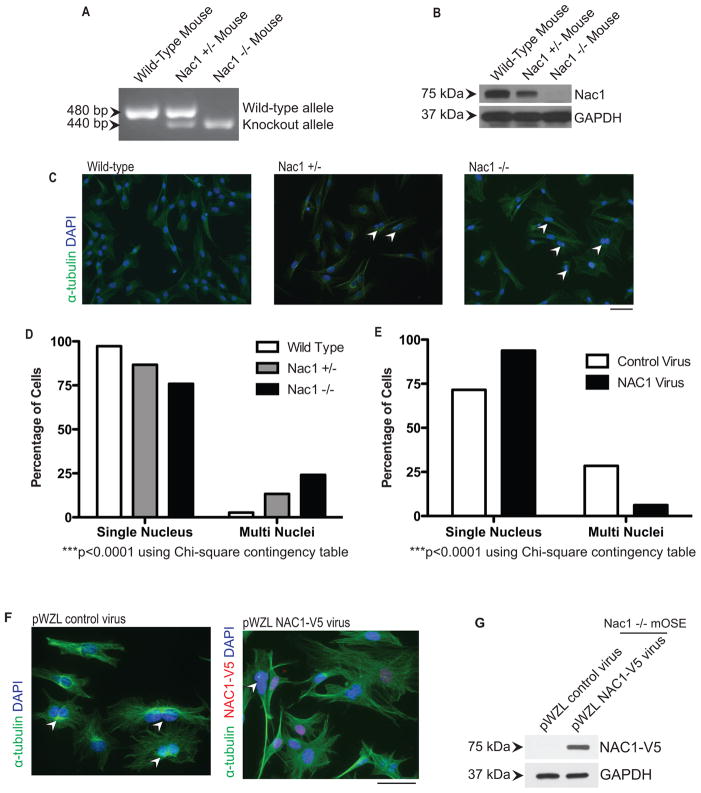
To determine if NAC1 was essential for effective cytokinesis, we re-introduced NAC1 expression in Nac1-null cells to determine whether ectopic NAC1 would prevent the multinucleated phenotype. For this purpose, we utilized a Nac1 homozygous knockout (Nac1−/−) mouse in which both copies of the BTB domain of the mouse Nacc1 gene were deleted (Fig. 4A). In addition, we bred Nac1 hemizygous knockout mice (Nac1+/−), and the cells isolated from those mice were analyzed in parallel. Western blot analysis demonstrated that Nac1−/− lung fibroblasts did not express a detectable level of Nac1 protein while lung fibroblasts from the Nac1+/− mice expressed detectable, albeit much less, Nac1 compared to cells isolated from wild type Nac1+/+ mice (Fig. 4B). More importantly, we observed that the percentage of multinucleated cells (2.7%) in Nac1+/+ (n= 930) was much lower than in Nac1−/− cells (23.7%) (n=1011), while heterozygous Nac1 cells (Nac1+/−) (n=1040) demonstrated an intermediate percentage (12.7%) of multinucleated cells (Fig. 4C and 4D).
Figure 4. Expression of NAC1 partially rescues multinucleation in Nac1−/− cells.
A. DNA gel electrophoresis validates the two knockout alleles (440bp) in Nac1−/− mice and one knockout and one wild type allele (480bp) in Nac1+/− mice. B. Western blot analysis shows differential protein expression levels of NAC1 in lung fibroblasts isolated from the respective genotypes. C. Immunofluorescence staining of alpha-tubulin facilitates the quantification of cells exhibiting multinucleation. Cells were counterstained with DAPI to reveal nuclei. Arrowheads indicate multinucleated cells. Scale bar, 50 μM. D. Number of multinucleated cells was presented as a percentage in primary lung fibroblast cultures established from different Nac1 genotype mice. There is a significant difference in the proportion of multinucleated cells between different Nac1 genotypes (***p< 0.0001, Chi-square contingency table). E. NAC1 was ectopically expressed in Nac1−/− cells, and its expression was associated with a significant decrease in the percentage of multinucleated cells as compared to control virus-treated group (***p< 0.0001, Chi-square contingency table). F. Double immunofluorescence staining of alpha-tubulin and V5 (to detect NAC1-V5) facilitates the quantification of cells exhibiting multinucleation after expression of NAC1. Cells were counterstained with DAPI to reveal nuclei. Arrowheads denote multinucleated cells. Scale bar, 50 μM. G. Western blot verified the expression of NAC1-V5 after viral transduction.
