Abstract
MicroRNAs have emerged as key players in the regulation of various biological processes in eukaryotes, including host-pathogen interactions. Recent studies suggest that viruses encode miRNAs to manipulate their host gene expression to ensure their effective proliferation, whereas the host limits virus infection by differentially expressing miRNAs that target essential viral genes. Here, we demonstrate that an insect virus, Bombyx mori nucleopolyhedrosis virus (BmNPV), modulates the small-RNA-mediated defense of its host, B. mori, by encoding an miRNA (bmnpv-miR-1) that downregulates the expression of the host GTP-binding nuclear protein Ran, an essential component of the exportin-5-mediated nucleocytoplasmic transport machinery mainly involved in small-RNA transport from the nucleus to the cytoplasm. We demonstrate the sequence-dependent interaction of bmnpv-miR-1 with Ran mRNA using cell culture and in vivo assays, including RNA interference (RNAi) of Ran. Our results clearly show that bmnpv-miR-1 represses Ran, leading to reduction in the host small-RNA population, and consequently, the BmNPV load increases in the infected larvae. Blocking of bmnpv-miR-1 resulted in higher expression levels of Ran and a decrease in BmNPV proliferation. In contrast, blockage of host miRNA, bmo-miR-8, which targets the immediate-early gene of the virus and whose production was repressed upon bmnpv-miR-1 and Ran dsRNA administration, resulted in a significant increase in the virus load in the infected B. mori larvae. The present study provides an insight into one of the evasion strategies used by the virus to counter the host defense for its effective proliferation and has relevance to the development of insect virus control strategies.
INTRODUCTION
MicroRNAs (miRNAs) are noncoding RNAs that regulate gene expression by binding to the complementary sequence of the target mRNAs. Canonically, miRNAs are transcribed by RNA polymerase II as long primary transcripts (pri-miRNAs), which are processed by a nuclear RNase III called Drosha to generate ∼70-nucleotide (nt) imperfectly complementary stem-loop precursor miRNAs (pre-miRNAs). The pre-miRNAs are then exported to the cytoplasm by RanGTP-dependent exportin-5 (6, 54) for further processing by the cytoplasmic RNase III Dicer into ∼22-nt miRNA-miRNA* duplexes, followed by incorporation of one of the strands into an RNA-induced silencing complex (RISC). Finally, the whole RISC assembly, loaded with miRNA, targets the specific mRNAs, leading to either their translational repression or degradation, depending upon the extent of complementarity between the miRNA and its target mRNA (3, 27).
Since the discovery of miRNAs (29, 40), a number of studies have been carried out to understand the role of miRNAs in almost all the biological processes in eukaryotes. Apart from eukaryotes, viruses have also been shown to encode miRNAs (18, 38). Recently, miRNAs have been shown to play a critical role in intricate host-virus interactions (13, 15). Its small size, nonantigenic nature, and target specificity make viral miRNA a potential weapon against the host defense machinery. Some of the well-known strategies adopted by viruses to achieve their establishment and persistent infection in the host cells are modulation of host immune response and escaping recognition by the host immune system (48, 51), antiapoptosis (9, 52), cell cycle arrest (14, 17), and mimicking of host miRNAs (16, 47, 56).
So far, a limited number of studies have been carried out on avoidance of host defenses by viral miRNAs, but they are confined to mammalian viruses. Insect viruses have hardly been the subject of such investigations, except for a recent report of autoregulation of viral gene expression by virus-encoded miRNAs in the Heliothis virescens ascovirus (21). Here, we show that Bombyx mori nucleopolyhedrosis virus (BmNPV) encodes an miRNA that modulates the host defense machinery by regulating the production of the host GTP-binding nuclear protein Ran (Ras-related nuclear protein). Ran is a 25-kDa protein that plays an important role in nucleocytoplasmic transport of various noncoding RNAs and proteins (5). During nuclear export of pre-miRNAs, exportin-5 binds to the pre-miRNAs in a RanGTP-dependent manner, and depletion of Ran results in a severe reduction of pre-miRNA export (6, 33, 50, 54). In humans, Ran also interacts with other transporters, like exportin-1 and exportin-t, which are involved in the transport of small nuclear RNAs (snRNAs) and tRNAs, respectively (44). In Drosophila, exportin-5 is known to transport both pre-miRNAs and tRNAs (44), whereas in B. mori, small-RNA export pathways and factors have yet to be investigated.
BmNPV, a double-stranded DNA (dsDNA) virus, belongs to the family Baculoviridae, which includes various pathogens of Lepidoptera, one of the largest and most economically important insect orders (41). BmNPV, a natural pathogen of the domesticated silk moth, B. mori, inflicts heavy larval mortality and thus causes severe economic damage to silk production. In our previous study, we discovered four BmNPV-encoded miRNAs, and in silico prediction of their targets showed Ran to be one of the potential targets of bmnpv-miR-1 (45). Here, we carried out functional analysis of bmnpv-miR-1 to show that it acts as a suppressor of Ran both in cell culture and in vivo in B. mori. We have found that bmnpv-miR-1 significantly represses the expression of Ran at both the transcript and protein levels, resulting in the impairment of RanGTP-mediated small-RNA transport in the host.
Further, we also analyzed the effect of bmnpv-miR-1-mediated downregulation of Ran on viral proliferation and found a significant increase in the viral load upon Ran knockdown. Consistent with this result, blocking of bmnpv-miR-1 by specific locked nucleic acid (LNA) resulted in higher Ran transcript levels and a decrease in the virus titer in BmNPV-infected larvae. Further, we assessed the virus load by blocking one of the host miRNAs, bmo-miR-8, which was suppressed by administration of bmnpv-miR-1 and Ran dsRNA, by specific LNA. The virus load was dramatically increased upon inhibition of this host miRNA. These results provide convincing evidence that BmNPV suppresses the small-RNA-mediated host defense to successfully proliferate in the host cells by employing bmnpv-miR-1. The present study thus provides insight into yet another layer of complex host-virus interactions mediated by a virus-encoded miRNA.
MATERIALS AND METHODS
In silico prediction of a bmnpv-miR-1 binding site on Ran transcript.
The targets of bmnpv-miR-1 were initially predicted by using the miRanda program (11), along with many screening parameters, as described in our previous report (45). The predicted binding site of bmnpv-miR-1 on the 3′ untranslated region (UTR) of Ran was reconfirmed by employing the RNAhybrid program, which calculates the minimum free energy of the miRNA-mRNA duplex.
B. mori strain and BmNPV infection.
The SBNP-1 strain of B. mori was provided by the Andhra Pradesh State Sericulture Research and Development Institute (APSSRDI), Hindupur, India. The larvae were reared at 27°C on fresh mulberry leaves. For infection study, fifth-instar larvae were fed per os with a purified suspension of BmNPV occlusion bodies (OBs) (20,000 OBs/larva).
RNA isolation and RT-qPCR analysis.
Total RNA was extracted from fat body tissues of uninfected and BmNPV-infected B. mori larvae using TRIzol reagent (Invitrogen), followed by DNase I treatment (Invitrogen). RNA (2 μg) was reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen) in a 20-μl reaction mixture. One microliter of the reverse transcription (RT) product was used to perform quantitative-PCR (qPCR) amplification on the RT-7500 system (Applied Biosystems) using primers specific to Ran, as described in the supplemental materials and methods. RT-qPCR data were analyzed by using the RT-7500 system (Applied Biosystems) software and were further verified by using the standard ΔΔCT method.
