Abstract
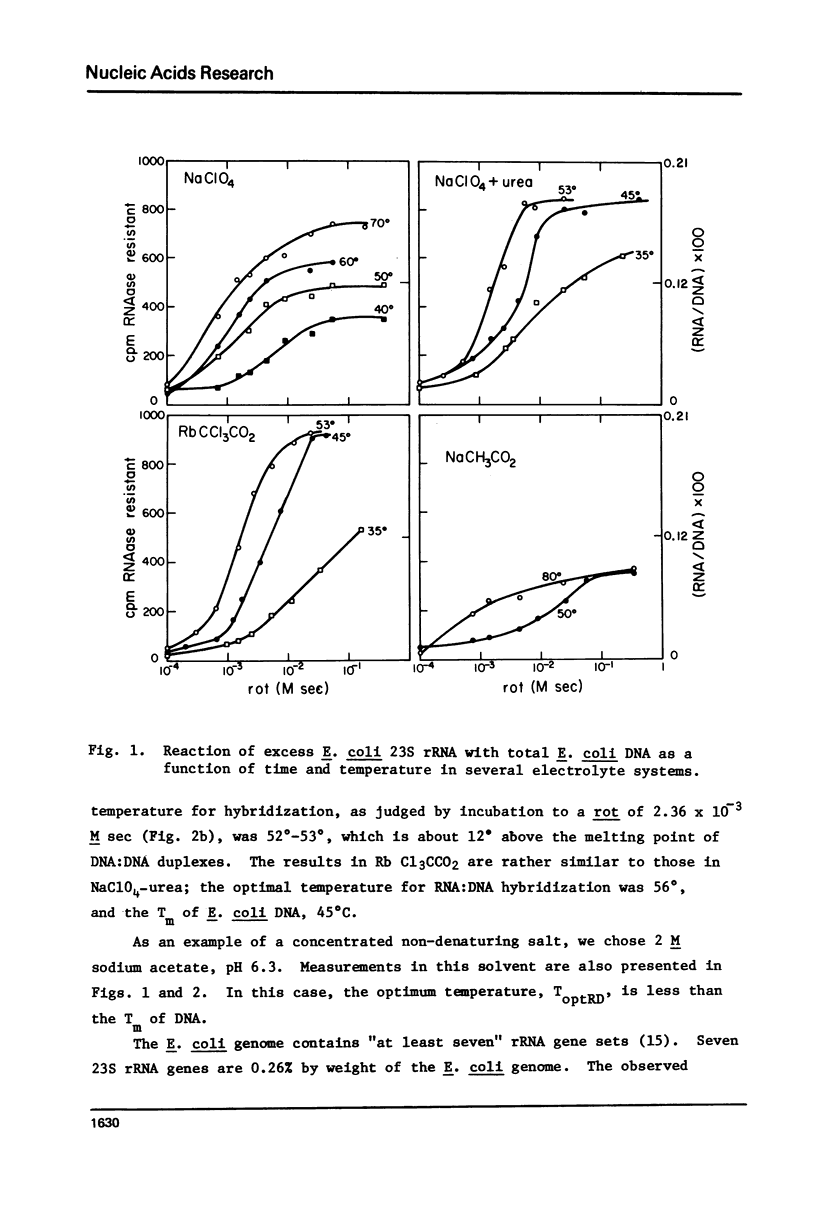
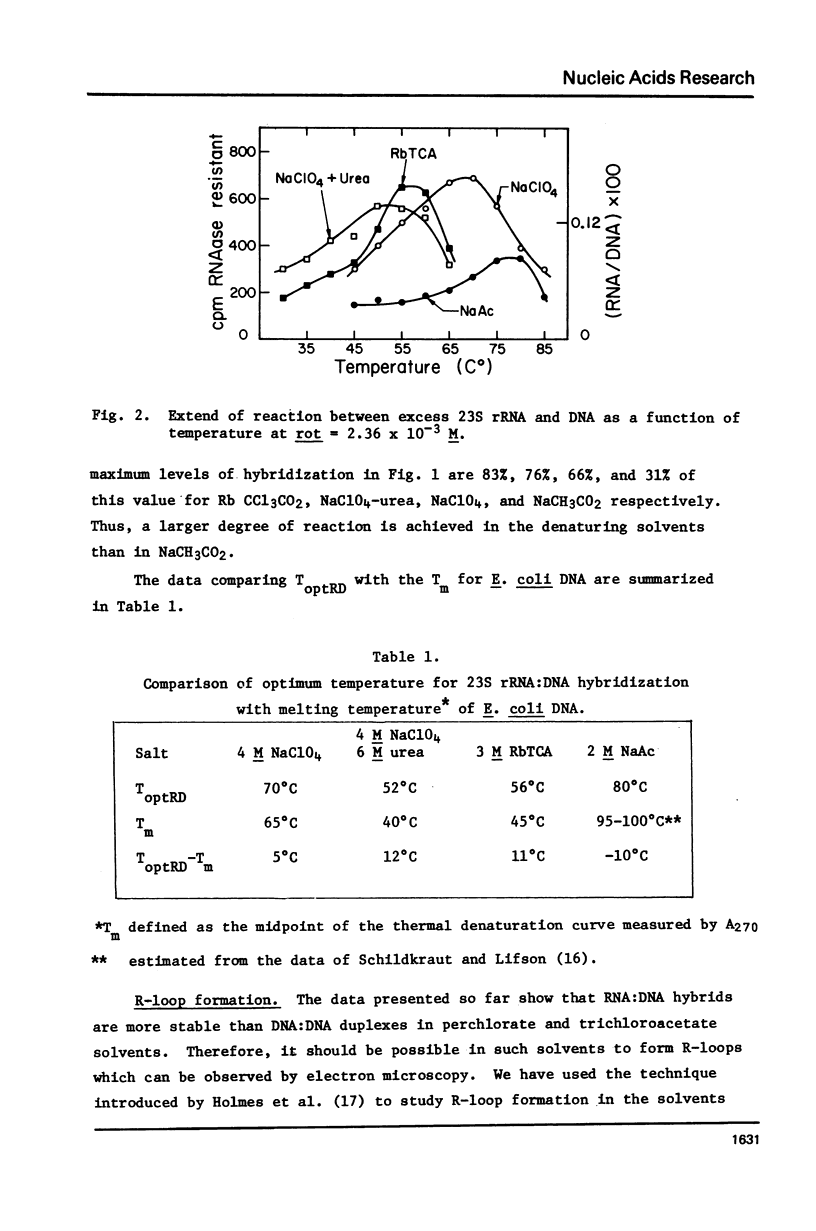
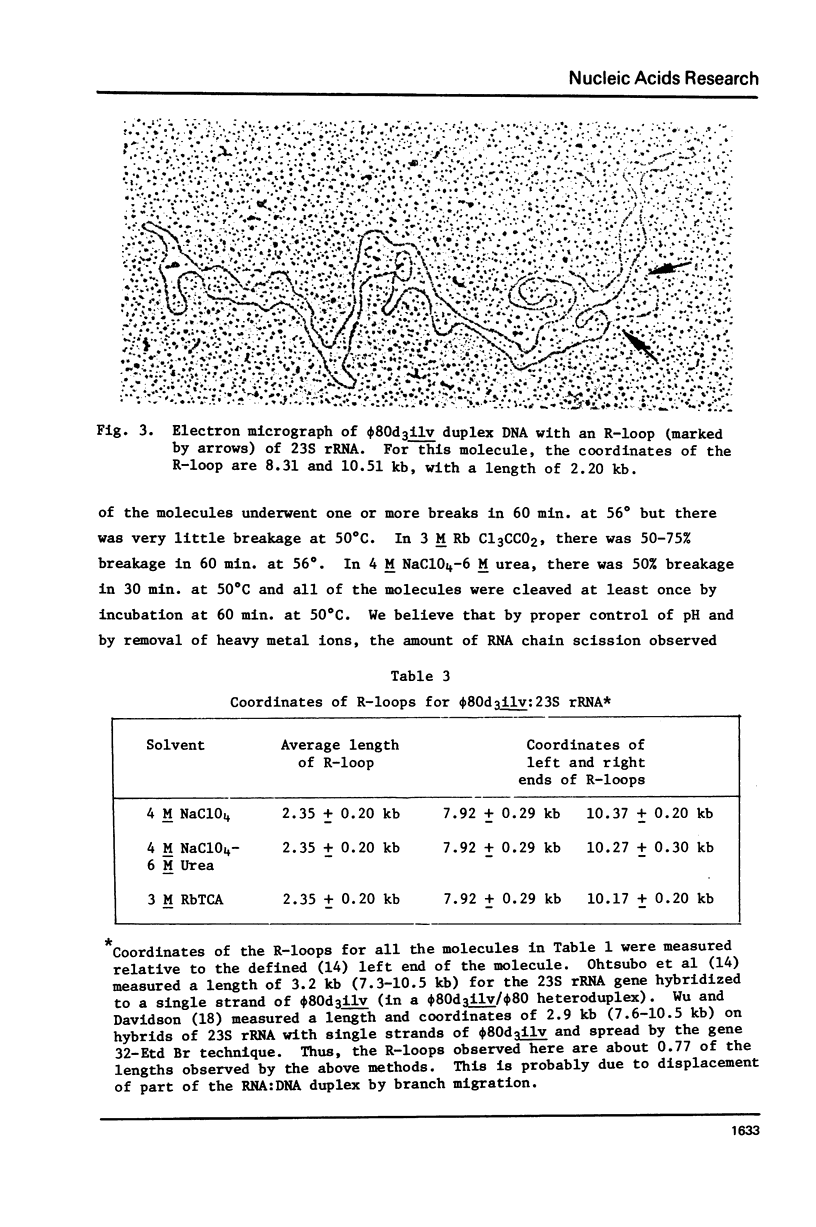
Rates of formation of RNA:DNA hybrids have been measured as a function of temperature and compared to DNA:RNA duplex denaturation temperatures in 4 M sodium perchlorate, 4 M NaClO4-6 M urea, and 3 M rubidium trichloracetate solvents. The usual bell shaped curves of reaction rate versus temperature were observed. The optimal temperatures for the RNA:DNA association reaction are 5 degrees to 12 degrees greater than the Tm's for DNA:DNA denaturation in these solvents, just as in formamide. R-loops of phi80d3ilv DNA with E. coli rRNA can be formed at high efficiency in these solvents.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Broker T. R., Soll L., Chow L. T. Underwound loops in self-renatured DNA can be diagnostic of inverted duplications and translocated sequences. J Mol Biol. 1977 Jul 15;113(4):579–589. doi: 10.1016/0022-2836(77)90223-6. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Bauer W. R. The properties of native and denatured DNA in buoyant rubidium trichloroacetate at neutral pH. Nucleic Acids Res. 1977 Jun;4(6):1891–1909. doi: 10.1093/nar/4.