To determine if the re-expression of Nac1 protein could rescue the multinucleation phenotype, NAC1 was re-introduced into Nac1−/− cells as a V5-NAC1 protein by a retroviral vector. We then quantified the number of multinucleated cells in cultures that were either treated with control virus (n=329) or with the NAC1 expression construct virus (n=304). We found that there was a significant reduction (28.3% to 6.25%) in the percentage of multinucleated cells after the introduction of the NAC1 expression vector (Fig. 4E). The successful introduction of NAC1 expression was confirmed by immunofluorescence detection of the V5 epitope (Fig. 4F) and by Western blot (Fig. 4G). A similar result could be obtained in mouse ovarian surface epithelial (mOSE) cells generated from the same strain of mice (Supplementary Fig. S5).
Cancer cells after profilin-1 silencing exhibit a similar multinucleation phenotype
The multinucleation phenotype observed in NAC1 inactivated or silenced cells is reminiscent of multinucleation reported in mouse chondrocytes after silencing of profilin-1 (19). In the current study, by silencing profilin-1 expression (Supplementary Fig. S6A), we were able to demonstrate the formation of multinucleated cells in SKOV3 cells (Fig. 5A) and HeLa cells (Supplementary Fig. S6B and S6C). The silencing of profilin-1 also caused a reduction in cellular proliferation in our previously examined panel of cancer cell lines, MCF7, HeLa, SKOV3 and TOV21G (Supplementary Fig. S6D). Profilin-1 is a ubiquitous actin monomer binding protein that is known to regulate actin polymerization during cytokinesis (22–24). The similarity of the phenotype observed in chondrocytes and tumor cells after profilin-1 silencing prompted us to investigate the possibility that NAC1 has a biological role in cytokinesis involving profilin-1/actin complexes. Given that the known role of NAC1 involved transcription regulation, it is possible that the multinucleated phenotype induced by NAC1 knockdown was mediated by down-regulation of profilin-1 expression. To test this possibility, we employed Western blot analysis but did not observe significant changes in the levels of profilin-1 protein expression between cells treated with NAC1 shRNA and control shRNA (Supplementary Fig. S2A: right panel), suggesting that the phenotype generated by the knockdown of NAC1 was not due to reduced profilin-1 levels.
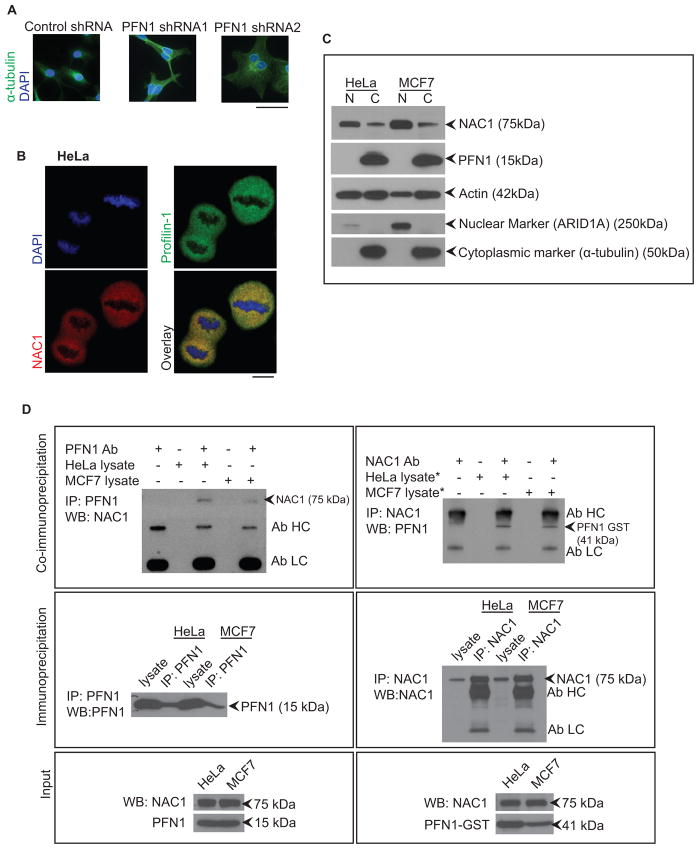
Figure 5. Silencing of profilin-1 causes a similar multinucleation phenotype, while NAC1 and profilin-1 can be isolated in a single protein complex.