Counting of OBs.
To determine the BmNPV load in B. mori larvae, OBs were isolated from the hemolymph of infected larvae by centrifugation (1,000 rpm for 1 min), followed by resuspending the viral pellet in 1× PBS. The OBs were counted using an automated cell counter (Countess; Invitrogen).
HeLa cell culture and luciferase assays.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (Thermo Scientific) containing 2 mM l-glutamine and 10% fetal bovine serum (FBS) (Invitrogen) at 37°C in 5% CO2. The luciferase construct was prepared using PmirGLO, a dual-luciferase miRNA target expression vector (Promega) fused with the bmnpv-miR-1 target site on the 3′ UTR of Ran, as detailed in the supplemental materials and methods. Around 105 HeLa cells/well were seeded in a 24-well plate a day prior to transfection. The cells were first transfected with 300 ng of PmirGLO-Ran–3′ UTR construct, and after 24 h, 200 nM perfectly complementary bmnpv-miR-1 duplexes was added. For miRNA inhibitor assays, the cells were cotransfected with bmnpv-miR-1 duplexes, along with its 200 nM antagomir (synthetic oligonucleotides antisense to miRNA), LNA-1, i.e., miRcury LNA knockdown probe (Exiqon), whereas scrambled LNA was used as a negative control. All the transfections were done using Lipofectamine (Invitrogen) according to the manufacturer's protocol. A luciferase assay was performed 48 h posttransfection using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Luciferase activity was normalized against the control plasmid harboring the unrelated insert (prophenoloxidase) without the bmnpv-miR-1 binding site. Assays were done independently three times in triplicate for each sample. The following miRNA duplexes and LNA sequences were used for transfections: bmnpv-miR-1, sense, 5′-AAAUGGGCGGCGUACAGCUGG-3′, and antisense, 5′-AGCUGUACGCCGCCCAUUUGG-3′; bmnpv-miR-3, sense, 5′-GAAAGCCAAACGAGGGCAGGCG-3′, and antisense, 5′-CCUGCCCUCGUUUGGCUUUCCG-3′; LNA-1, 5′-CCAGCTGTACGCCGCCCATTT-3′; and scrambled LNA, 5′-CATGAGCTGACCGGAACAGCT-3′.
Ran 3′ UTR mutational analysis.
To perform mutational analysis of the bmnpv-miR-1 binding site on Ran mRNA, we designed oligonucleotides with either the intact (wild-type [WT]) or mutated (Mut) target site of bmnpv-miR-1 (Fig. 1C). The restriction sites for SacI and XhoI were added to these oligonucleotides at their 5′ and 3′ ends, respectively, and were digested with the same enzymes and ligated to the SacI- and XhoI-digested PmirGLO vector. The mutations in the target site were confirmed by sequencing. Transfections and luciferase assays were done in HeLa cells as described previously.
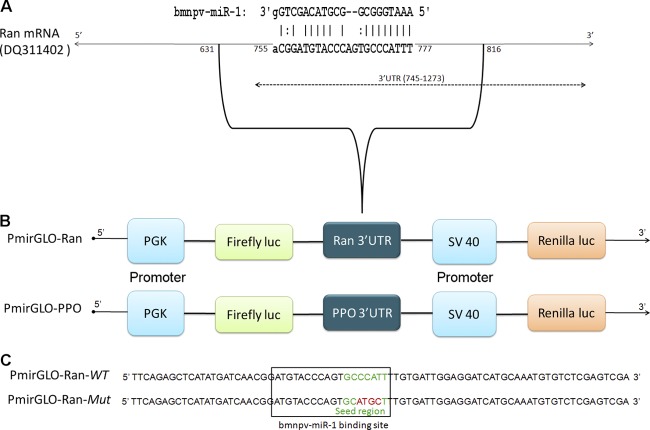
Fig 1.
Schematic representation of the bmnpv-miR-1 binding site on Ran mRNA and the luciferase constructs. (A) Ran mRNA showing the putative binding site of bmnpv-miR-1. (B) Luciferase reporter constructs containing the Ran 3′ UTR (as shown in panel A) and the 3′ UTR of the unrelated target PPO (prophenoloxidase) fused at the 3′ end of the firefly luciferase (luc) gene. The firefly and Renilla luciferase genes are driven by human phosphoglycerate kinase (PGK) and simian virus 40 (SV40) promoters, respectively. (C) The WT or the Mut 3′ UTR of Ran encompassing the binding site of bmnpv-miR-1 was cloned into the PmirGLO vector downstream of the firefly luciferase gene (PmirGLO-Ran-WT and PmirGLO-Ran-Mut, respectively). The 4 mutated nucleotides (PmirGLO-Ran-Mut) in the seed region of the bmnpv-miR-1 binding site are highlighted in red.
BmN cell culture and transfections.
BmN cells were cultured in TC-100 insect medium (PAN Biotech GmbH) supplemented with 10% FBS at 27°C. Around 106 cells were plated in a six-well plate overnight prior to transfection; 1 pmol of bmnpv-miR-1 perfect duplexes and small interfering RNA (siRNA) duplexes against green fluorescent protein (GFP) (sense, 5′ GCUACCUGUUCCAUGGCCATT 3′, and antisense, 5′ UGGCCAUGGAACAGGUAGCTT 3′) were transfected using Cellfectin (Invitrogen). After 72 h of transfection, the cells were harvested, and RNA was isolated by the TRizol method (Invitrogen).
In vivo assay.
bmnpv-miR-1 was administered to B. mori larvae (5th instar, 2nd day) by injecting 100 pmol of bmnpv-miR-1 perfect duplexes with 100 pmol of specific or nonspecific LNA as an antagomir. Larvae injected only with 10 μl diethyl pyrocarbonate (DEPC)-treated water were taken as the negative control. After 60 h, fat body tissues were extracted from the injected larvae, and RNA was isolated. For blocking the endogenous expression of bmnpv-miR-1, 100 pmol (each) of specific LNA of bmnpv-miR-1 (LNA-1) and scrambled LNA was injected 24 h post-BmNPV infection into larvae on the 2nd day of the 5th instar. Cellular miRNA (bmo-miR-8) inhibition was also done by injecting 100 pmol LNA specific to bmo-miR-8 (5′-GACATCTTTACCTGACAGTATTA-3′) into BmNPV-infected larvae on the 2nd day of the 5th instar. The scrambled LNA (5′-GTGTAACACGTCTATACGCCTA-3′) was used as a control. Three independent experiments were carried out in three replicates each with a set of 3 larvae.
RNA interference (RNAi) analysis.
Double-stranded RNAs for Ran, Dicer-2, and GFP were synthesized by using an in vitro transcription kit (Megascript; Ambion) as described in the supplemental materials and methods. Ten micrograms of dsRNA was injected into 2-day-old 5th-instar larvae at the fourth thoracic abdominal leg. GFP dsRNA-injected larvae at the same developmental stage were maintained as negative controls. After 3 days of injection, fat body tissues were extracted, and RNA was isolated from both injected and control larvae.
Northern blot analysis.