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. T., Hain T. C., Hutton J. R., Wetmur J. G. Effects of microscopic and macroscopic viscosity on the rate of renaturation of DNA. Biopolymers. 1974;13(9):1847–1858. doi: 10.1002/bip.1974.360130915. [DOI] [PubMed] [Google Scholar]
- Friedrich R., Feix G. RNA-RNA hybridization in aqueous solutions containing formamide. Anal Biochem. 1972 Dec;50(2):467–476. doi: 10.1016/0003-2697(72)90056-5. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Cohn R. H., Kedes L. H., Davidson N. Positions of sea urchin (Strongylocentrotus purpuratus) histone genes relative to restriction endonuclease sites on the chimeric plasmids pSp2 and pSp17. Biochemistry. 1977 Apr 5;16(7):1504–1512. doi: 10.1021/bi00626a040. [DOI] [PubMed] [Google Scholar]
- Hutton J. R., Wetmur J. G. Renaturation of bacteriophage phiX174 DNA-RNA hybrid: RNA length effect and nucleation rate constant. J Mol Biol. 1973 Jul 15;77(4):495–500. doi: 10.1016/0022-2836(73)90218-0. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Schlessinger D., Wellauer P. K. 30 S pre-ribosomal RNA of Escherichia coli and products of cleavage by ribonuclease III: length and molecular weight. J Mol Biol. 1974 Jul 15;86(4):741–747. doi: 10.1016/0022-2836(74)90350-7. [DOI] [PubMed] [Google Scholar]
- Nomura M., Morgan E. A. Genetics of bacterial ribosomes. Annu Rev Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- Otsubo E., Soll L., Deonier R. C., Lee H. J., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. VIII. The structure of bacteriophage phi 80d-3ilv+su+7, including the mapping of the ribosomal RNA genes. J Mol Biol. 1974 Nov 15;89(4):631–646. doi: 10.1016/0022-2836(74)90040-0. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Jr, Kelly R. B., Sinsheimer R. L. Denaturation of RNA with dimethyl sulfoxide. Biopolymers. 1968 Jun;6(6):793–807. doi: 10.1002/bip.1968.360060604. [DOI] [PubMed] [Google Scholar]
- TS'O P. O., HELMKAMP G. K., SANDER C. Interaction of nucleosides and related compounds with nucleic acids as indicated by the change of helix-coil transition temperature. Proc Natl Acad Sci U S A. 1962 Apr 15;48:686–698. doi: 10.1073/pnas.48.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wu M., Davidson N. Use of gene 32 protein staining of single-strand polynucleotides for gene mapping by electron microscopy: application to the phi80d3ilvsu+7 system. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4506–4510. doi: 10.1073/pnas.72.11.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]