A. Silencing of profilin-1 using lentiviral shRNA causes a similar multinucleation phenotype as NAC1 silencing in SKOV3 cells. Cells were counterstained with DAPI. Scale bar, 50 μM. B. NAC1 and profilin-1 co-localize in the same cellular compartment during cell division. Images were obtained using confocal microscopy. Scale bar, 10 μM. C. Western blot analysis of proteins localized in nuclear and cytoplasmic compartments of interphase cells (data are from unsynchronized cells). D. Reciprocal protein co-immunoprecipitation experiments showed that NAC1 complexed with profilin-1 in HeLa and MCF7 cells expressing abundant NAC1. *asterisk denotes lysates that were transfected with the profilin-1-GST expression construct.
Next, using immunofluorescence microscopy we found that NAC1 and profilin-1 co-localized during cell division in HeLa cells (Fig. 5B). Nuclear and cytoplasmic protein extracts from HeLa and MCF7 cells were used for Western blot analysis to analyze the distribution of the proteins in the cellular compartments. Interestingly, NAC1 was mainly found in the nuclear extract during interphase, with low levels in the cytoplasm, while profilin-1 was almost exclusively detected in the cytoplasmic extract (Fig. 5C). When we mitotically arrested cells with the microtubule-depolymerizing drug, nocodazole, and examined NAC1 levels by Western blot in the cytoplasmic fraction, we found that there was a significant amount of NAC1 in the cytoplasmic fraction as compared to the chromosomal fraction (Supplementary Fig. S6E). This cell cycle dependent localization of NAC1 was previously described in an earlier publication (18). Thus, during cell division NAC1 abundantly accumulated in the cytoplasm where it may colocalize with profilin-1.
Accordingly, co-immunoprecipitation experiments were conducted to determine whether NAC1 and profilin-1 interacted in vivo. We found that endogenous NAC1 and profilin-1 could be co-immunoprecipitated from lysates prepared from HeLa and MCF7 cells (Fig. 5D). Both HeLa and MCF7 cells have abundant NAC1 expression as previously documented (Supplementary Fig. S2E). Reciprocal co-immunoprecipitation experiments using anti-profilin-1 for pull-down and anti-NAC1 for Western blot detection confirmed that NAC1 and profilin-1 were constituents of the same protein complexes.
NAC1 interacts directly with actin monomers
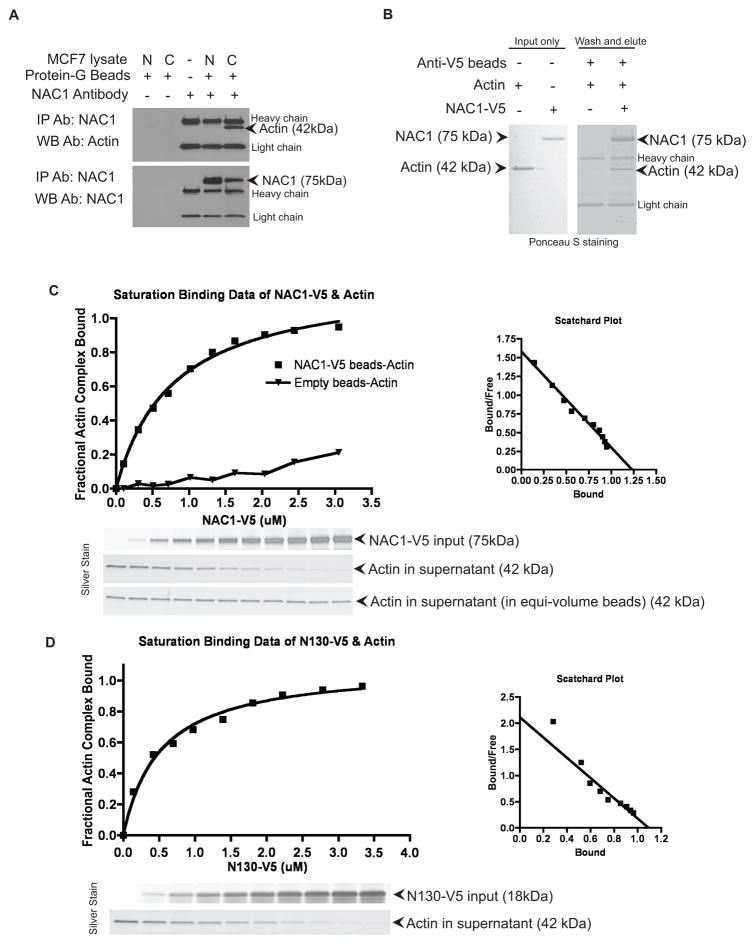
As profilin-1 is an actin binding protein and plays a pivotal role in actin polymerization (23), we investigated the possibility that NAC1 also interacted with actin. Co-immunoprecipitation experiments using cytoplasmic extracts of MCF7 cells demonstrated that NAC1 co-immunoprecipitated with actin monomers in cytoplasmic but not in nuclear lysates (Fig. 6A). To further determine if the NAC1-actin interaction was direct or depended on profilin-1, we performed an in vitro binding assay using NAC1-V5, profilin-1, and actin recombinant proteins. After incubating NAC1-V5 and