For host miRNA detection by Northern blotting, 20 μg of total RNA (for bmnpv-miR-1, 50 μg of total RNA was used) was resolved on a 15% denaturing polyacrylamide gel and transferred to a nylon Hybond+ membrane (Amersham) using a semidry transfer cell (Trans-blot SD; Bio-Rad). UV-cross-linked membranes were hybridized overnight at 37°C in Ultrahyb-Oligo hybridization buffer (Ambion) with γ32P-ATP (specific activity, >1 × 108 cpm/pmol)-labeled probes (the sequences of DNA probes are given in Table S1 in the supplemental material) and scanned using a FLA-9000 Starion phosphorimager (Fujifilm Global).
Western blot assay.
Fifty micrograms of protein from samples administered Ran knockdown and bmnpv-miR-1 was resolved on a 12% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membranes with a semidry transfer cell (Trans-blot SD; Bio-Rad), followed by blocking in NAP-Blocker (G-Biosciences) overnight at 4°C. The membrane was then incubated for 2 h at room temperature with polyclonal antibody (1:1,000 dilution) generated in rabbits against Ran (SCL Group). Detection was done with donkey anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (1/2,000 dilution) and ECL detection reagent (Amersham).
Statistical analysis.
We carried out three independent experiments, each with three replicates for every assay. The results obtained were analyzed using a one-way analysis of variance (ANOVA). This was followed by a post hoc test, i.e., Tukey's test for pairwise comparison, between each pair of treatments. All the tests were performed at a P value of <0.05. In all the histograms representing qPCR, the results were normalized against control samples.
RESULTS AND DISCUSSION
In silico prediction of Ran as a target of bmnpv-miR-1.
Ran was predicted to be one of the potential targets of bmnpv-miR-1 in our previous study (45). The viral miRNA bmnpv-miR-1 was found to have a binding site in the 3′ UTR of Ran transcript with complete complementarity in the seed region (24, 30), i.e., consecutive Watson-Crick matches from positions 2 to 8 at the 5′ end of the miRNA (Fig. 1A). The thermodynamic stability of the bmnpv-miR1–Ran mRNA duplex was further confirmed using another target prediction program, RNAhybrid (39), which gave a value of −26.7 kcal/mol as the minimum free energy of hybridization.
bmnpv-miR-1 negatively regulates Ran expression by binding to the 3′ UTR of Ran mRNA.
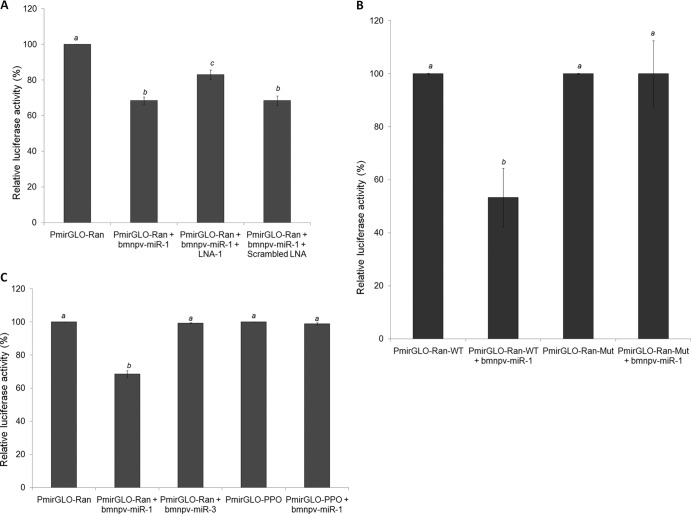
To determine whether bmnpv-miR-1 binds to its predicted target site on the 3′ UTR of the Ran mRNA sequence specifically and regulates its expression (Fig. 1A), we constructed a dual-luciferase reporter vector (firefly and Renilla) by inserting 186 bases (631 to 816) of Ran cDNA sequence comprising the bmnpv-miR-1 binding site downstream of the firefly luciferase gene (Fig. 1B). Renilla luciferase served as an endogenous control. The vector was transfected into HeLa cells, and bmnpv-miR-1 duplexes were added after 24 h. Luciferase activity was found to be reduced by >36% in these cells, which was significantly rescued when bmnpv-miR-1 was blocked by its specific antagomir LNA (LNA-1). The addition of scrambled LNA sequences, on the other hand, did not alter luciferase activity (Fig. 2A). To confirm that the inhibitory effect on luciferase activity was due to sequence-specific binding of bmnpv-miR-1 to its target site on Ran, we mutated 4 nucleotides on the binding site of Ran mRNA corresponding to the seed region of bmnpv-miR-1 (Fig. 1C) and analyzed the interaction using a luciferase assay. For the luciferase assay, the HeLa cells were transfected either with the PmirGLO-Ran-WT or with the PmirGLO-Ran-Mut construct, followed by bmnpv-miR-1 transfection. We did not observe any significant reduction in luciferase activity in the cells transfected with PmirGLO-Ran-Mut, whereas >50% repression was observed in the cells harboring PmirGLO-Ran-WT (Fig. 2B). We also analyzed the effect of an unrelated miRNA (bmnpv-miR-3) on Ran and the bmnpv-miR-1 effect on an unrelated target (prophenoloxidase 3′ UTR) (Fig. 1B) by luciferase assay. As shown in Fig. 2C, unrelated miRNA did not alter the luciferase activity of a reporter vector carrying a bmnpv-miR-1 binding site. Similarly, upon introduction of bmnpv-miR-1 duplexes, no effect was observed in the luciferase level of the reporter vector lacking a bmnpv-miR-1 binding site (carrying the prophenoloxidase 3′ UTR). From these results, we conclude that bmnpv-miR-1 indeed binds sequence specifically to its target site located in the 3′ UTR of the Ran mRNA and represses its translation.
Fig 2.
bmnpv-miR-1 negatively regulates Ran expression by binding to its 3′ UTR in HeLa cells. (A) Luciferase assay showing that bmnpv-miR-1 represses luciferase activity when it binds to the Ran 3′ UTR and that this repression is rescued when the cells are treated with a bmnpv-miR-1-specific antagomir, LNA-1, but the expression remains repressed upon scrambled-LNA treatment. (B) Mutation in the seed region of the bmnpv-miR-1 binding site on the 3′ UTR of Ran mRNA leads to derepression of luciferase activity. (C) To determine the specificity of bmnpv-miR-1–Ran 3′ UTR interaction, a PmirGLO-Ran construct was transfected with an unrelated BmNPV miRNA (bmnpv-miR-3). Also, bmnpv-miR-1 was subjected to an unrelated target site containing the reporter vector (PmirGLO-PPO). In both cases, no significant change was observed in luciferase activity. The assays were done independently three times in triplicate for each sample. Significant differences (P values of <0.05) are denoted by different letters above the error bars. The data are presented as means and standard deviations (SD) (n = 3).
Ran and bmnpv-miR-1 levels are inversely correlated in BmNPV-infected larvae.
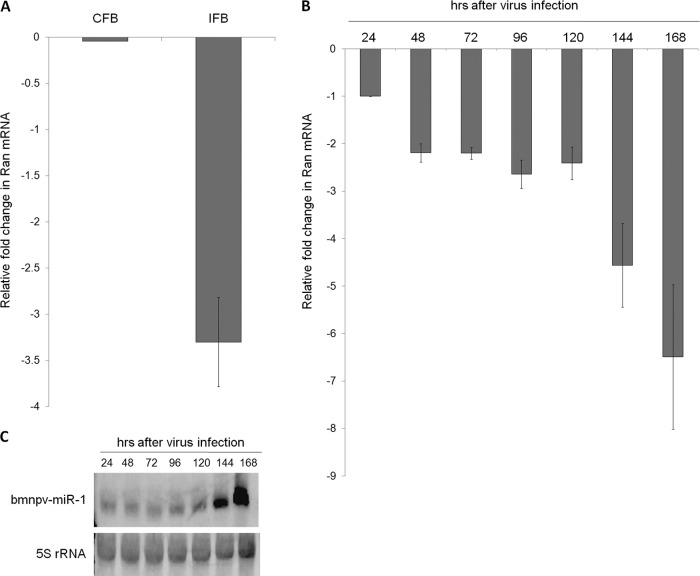
In order to determine the expression level of Ran upon BmNPV infection, we first analyzed the transcript levels of Ran upon BmNPV infection. RT-qPCR was carried out using RNA isolated from the fat body tissues from uninfected larvae and larvae 4 days post-BmNPV infection. The Ran transcript level was found to decrease >3-fold upon BmNPV infection (Fig. 3A). Then, we analyzed the expression of Ran in BmNPV-infected samples at different time points after infection by RT-qPCR, and we observed a gradual decrease in Ran expression with the increase of BmNPV infection (Fig. 3B). To establish that downregulation of Ran upon BmNPV infection is due to bmnpv-miR-1, we followed bmnpv-miR-1 expression, as determined by Northern blotting, and noted that its expression was enhanced in the late stages of infection (Fig. 3C). This contradiction may be attributed to the saturation of Ran-mediated transport by pre-bmnpv-miR-1 (see below). These results indicate that Ran and bmnpv-miR-1 expression levels are inversely correlated.
Fig 3.
Expression of Ran decreases as the bmnpv-miR-1 level increases post-BmNPV infection in B. mori larvae. (A) RT-qPCR analysis of Ran transcripts from control fat body (CFB) tissues and BmNPV IFB tissues 4 days postinfection. (B) Ran transcript levels upon BmNPV infection at different time points examined by RT-qPCR in larvae. Each of the reactions was performed in triplicate three times independently, and the results were normalized against endogenous 18S rRNA. The data are presented as means ± SD (n = 3). (C) bmnpv-miR-1 expression analyzed by Northern blotting in BmNPV-infected RNA samples used in the experiment shown in panel B. The blots were probed with bmnpv-miR-1 (top) and 5S rRNA (bottom), an endogenous control.
bmnpv-miR-1 downregulates Ran in B. mori larvae and cells.
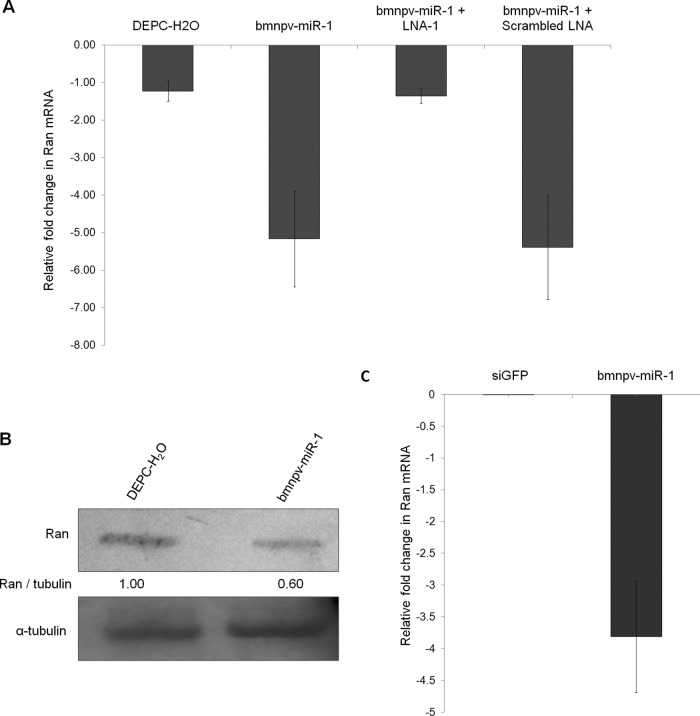
To check the effect of bmnpv-miR-1 on Ran expression in B. mori larvae, double-stranded bmnpv-miR-1 perfect duplexes with or without its LNA were administered to the larvae, and the Ran transcript and protein levels in the fat body tissues were determined by RT-qPCR and Western blotting, respectively. The results showed a >4-fold decrease in the Ran transcript level in the larvae administered bmnpv-miR-1 compared to that in the control larvae (administered DEPC-H2O) (Fig. 4A), whereas no significant change was observed in the expression level of Ran in the larvae treated with both bmnpv-miR-1 and its specific LNA (LNA-1) (Fig. 4A). However, no inhibition of bmnpv-miR-1 was observed in larvae administered scrambled LNA. Inhibition of Ran expression by bmnpv-miR-1 was also confirmed at the protein level by Western blotting (Fig. 4B). bmnpv-miR-1-mediated negative regulation of Ran was also observed in B. mori cells (the BmN cell line) by analyzing the transcript level of Ran upon transfection of bmnpv-miR-1 (Fig. 4C). Taken together, these results permit us to conclude that bmnpv-miR-1 effectively represses the expression of Ran in B. mori larvae, as well as in cells.
Fig 4.
bmnpv-miR-1 downregulates Ran expression in B. mori larvae and in BmN cells. (A) RT-qPCR analysis of Ran transcripts in the fat body tissues of B. mori larvae administered bmnpv-miR-1 with or without its specific LNA. Larvae administered DEPC-H2O were used as a negative control. (B) Western blots showing a decrease in the protein level of Ran in larvae administered bmnpv-miR-1 compared to control larvae (DEPC-H2O administration). The band ratio was determined by densitometry and normalized against internal control α-tubulin. (C) Ran transcript analysis by RT-qPCR in BmN cells upon bmnpv-miR-1 transfection. For transfection, siRNA against GFP (siGFP) was used as a negative control. For RT-qPCR analysis, three independent experiments were carried out in three replicates, each with a set of 3 larvae, and the results were normalized against endogenous 18S rRNA. The data are presented as means ± SD (n = 3).
Abolition of Ran function by RNAi results in suppression of the host small-RNA population.