actin, we observed that actin could be co-eluted with NAC1 on anti-V5 agarose beads. In the absence of NAC1-V5 protein, however, actin was not retained on the anti-V5 beads and was undetectable in the eluted protein fraction (Fig. 6B). We also conducted quantitative binding assays using a method previously described (20) to characterize the protein interactions (actin standard curve documented in Supplementary Fig. S7A). A fixed amount of actin or profilin-1 protein was incubated with increasing amounts of NAC1-V5-beads. After incubation, the amount of unbound actin or profilin-1 in the supernatant was measured and used to determine the bound fraction. We found that recombinant NAC1 was unable to demonstrate a dose-dependent co-binding of profilin-1 in the binding affinity assay (Supplementary Fig. S7B). On the other hand, we were able to demonstrate the dose-dependent binding of NAC1 and actin in the binding affinity assay (Fig. 6C). Taken together, these results indicated a direct protein-protein interaction between NAC1 and actin, but not between NAC1 and profilin-1, although in vivo co-immunoprecipitation reactions (Fig. 5D) demonstrated NAC1 and profilin-1 are in the same protein complexes.
Figure 6. NAC1 directly binds to actin monomers.
A. NAC1 co-immunoprecipitates with actin monomers in cytoplasmic extract (C) but not in nuclear extract (N) prepared from MCF7 cells. IP, immunoprecipitation; WB, Western blot. Ab, antibody. B. Recombinant protein co-immunoprecipitation confirmed that NAC1 and actin interactions are direct. Recombinant actin co-immunoprecipitated with NAC1-V5 that was immobilized on anti-V5 agarose beads. Actin protein was unable to be retained in the co-immunoprecipitation reaction in the absence of NAC1-V5. C. Analysis of NAC1-V5, and D. analysis of N130-V5 and actin binding affinity using a quantitative anti-V5 pull-down assay. NAC1-V5 and N130-V5 bound to actin in a saturable dose-dependent manner, Kd ~0.78 μM and ~0.52 μM, respectively. Scatchard plots of the binding isotherms were derived.
The quantitative binding assays performed found that NAC1-V5 and actin interacted with a Kd of 0.78 μM (Fig. 6C). As a negative control, an irrelevant protein of similar molecular weight as NAC1, Notch3-ICD-V5 was analyzed in parallel to demonstrate the assay specificity (Supplementary Fig. S7C). To determine the domain of the NAC1 protein that was involved in the direct interaction with actin, we generated another recombinant NAC1 protein composed of only the BTB domain (N130-V5). We carried out a similar quantitative binding assay between N130-V5 and actin. These proteins bound with a Kd of 0.52 μM (Fig. 6D). To further support the finding that the BTB domain of NAC1 was responsible for its interaction with the actin, we used a series of NAC1 deletion mutants (Supplementary Fig. S8A), and were able to confirm that the BTB domain of NAC1 (Supplementary Fig. S8C1) was responsible for its interaction with the profilin-1/actin complex.
NAC1 is able to modulate binding between profilin-1 and actin monomers
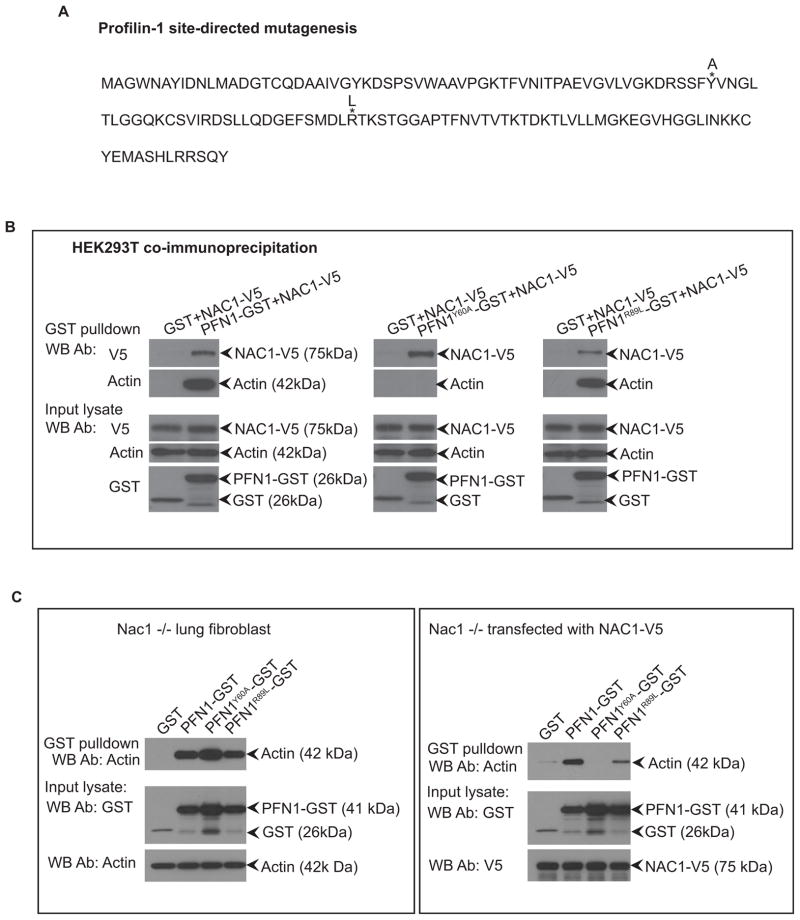
Because the interaction between actin and profilin-1 is known to be critical for cytokinesis (22), our results suggested that the interaction between NAC1 and actin could modulate the binding between profilin-1 and actin. We employed Nac1−/− lung fibroblasts as a cellular system to test this hypothesis. Due to the relatively high binding affinity between actin and profilin-1, we created a profilin-1 mutant, PFN1Y60A, which has been demonstrated to exhibit reduced actin binding activity (25). We also generated PFN1R89L, a control that does not affect the binding between profilin-1 and actin (26) (Fig. 7A). As verified by our protein co-immunoprecipitation experiments, PFN1Y60A exhibited diminished binding to actin while mutant PFN1R89L did not (Fig. 7B). We expressed wild type and mutant profilin-1 in Nac1−/− lung fibroblasts previously transfected with a NAC1-V5 expression vector or an empty vector. We observed that in the presence of NAC1-V5, the actin binding activity of profilin-1 decreased, particularly for PFN1Y60A in which actin-binding activity was totally abrogated (Fig. 7C: right panel). Conversely, in fibroblasts without NAC1 expression, PFN1Y60A mutant co-immunoprecipitated with actin (Fig. 7C: left panel), suggesting that the presence of NAC1 inhibited the interaction between profilin-1 and actin.
Figure 7. Protein interactions between profilin-1 and actin can be modulated by the presence of NAC1.
A. Mutant profilin-1 was generated with amino acid changes denoted by asterisk* and the annotated replacement amino acids. B. HEK293T cells were transfected with NAC1-V5 and profilin-1-GST constructs, and the lysates were collected for immunoprecipitation of the GST proteins and immunoblotted for either NAC1 or actin. PFN1Y60A mutant significantly reduces the binding of profilin-1 to actin. C. Interaction of profilin-1 and actin could be modulated by the presence of NAC1. Nac1−/− lung fibroblast cells were co-transfected with the profilin-1-GST constructs in the presence or absence of the NAC1 expression plasmid. Cell lysates were immunoprecipitated for the profilin-1-GST proteins using glutathione-agarose beads and immunoblotted with an anti-actin antibody. Presence of NAC1 protein reduces the binding of profilin-1 and actin, especially for PFN1Y60A mutant where actin binding is almost completely abrogated.