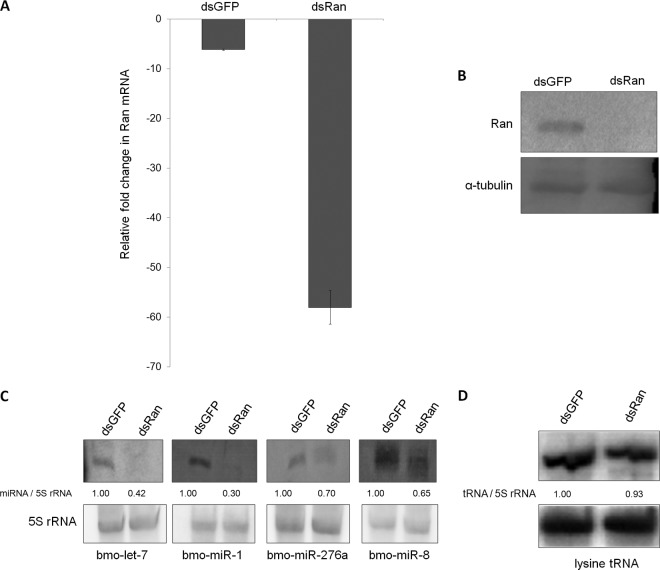
Ran is known to be involved in pre-miRNA transport from the nucleus to the cytoplasm by binding and inducing a conformational change in the key export protein, exportin-5 (33). In B. mori, the function of Ran has not yet been demonstrated, and hence, in order to establish its role in pre-miRNA transport, we carried out dsRNA-mediated knockdown of Ran and analyzed its effects on the expression of the host miRNAs. As a result of RNAi, the Ran transcript level was markedly reduced (Fig. 5A). Western blot analysis also confirmed the knockdown of Ran (Fig. 5B). Next, we assessed the effect of Ran repression on host miRNA expression. Northern blot analysis was carried out for 4 randomly selected, highly expressed known host miRNAs, i.e., bmo-let-7, bmo-miR-1, bmo-miR-276a, and bmo-miR-8 (22). As expected, the expression levels of all 4 miRNAs were found to be reduced compared with their respective GFP dsRNA-injected control larvae (Fig. 5C). As a positive control for miRNA processing, we performed knockdown of Dicer-1 (49) in B. mori larvae and checked the expression levels of two of these miRNAs (bmo-miR-1 and bmo-miR-8) by Northern blotting, but unexpectedly, we could not see any changes in the levels of these miRNAs, although Dicer-1 knockdown was efficient (data not shown). Then, we analyzed the host miRNA expression upon Dicer-2 (NM_001193614) knockdown, and we found reduced levels of host miRNA expression (see Fig. S1 in the supplemental material) similar to that observed in Ran knockdown larvae, clearly implying a role for Ran in miRNA biogenesis.
Fig 5.
Functional analysis of Ran by RNAi in B. mori larvae. (A) dsRNA-mediated knockdown of Ran in larvae analyzed by RT-qPCR. Larvae injected with GFP dsRNA were used as a negative control for all the knockdown experiments. Three independent experiments were carried out in three replicates each with a set of 3 larvae, and the results were normalized against endogenous 18S rRNA. The data are presented as means ± SD (n = 3). (B) Western blot showing a marked decrease in the Ran protein level upon dsRNA injection in larvae. α-Tubulin was used as an internal control. (C) Downregulation of host miRNAs upon Ran knockdown as determined by Northern blot analysis. (D) Northern blot showing expression of lysine tRNA in Ran knockdown larvae. The probes used are shown beneath each of the blots. The band ratio was obtained by densitometry and normalized against endogenous control 5S rRNA.
In addition to pre-miRNAs, Ran is also known to transport other small RNAs, like tRNAs (2, 26). Hence, we checked the effect of Ran knockdown on lysine tRNA expression and did not find any significant change. This might be due to the fact that maturation of tRNAs occurs in the nucleus, and if their transport is abrogated, the tRNAs may accumulate in the nucleus, which in turn may inhibit tRNA production; consequently, the amount of tRNAs may decrease to some extent (Fig. 5D). However, Dicer-2 knockdown did not alter the expression of lysine tRNA (see Fig. S1 in the supplemental material), as it is known to specifically process stem-loop pre-miRNAs and dsRNAs into miRNAs and siRNAs, respectively, and not tRNAs (34).
bmnpv-miR-1 inhibits Ran function.
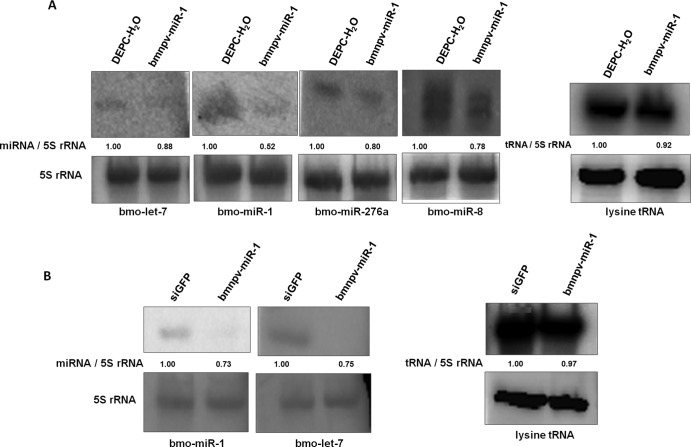
To demonstrate that bmnpv-miR-1-mediated downregulation of Ran mRNA indeed results in impairment of its function, we checked, by Northern blot analysis, the expression levels of 4 host miRNAs that we had selected earlier, as well as lysine tRNA, in larvae administered bmnpv-miR-1. Again, we noticed a reduction in the expression levels of all 4 host miRNAs, but the lysine tRNA expression level remained unchanged (Fig. 6A). The experiments carried out in the BmN cell line also yielded results consistent with the in vivo experiments (Fig. 6B). Expression of these 4 host miRNAs and lysine tRNA were also analyzed 4 days after bmnpv-miR-1 administration, and we found results for all 4 miRNAs similar to those we observed after 60 h of treatment (see Fig. S2 in the supplemental material), whereas no noticeable change was observed in the expression level of lysine tRNA (see Fig. S3 in the supplemental material). Together, these results suggest that BmNPV utilizes its miRNA to repress the host miRNAome. Then, we asked what advantage the virus derives by suppressing the production of host miRNAs.
Fig 6.
bmnpv-miR-1-mediated repression of Ran hampers small-RNA transport in B. mori larvae and in BmN cells. (A) Northern blot analysis of B. mori miRNAs and lysine tRNA expression in larvae administered bmnpv-miR-1. Larvae administered DEPC-H2O were used as a negative control. (B) bmnpv-miR-1 effects on host miRNAs, bmo-miR-1, bmo-let-7, and lysine tRNA were analyzed by Northern blotting. siGFP- and bmnpv-miR-1-transfected BmN cells are represented as siGFP and bmnpv-miR-1, respectively. The probes used are shown beneath each of the blots. The band ratio was determined by densitometry and normalized against endogenous control 5S rRNA.
Blocking of a cellular miRNA, bmo-miR-8, results in a larger BmNPV load.
To answer the above question and to further understand the functional importance of bmnpv-miR-1-mediated Ran inhibition in the host-pathogen interaction, we looked for the targets of the 4 selected host miRNAs, which were found to be repressed upon inhibition of Ran by bmnpv-miR-1 and Ran dsRNA (Fig. 5C and 6A), both in the virus (trans) and in the host (cis), using the miRanda program (as described in reference 45). The putative functions of some of these targets are listed in Table S2 in the supplemental material. The known biological functions of these targets clearly indicate that the role of host miRNAs is not restricted to regulating its own immune genes but also includes various viral genes required for viral entry and establishment inside the host, as demonstrated in various mammalian systems (20, 28, 36, 37, 55).