Discussion
The results from the current study unveil a novel mechanism by which cancer cells facilitate their incessant cellular divisions, which is the cardinal feature of neoplastic diseases. Our results indicate that abundant NAC1 proteins in cancer cells promote cytokinesis through modulating the interaction of actin and profilin-1, ensuring an effective completion of cellular division in highly proliferative tumors. The observation that NAC1 overexpressing tumor cells develop molecular dependence on NAC1 protein (7–9) promises future exploitation by targeting NAC1 as a cytostatic cancer therapy aimed at compromising cytokinesis in cancer cells.
Cytokinesis is a highly complex event involving a cascade of sequential and highly coordinated molecular processes (27). In eukaryotic cells, this is made possible by chromosomal segregation and Rho GTPase signaling followed by the effective assembly and constriction of contractile rings by non-muscle myosin II, actin, and its binding partners before actual cellular division occurs (27–29). For example, disruption of Rho GTPase signaling, which is known to regulate the actin cytoskeleton, can result in the misalignment of the cleavage furrow and contractile ring (30). Cytokinesis failure can also result from the slow ring closure of the contractile apparatus due to inadequate constriction, leading to a cytoplasmic connection between daughter cells that precedes furrow regression (31). As shown in this study, inactivation or silencing of NAC1 did not cause detectable aberrations in early cytokinetic events such as chromosome separation or formation and positioning of cleavage furrow and contractile ring; however, NAC1 inactivation or silencing resulted in the inability of the dividing cells to generate the force required to constrict the contractile actomyosin ring or to generate sufficient traction to move the dividing cells apart to make the “final cut”, impeding abscission and resulting in incomplete partitioning of the cytoplasm of the cell (29). This finding is consistent with previous studies demonstrating that traction force is required for complete cytokinesis in dividing cells in Dictyostelium and mouse chondrocytes (19, 27).
The actin binding property of NAC1 and its ability to modulate the interaction between profilin-1 and actin may account for the defective cytokinesis observed in tumor cells after NAC1 inactivation or knockdown. Since actin structures have been shown to be important for the regulation of cell shape and movement (Pollard and Cooper, 2009), the demonstration that NAC1 is an actin binding protein could also explain previously reported findings that reducing NAC1 expression by RNA interference decreased cellular motility (10, 32). Apropos to these observations, it has been well established that profilin-1, which is a ubiquitous actin monomer-binding protein, regulates actin polymerization in response to various extracellular signals by catalyzing ADP-to-ATP exchange on G-actin and escorting actin monomers to the growing barbed ends of actin filaments (23, 24). Profilin-1 has been thought to be a key component of the core cytokinesis machinery (19, 22, 33). The fact that NAC1 and profilin-1 co-exist in the same actin complex and that the same phenotype, i.e., incomplete cytokinesis followed by multinucleation, is produced by knockdown of either gene argues that NAC1 plays a role similar to profilin-1 in regulating actin polymerization. Although NAC1 may finely tune the function of profilin-1 in regulating actin dynamics, it remains unclear how NAC1 directly affects actin assembly independent of profilin-1.
Our results demonstrate that NAC1 represents a rate-limiting factor in cytokinesis in cancer cells; that NAC1 gene amplification and upregulation provides tumor cells with a sufficient amount of NAC1 for sustaining numerous cytokinesis events, but results in the development of molecular dependence on NAC1. Thus, inactivation or knockdown of NAC1 would not allow completion of cytokinesis in those cancer cells. This indicates the possibility of targeting NAC1 in cancer cells for maintenance cancer therapy. This is important because the conventional cytotoxic chemotherapy is always complicated by its cytotoxic effects on normal cells Unlike conventional cytotoxic drugs, NAC1-targeted anti-tumor strategies involving pharmacological compounds that disrupt protein-protein interactions between the well-characterized BTB domains of NAC1 (34) and actin are likely to have minimal cytotoxicity for normal cells because NAC1 is preferentially overexpressed only in cancer cells.
This finding is of great interest in light of the lack of actin-targeting agents in the arsenal of anti-cancer drugs, a consequence of the fact that the known reagents target the actin assembly in both cancer and normal cells (35). That disrupting NAC1 BTB domain protein interactions as a cancer chemotherapeutic strategy is feasible is suggested by the use of peptidomimetic and small molecule inhibitors to disrupt the BTB interacting domain of BCL6, which has been shown to be effective in suppressing tumor growth in B cell lymphomas (36–38). Triggering cytokinesis failure in cancer cells could give rise to either chromosomal instability or apoptosis. However, it is expected that in the presence of the functional p53, cells that were treated with drugs blocking cytokinesis will acquire extreme polyploidy and undergo rapid cell death or senescence (31, 39).
In summary, our findings provide cogent evidence that NAC1 is a member of the actin-binding protein family. The potential involvement of NAC1 in regulating actin organization during late stage cytokinesis represents a functional property well beyond its canonical function as a transcriptional regulator. By highlighting the participation of NAC1 in regulation of the actin assembly in cancer cells, our results should have profound implications on the study of actin biology in cancer, and should advance our basic understanding of the process of cytokinesis.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Bin Guan, Alexander Stoeck, Xu Chen, and Pei-Hsun Wu for helpful discussions.
Grant support
This study was supported by an NIH/NCI grants RO1CA103937, RO1CA129080, RO1CA148826, and U54CA143868.
Footnotes
The authors declare no conflict of interest
Author contributions
K.L.Y conceived experiments, analyzed data and co-wrote the manuscript. S.F. developed biophysical methods of analysis. M.M.T. initiated protein interaction studies for screening candidate genes. N.J. and K.N. built initial constructs and validated them in vitro. J.W. contributed the Nac1 knockout mouse. D.W. co-supervised the project. T.L.W and I.M.S co-supervised the project and co-wrote the manuscript.
References
- 1.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 2.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–77. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 3.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–8. [PubMed] [Google Scholar]
- 4.Cha XY, Pierce RC, Kalivas PW, Mackler SA. NAC-1, a rat brain mRNA, is increased in the nucleus accumbens three weeks after chronic cocaine self-administration. J Neurosci. 1997;17:6864–71. doi: 10.1523/JNEUROSCI.17-18-06864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–8. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 6.Ma T, Wang Z, Guo Y, Pei D. The C-terminal pentapeptide of Nanog tryptophan repeat domain interacts with Nac1 and regulates stem cell proliferation but not pluripotency. J Biol Chem. 2009;284:16071–81. doi: 10.1074/jbc.M109.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama K, Nakayama N, Davidson B, Sheu JJ, Jinawath N, Santillan A, et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proc Natl Acad Sci U S A. 2006;103:18739–44. doi: 10.1073/pnas.0604083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakayama K, Nakayama N, Wang TL, Shih IM. NAC-1 controls cell growth and survival by repressing transcription of Gadd45GIP1, a candidate tumor suppressor. Cancer Res. 2007;67:8058–64. doi: 10.1158/0008-5472.CAN-07-1357. [DOI] [PubMed] [Google Scholar]
- 9.Jinawath N, Vasoontara C, Yap KL, Thiaville MM, Nakayama K, Wang TL, et al. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene. 2009;28:1941–8. doi: 10.1038/onc.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama K, Rahman MT, Rahman M, Yeasmin S, Ishikawa M, Katagiri A, et al. Biological role and prognostic significance of NAC1 in ovarian cancer. Gynecol Oncol. 2010;119:469–78. doi: 10.1016/j.ygyno.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 11.Shih IM, Nakayama K, Wu G, Nakayama N, Zhang J, Wang TL. Amplification of the ch19p13. 2 NACC1 locus in ovarian high-grade serous carcinoma. Mod Pathol. 2011;24:638–45. doi: 10.1038/modpathol.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeasmin S, Nakayama K, Rahman MT, Rahman M, Ishikawa M, Katagiri A, et al. Biological and clinical significance of NAC1 expression in cervical carcinomas: a comparative study between squamous cell carcinomas and adenocarcinomas/adenosquamous carcinomas. Hum Pathol. 2011;43:506–19. doi: 10.1016/j.humpath.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Yeasmin S, Nakayama K, Ishibashi M, Katagiri A, Iida K, Purwana IN, et al. Expression of the bric-a-brac tramtrack broad complex protein NAC-1 in cervical carcinomas seems to correlate with poorer prognosis. Clin Cancer Res. 2008;14:1686–91. doi: 10.1158/1078-0432.CCR-07-4085. [DOI] [PubMed] [Google Scholar]