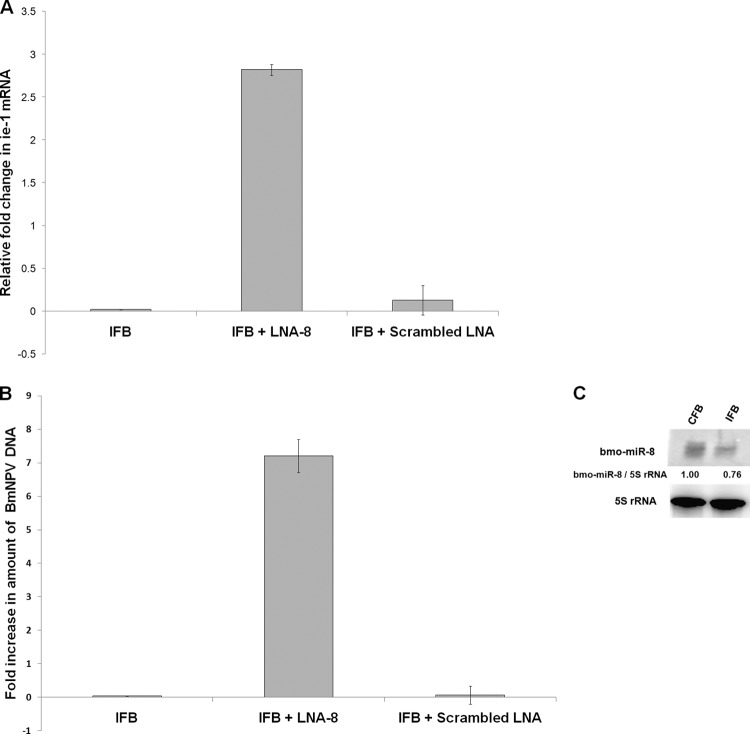
To discover the direct role of cellular miRNAs in viral proliferation, we analyzed the effect of one of the host miRNAs, bmo-miR-8, which was found to be repressed in bmnpv-miR-1 and Ran knockdown samples on virus proliferation. bmo-miR-8 has multiple putative binding sites on BmNPV immediate-early gene (ie-1) mRNA, as well as on other important genes of BmNPV. The ie-1 gene encodes a trans-activator protein required for initial establishment of the virus in host cells. On blocking of bmo-miR-8 by specific LNA, we observed an ∼3-fold increase in the ie-1 transcript level and an ∼8-fold increase in BmNPV accumulation in fat body tissues of infected larvae, as quantified by RT-qPCR and qPCR, respectively, whereas no such effect was seen in the sample administered scrambled LNA (Fig. 7A and B). The marked effect of bmo-miR-8 on the increase of the virus load might be because of its multiple targets in the viral genome. As expected, bmo-miR-8 expression was found to be repressed upon BmNPV infection (Fig. 7C). Taken together, these results suggest that in order to counterattack the miRNA-mediated host defense, the virus uses its miRNA resources to block the host miRNAome by modulating some of the key components of the miRNA biogenesis pathway in its own favor. Further, the functional basis of Ran inhibition by bmnpv-miR-1 was determined by analyzing the virus load upon repression of Ran by bmnpv-miR-1 and Ran dsRNA administration in BmNPV-infected B. mori larvae.
Fig 7.
Inhibition of cellular miRNA, bmo-miR-8, results in greater BmNPV proliferation in B. mori larvae. (A) Transcript analysis of BmNPV ie-1 by RT-qPCR in BmNPV-infected larvae administered LNA-8 and scrambled LNA. (B) BmNPV loads in BmNPV-infected larvae administered LNA-8 and scrambled LNA were determined by qPCR of viral DNA using BmNPV ie-1-specific primers. For qPCR analysis, three independent experiments were carried out in three replicates each with a set of 3 larvae, and the results were normalized against endogenous 18S rRNA. The data are presented as means ± SD (n = 3). (C) Northern blots showing reduction in expression of bmo-miR-8 upon BmNPV infection in B. mori larvae. The band ratio was determined by densitometry and normalized against endogenous 5S rRNA.
Inhibition of bmnpv-miR-1 effectively suppresses BmNPV proliferation.
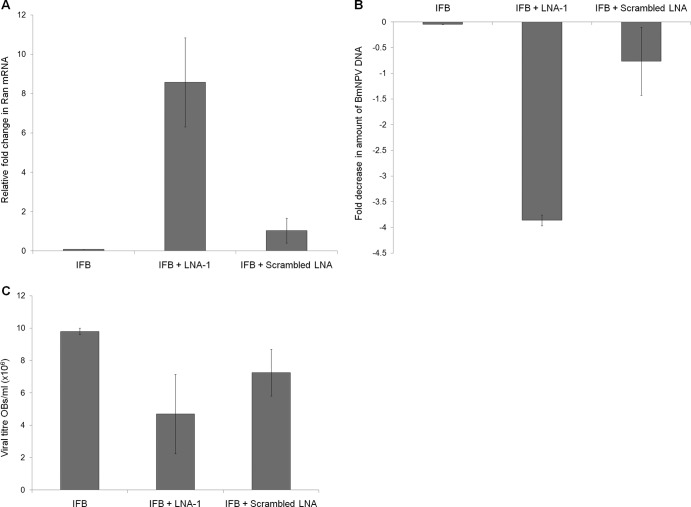
To confirm the role of bmnpv-miR-1 in BmNPV proliferation, we first blocked the endogenous expression of bmnpv-miR-1 by injecting its antagomir, LNA, into the BmNPV-infected larvae and then measured Ran expression and the viral titer in the fat body tissues by RT-qPCR and qPCR, respectively, and the results were normalized against infected fat body (IFB) samples without LNA treatment. The viral titer was determined by quantifying the viral DNA levels using the ie-1 gene of BmNPV and also by scoring OBs in BmNPV-infected larvae injected with the specific (LNA-1) and scrambled LNA. We found a significant increase in the expression of Ran in the larvae administered LNA-1 (Fig. 8A) and a consequent decrease in the viral load (Fig. 8B and C). No such change was observed either in Ran expression levels or on viral accumulation in the infected larvae treated with scrambled LNA (Fig. 8B and C).
Fig 8.
Blocking of bmnpv-miR-1 by LNA leads to a decrease in BmNPV accumulation in B. mori larvae. (A) RT-qPCR analysis shows an increase in Ran expression upon inhibition of bmnpv-miR-1 by specific LNA (LNA-1), while scrambled LNA has no effect compared to the respective control samples. (B) BmNPV titers, determined by qPCR using ie-1 of BmNPV, showed drastic reduction in the virus load when bmnpv-miR-1 was blocked by specific LNA (LNA-1), whereas no significant decrease in the virus load was observed in larvae administered scrambled LNA compared to control samples. Three independent experiments were carried out in three replicates each with a set of 3 larvae, and the results were normalized against the constitutively expressed 18S rRNA gene of B. mori. (C) The virus load was determined by scoring the OBs from hemolymph of BmNPV-infected larvae administered LNA. Three independent experiments were performed with a set of 3 larvae. The data are presented as means ± SD (n = 3).
BmNPV proliferates upon Ran knockdown.