- 14.Ishibashi M, Nakayama K, Yeasmin S, Katagiri A, Iida K, Nakayama N, et al. Expression of a BTB/POZ protein, NAC1, is essential for the proliferation of normal cyclic endometrial glandular cells and is up-regulated by estrogen. Clin Cancer Res. 2009;15:804–11. doi: 10.1158/1078-0432.CCR-08-2134. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa M, Nakayama K, Yeasmin S, Katagiri A, Iida K, Nakayama N, et al. NAC1, a potential stem cell pluripotency factor expression in normal endometrium, endometrial hyperplasia and endometrial carcinoma. Int J Oncol. 2010;36:1097–103. doi: 10.3892/ijo_00000591. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Cheng Y, Ren X, Zhang L, Yap KL, Wu H, et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2012;31:1055–64. doi: 10.1038/onc.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueda SM, Yap KL, Davidson B, Tian Y, Murthy V, Wang TL, et al. Expression of Fatty Acid Synthase Depends on NAC1 and Is Associated with Recurrent Ovarian Serous Carcinomas. J Oncol. 2010;2010:285191. doi: 10.1155/2010/285191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu PH, Hung SH, Ren T, Shih Ie M, Tseng Y. Cell cycle-dependent alteration in NAC1 nuclear body dynamics and morphology. Phys Biol. 2011;8:015005. doi: 10.1088/1478-3975/8/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bottcher RT, Wiesner S, Braun A, Wimmer R, Berna A, Elad N, et al. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J. 2009;28:1157–69. doi: 10.1038/emboj.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21:4061–7. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoeck A, Shang L, Dempsey PJ. Sequential and gamma-secretase-dependent processing of the betacellulin precursor generates a palmitoylated intracellular-domain fragment that inhibits cell growth. J Cell Sci. 2010;123:2319–31. doi: 10.1242/jcs.060830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korenbaum E, Nordberg P, Bjorkegren-Sjogren C, Schutt CE, Lindberg U, Karlsson R. The role of profilin in actin polymerization and nucleotide exchange. Biochemistry. 1998;37:9274–83. doi: 10.1021/bi9803675. [DOI] [PubMed] [Google Scholar]
- 24.Paavilainen VO, Bertling E, Falck S, Lappalainen P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004;14:386–94. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Schluter K, Schleicher M, Jockusch BM. Effects of single amino acid substitutions in the actin-binding site on the biological activity of bovine profilin I. J Cell Sci. 1998;111 (Pt 22):3261–73. doi: 10.1242/jcs.111.22.3261. [DOI] [PubMed] [Google Scholar]
- 26.Wittenmayer N, Jandrig B, Rothkegel M, Schluter K, Arnold W, Haensch W, et al. Tumor suppressor activity of profilin requires a functional actin binding site. Mol Biol Cell. 2004;15:1600–8. doi: 10.1091/mbc.E03-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–6. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–9. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 29.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–60. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–8. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Lacroix B, Maddox AS. Cytokinesis, ploidy and aneuploidy. J Pathol. 2012;226:338–51. doi: 10.1002/path.3013. [DOI] [PubMed] [Google Scholar]
- 32.Tsunoda K, Oikawa H, Tada H, Tatemichi Y, Muraoka S, Miura S, et al. Nucleus accumbens-associated 1 contributes to cortactin deacetylation and augments the migration of melanoma cells. J Invest Dermatol. 2011;131:1710–9. doi: 10.1038/jid.2011.110. [DOI] [PubMed] [Google Scholar]
- 33.Edamatsu M, Hirono M, Watanabe Y. Tetrahymena profilin is localized in the division furrow. J Biochem. 1992;112:637–42. doi: 10.1093/oxfordjournals.jbchem.a123952. [DOI] [PubMed] [Google Scholar]
- 34.Stead MA, Carr SB, Wright SC. Structure of the human Nac1 POZ domain. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65:445–9. doi: 10.1107/S1744309109012214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer. 2011;11:177–87. doi: 10.1038/nrc3003. [DOI] [PubMed] [Google Scholar]
- 36.Polo JM, Dell’Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–35. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 37.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–11. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, et al. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113:3397–405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzywicka-Racka A, Sluder G. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J Cell Biol. 2011;194:199–207. doi: 10.1083/jcb.201101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.