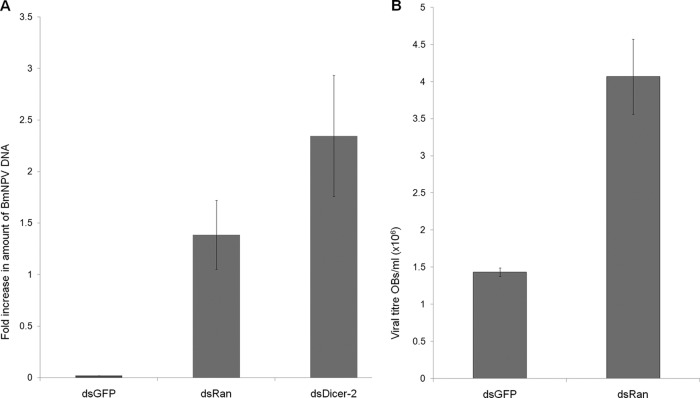
To check viral proliferation upon Ran knockdown, dsRNAs against GFP and Ran were injected into the BmNPV-infected larvae, and ie-1 expression was quantified by qPCR. As shown in Fig. 9A, a very high viral load was observed in the larvae injected with Ran dsRNA compared to that seen in the control larvae, and a similar result was also observed when OBs were scored in the same samples (Fig. 9B). To further confirm that the higher virus load upon Ran knockdown is due to inhibition of the miRNAome of the host and not to some other function of Ran, we also checked the virus titer in the Dicer-2 knockdown larvae. We noticed a significant increase in the virus load in the Dicer-2 knockdown larvae, as was also observed upon Ran knockdown (Fig. 9B), suggesting that the higher virus accumulation is due to suppression of the host miRNAs. No observable difference was noticed in the phenotypes of Ran and Dicer-2 knockdown larvae (see Fig. S4 in the supplemental material). Interestingly, blocking of bmnpv-miR-1 resulted in a marked decrease (∼8-fold) in the BmNPV load (Fig. 8B), whereas knockdown of Ran and Dicer-2 larvae showed only ∼1.5 and 2.5-fold increases in virus accumulation (Fig. 9B), respectively. This may possibly be due to bmnpv-miR-1 targeting other host genes that are involved in host defense. Based on these results, we conclude that BmNPV-encoded miRNA, bmnpv-miR-1, combats the host small-RNA-mediated antiviral attack by suppressing the expression of Ran, an important component of the small-RNA transport machinery of the cell, as proposed in the model shown in Fig. 10.
Fig 9.
B. mori miRNA suppression increases BmNPV proliferation in larvae. (A) Quantitative-PCR analysis of ie-1 of BmNPV to determine the viral load in Ran and Dicer-2 knockdown larvae. Larvae injected with GFP dsRNA were used as a negative control. Three independent experiments were carried out in three replicates each with a set of 3 larvae, and the results were normalized against endogenous 18S rRNA. (B) The virus load was determined by scoring the OBs from hemolymph of Ran knockdown BmNPV-infected larvae. Three independent experiments were performed with a set of 3 larvae. The data are presented as means ± SD (n = 3).
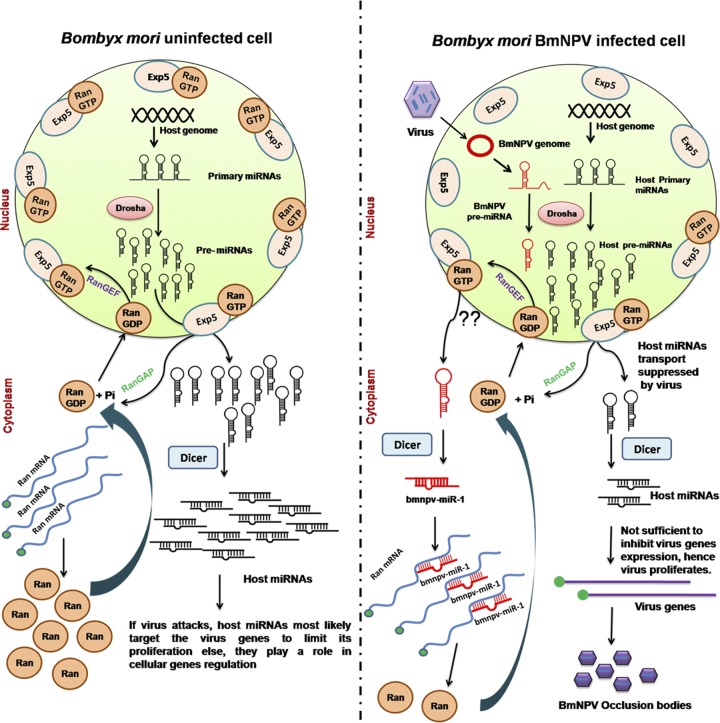
Fig 10.
The proposed model for bmnpv-miR-1-mediated evasion of the host defense response mounted by small RNAs for effective proliferation of BmNPV in infected larvae. The model proposed here shows that BmNPV, upon infection of silkworm larvae, encodes an miRNA, bmnpv-miR-1, which represses Ran, an important component of small-RNA export from the nucleus to the cytoplasm, by binding its 3′ UTR. As a result, the small-RNA population is reduced in the cytoplasm, leading to proliferation of the virus.
Viruses are known to evade host defenses in different ways. Recently, virus-derived miRNAs have emerged as potential modulators of the host immune system. Many mammalian viral miRNAs have been shown to regulate both the host's and their own sets of genes, which otherwise are obstacles to virus entry or proliferation inside the host (15). Similarly, there are many reports of cellular miRNAs of the host modulating the expression of various viral genes (20, 28, 36, 37, 55); hence, it is likely that B. mori miRNAs can directly or indirectly inhibit BmNPV proliferation. In the present study, we have shown that Ran is involved in small-RNA trafficking in B. mori and that BmNPV-encoded miRNA, bmnpv-miR-1, controls Ran expression by destabilizing its mRNA, followed by translation repression that leads to suppression of cellular miRNA production, which in turn results in enhanced virus proliferation.
The contradiction that we came across in our study is a higher level of expression of bmnpv-miR-1 in the late stage of infection despite effective repression of Ran (Fig. 3B and C). The pertinent question here is, when the bmnpv-miR-1 pre-miRNA is not transported out of the nucleus as a result of Ran repression, how such a high load of processed bmnpv-miR-1 could still be seen in the cell. This result suggests that BmNPV may deploy a variety of strategies to effectively replicate in the host cell. One such strategy we propose here is that the baculovirus may not fully block RanGTP-mediated transport by its bmnpv-miR-1, and as a result, transport of host pre-miRNAs in small quantities may still occur in the infected larvae. This strategy allows the virus to proliferate in the host cell by keeping the small-RNA-mediated host defense in check and, at the same time, keeping the larvae viable to enhance its proliferation. During the late stage of infection, a massive quantity of virus is produced in the host cell, giving rise to a larger amount of virus-derived pre-bmnpv-miR-1, which saturates the Ran-mediated transport machinery, resulting in higher-level transport of pre-bmnpv-miR-1 into the cytoplasm and, at the same time, limiting the transport of host pre-miRNAs. These speculations draw credence from recent studies that have uncovered ingenious and complex strategies employed by the viruses to enhance their accumulation in host cells. For example, adenovirus-derived small RNAs massively produced in the late stage of infection are able to deplete Dicer expression by saturating exportin-5 and abrogating the nuclear export of exportin-5-dependent Dicer mRNA. As a result, maturation of small RNA is terminated by the virus using this strategy (1, 4, 7, 31). Similarly, influenza virus-encoded NS1 protein has been shown to inhibit the host mRNA export pathway to produce greater permissiveness by host cells for influenza virus replication (32). It is also possible that BmNPV employs alternative strategies, including utilization of an mRNA transport pathway that is independent of Ran (23). In our previous study (45), we showed that bmnpv-miR-1 is derived from the coding region of cathepsin, which is expressed heavily during the late stage of infection, as it is required for liquefaction of infected larvae (19), and hence, bmnpv-miR-1 may escape Drosha cleavage and be transported as mRNA to be processed subsequently by canonical or noncanonical pathways in the cytoplasm to give rise to mature bmnpv-miR-1 (8, 10, 43, 46, 53).
The important question that remains to be addressed further is how the virus controls the expression of bmnpv-miR-1. Ran has been shown to be an important component of nucleocytoplasmic transport of small RNAs, as well as proteins (5, 6, 33, 50, 54), and its complete depletion may be lethal, but we did not observe such a drastic effect in either bmnpv-miR-1- or dsRNA-mediated knockdown of Ran, although the treated larvae showed stunted growth (see Fig. S4 in the supplemental material). These observations point to the role of miRNAs as fine-tuners of gene expression that may not act as phenotype switchers (12, 25, 35, 42). Since viruses do not possess any miRNA biogenesis component in their genomes, it is interesting to investigate how virus generates its own miRNAs when it blocks its host's miRNA transport.
Can we generalize this strategy to other viruses, especially latent viruses? As miRNAs are also required for the regulation of many cellular genes, which are important for the survival of the host, inhibition of miRNA processing would have a serious effect on these genes and might lead to cell death. Hence, in cases of persistent or latent infection, instead of blocking miRNA biogenesis, the virus may try to escape the silencing machinery. The answers to all these questions might also reveal some of the very complicated aspects of virus-governed fine-tuning of host genes.
In summary, our results revealed one of the strategies employed by the virus to modulate miRNA-mediated antiviral host defense by producing an miRNA that suppresses Ran, an important component of miRNA biogenesis. Our results thus provide useful insights into the multilayered complexity of intricate host-virus interactions and have implications for future applications in insect virus control and possibly a novel vision for therapeutic strategies.
Supplementary Material
ACKNOWLEDGMENTS
J.N. acknowledges financial support from the Department of Biotechnology, Government of India, under the Centre of Excellence for Genetics and Genomics of Silkmoths (grant number BT/01/COE/05/12). C.P.S. and J.S. are recipients of senior research fellowships from the Council of Scientific and Industrial Research, India, and the Department of Biotechnology, Government of India, respectively.
Footnotes
Published ahead of print 16 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Andersson MG, et al. 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 79:9556–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arts GJ, Fornerod M, Mattaj IW. 1998. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8:305–314 [DOI] [PubMed] [Google Scholar]
- 3. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 4. Bennasser Y, et al. 2011. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat. Struct. Mol. Biol. 18:323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bischoff FR, Ponstingl H. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354:80–82 [DOI] [PubMed] [Google Scholar]
- 6. Bohnsack MT, Czaplinski K, Gorlich D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10:185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carnero E, Sutherland JD, Fortes P. 2011. Adenovirus and miRNAs. Biochim. Biophys. Acta 1809:660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465:584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choy EY, et al. 2008. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 205:2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cullen BR. 2011. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25:1881–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enright AJ, et al. 2003. MicroRNA targets in Drosophila. Genome Biol. 5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flynt AS, Lai EC. 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghosh Z, Mallick B, Chakrabarti J. 2009. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 37:1035–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gottwein E, Cullen BR. 2010. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol. 84:5229–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gottwein E, Cullen BR. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gottwein E, et al. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grey F, et al. 2010. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog. 6:e1000967 doi:10.1371/journal.ppat.1000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hawtin RE, et al. 1997. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238:243–253 [DOI] [PubMed] [Google Scholar]
- 20. Hussain M, Asgari S. 2010. Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J. Virol. 84:612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hussain M, Taft RJ, Asgari S. 2008. An insect virus-encoded microRNA regulates viral replication. J. Virol. 82:9164–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jagadeeswaran G, et al. 2010. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics 11:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kohler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761–773 [DOI] [PubMed] [Google Scholar]
- 24. Krek A, et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500 [DOI] [PubMed] [Google Scholar]
- 25. Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11:597–610 [DOI] [PubMed] [Google Scholar]
- 26. Kutay U, et al. 1998. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1:359–369 [DOI] [PubMed] [Google Scholar]
- 27. Lau NC, Lim LP, Weinstein EG, Bartel DP. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858–862 [DOI] [PubMed] [Google Scholar]
- 28. Lecellier CH, et al. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 [DOI] [PubMed] [Google Scholar]
- 29. Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854 [DOI] [PubMed] [Google Scholar]
- 30. Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 [DOI] [PubMed] [Google Scholar]
- 31. Lu S, Cullen BR. 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 78:12868–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu Y, Qian XY, Krug RM. 1994. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 8:1817–1828 [DOI] [PubMed] [Google Scholar]
- 33. Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. 2004. Nuclear export of microRNA precursors. Science 303:95–98 [DOI] [PubMed] [Google Scholar]
- 34. Macrae IJ, et al. 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311:195–198 [DOI] [PubMed] [Google Scholar]
- 35. Mukherji S, et al. 2011. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 43:854–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nuovo GJ, et al. 2010. Strong inverse correlation between microRNA-125b and human papillomavirus DNA in productive infection. Diagn. Mol. Pathol. 19:135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pedersen IM, et al. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pfeffer S, et al. 2004. Identification of virus-encoded microRNAs. Science 304:734–736 [DOI] [PubMed] [Google Scholar]
- 39. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reinhart BJ, et al. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906 [DOI] [PubMed] [Google Scholar]
- 41. Rohrmann GF. 2008. Baculovirus molecular biology. National Library of Medicine, NCBI, Bethesda, MD [Google Scholar]
- 42. Sevignani C, Calin GA, Siracusa LD, Croce CM. 2006. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm. Genome 17:189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shapiro JS, Varble A, Pham AM, Tenoever BR. 2010. Noncanonical cytoplasmic processing of viral microRNAs. RNA 16:2068–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shibata S, et al. 2006. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 34:4711–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singh J, Singh CP, Bhavani A, Nagaraju J. 2010. Discovering microRNAs from Bombyx mori nucleopolyhedrosis virus. Virology 407:120–128 [DOI] [PubMed] [Google Scholar]
- 46. Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Skalsky RL, et al. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836–12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stern-Ginossar N, et al. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swevers L, Liu J, Huvenne H, Smagghe G. 2011. Search for limiting factors in the RNAi pathway in silkmoth tissues and the Bm5 cell line: the RNA-binding proteins R2D2 and Translin. PLoS One 6:e20250 doi:10.1371/journal.pone.0020250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X, et al. 2011. Dynamic mechanisms for pre-miRNA binding and export by Exportin-5. RNA 17:1511–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xia T, et al. 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68:1436–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu S, Xue C, Li J, Bi Y, Cao Y. 2011. Marek's disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J. Virol. 85:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang JS, Lai EC. 2011. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell 43:892–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang GL, et al. 2010. Suppression of hepatitis B virus replication by microRNA-199a–3p and microRNA-210. Antiviral Res. 88:169–175 [DOI] [PubMed] [Google Scholar]
- 56. Zhao Y, et al. 2009. A functional microRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J. Virol. 83:489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.