Abstract
Avocado sunblotch viroid, peach latent mosaic viroid, chrysanthemum chlorotic mottle viroid, and eggplant latent viroid (ELVd), the four recognized members of the family Avsunviroidae, replicate through the symmetric pathway of an RNA-to-RNA rolling-circle mechanism in chloroplasts of infected cells. Viroid oligomeric transcripts of both polarities contain embedded hammerhead ribozymes that, during replication, mediate their self-cleavage to monomeric-length RNAs with 5′-hydroxyl and 2′,3′-phosphodiester termini that are subsequently circularized. We report that a recombinant version of the chloroplastic isoform of the tRNA ligase from eggplant (Solanum melongena L.) efficiently catalyzes in vitro circularization of the plus [(+)] and minus [(−)] monomeric linear replication intermediates from the four Avsunviroidae. We also show that while this RNA ligase specifically recognizes the genuine monomeric linear (+) ELVd replication intermediate, it does not do so with five other monomeric linear (+) ELVd RNAs with their ends mapping at different sites along the molecule, despite containing the same 5′-hydroxyl and 2′,3′-phosphodiester terminal groups. Moreover, experiments involving transient expression of a dimeric (+) ELVd transcript in Nicotiana benthamiana Domin plants preinoculated with a tobacco rattle virus-derived vector to induce silencing of the plant endogenous tRNA ligase show a significant reduction of ELVd circularization. In contrast, circularization of a viroid replicating in the nucleus occurring through a different pathway is unaffected. Together, these results support the conclusion that the chloroplastic isoform of the plant tRNA ligase is the host enzyme mediating circularization of both (+) and (−) monomeric linear intermediates during replication of the viroids belonging to the family Avsunviroidae.
INTRODUCTION
Viroids are plant pathogens constituted by a small circular noncoding RNA (12, 18, 19, 45). Most viroid species are gathered within the family Pospiviroidae and characteristically contain a central conserved region (CCR) in the middle of their molecules, which are predicted to fold in rod- or quasi-rod-like minimum free-energy conformations, and replicate in the nuclei of infected cells. However, four viroid species—Avocado sunblotch viroid (ASBVd), Peach latent mosaic viroid (PLMVd), Chrysanthemum chlorotic mottle viroid (CChMVd), and Eggplant latent viroid (ELVd)—that do not contain a CCR in their molecules are grouped into the family Avsunviroidae (15, 17). These four species contain hammerhead ribozymes embedded in both polarities of their RNA strands that catalyze self-cleavage of the oligomeric viroid RNA intermediates resulting from replication that occurs in the chloroplasts of infected cells (17). In this family, replication follows the symmetric variant of an RNA-based rolling-circle mechanism (4, 9, 25). In this variant, the circular viroid strand of plus [(+)] polarity—which is arbitrarily assigned to the viroid RNA strand most abundant in the infected tissue—is reiteratively transcribed by an RNA polymerase to produce oligomeric strands of complementary or minus [(−)] polarity. These RNAs are self-cleaved by the ribozymes to produce monomeric linear RNAs that are circularized. Then, in a second and symmetrical part of the cycle, the monomeric (−) circular RNA is transcribed to oligomers that are self-cleaved and the resulting linear monomers subsequently circularized to finally produce the monomeric (+) circular RNA.
The effect of the inhibitor tagetitoxin on RNA synthesis in chloroplastic preparations of ASBVd-infected avocado (Persea americana Mill.) leaves suggests that the nucleus-encoded chloroplastic RNA polymerase (NEP), and not the plastid-encoded RNA polymerase (PEP), is the enzyme that transcribes the viroid RNAs in the infected tissue (37). This notion is sustained by the intense PLMVd replication in peach [Prunus persica (L.) Batsch] leaves expressing a PLMVd-incited albinism in which PEP-dependent transcription is basically absent (41). Consistent with this view, ASBVd double-stranded RNAs, regarded as the replication intermediates, have been detected in chloroplasts of infected cells (36). Concerning the second replication step, characterization of the termini of linear ASBVd and CChMVd strands accumulating in infected tissues supports the hypothesis that hammerhead ribozymes mediate the self-cleavage in vivo of the oligomeric RNAs (9, 32, 35). Moreover, covariations that preserve the stability of the hammerhead structures are frequently found in natural sequence variants of PLMVd (2, 23) and CChMVd (11, 35), thus reinforcing their physiological role. In contrast, there is a lack of experimental evidence regarding the host factor mediating the subsequent circularization of monomeric linear viroid RNA intermediates.
The RNA circularizing activity during the replication of the Avsunviroidae could also reside in the hammerhead ribozyme, which can catalyze not only RNA cleavage but also ligation (24, 38), especially when their tertiary stabilizing motifs (10, 27) are preserved (5). However, the low efficiency of the ligation reaction (at least in vitro) does not explain the high ratio of circular to total monomeric viroid RNA accumulating in ASBVd-infected avocado and ELVd-infected eggplant (Solanum melongena L.) tissues (9, 15). Also, our previous study about ELVd processing in transplastomic lines of the green alga Chlamydomonas reinhardtii P.A. Dangeard expressing different ELVd mutant forms indicates that the viroid sequence requirements for cleavage and ligation are different. More specifically, a quasi-double-stranded structure in the central part of the ELVd molecule (folded in the predicted minimum free-energy conformation) containing the ligation site in an internal loop motif seems involved in ELVd circularization in a chloroplastic context (33). This result suggested that an RNA ligase activity of chloroplastic localization, able to recognize the viroid ligation conformation, likely mediates ELVd circularization (33). A candidate for providing this activity is tRNA ligase, a conserved enzyme involved in maturation of nuclear tRNAs (1, 13, 31, 46) that in plants is targeted to chloroplast, in addition to the nucleus and cytoplasm (14). Supporting this hypothesis, tRNA ligase shows strong specificity toward 5′-hydroxyl and 2′,3′-cyclic phosphodiester RNA terminal groups (29, 39, 44), which are produced by the hammerhead ribozymes (26, 40).
In this study, we have examined the ability of the chloroplastic isoform of the plant tRNA ligase to mediate circularization of viroid RNA during the replication of members of the family Avsunviroidae. For this purpose, we cloned the eggplant tRNA ligase, purified a recombinant version of this protein, and analyzed its ability to circularize in vitro different monomeric linear ELVd RNAs, as well as the monomeric linear replication intermediates of the other Avsunviroidae. The effects of silencing the endogenous tRNA ligase in planta provide additional evidence supporting the role of this enzyme in the replication of the Avsunviroidae.
MATERIALS AND METHODS
RT and PCR amplification of tRNA ligase cDNAs.
Eggplant (cv. Black Beauty) leaf tissue was homogenized with 5 volumes of buffer consisting of 0.1 M Tris-HCl (pH 9.0), 0.1 M NaCl, 10 mM EDTA, 0.1 M 2-mercaptoethanol, and 5 M urea. The clarified extract was fractionated with buffered (pH 8.0) phenol-chloroform (1:1), and RNAs in the aqueous phase were precipitated with isopropanol. RNAs were further purified by chromatography using a silica gel spin column (Centrifuge Plant RNA minikit; Nedken). Aliquots of the RNA preparation were subjected to reverse transcription (RT) using 5 pmol of complementary primers in 10-μl reaction mixtures containing 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, 10 mM dithiothreitol (DTT), 0.5 mM each deoxynucleoside triphosphate (dNTP), 10 U RNase inhibitor, and 50 U of Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Fermentas) for 45 min at 42°C, followed by 10 min at 50°C and 5 min at 60°C.
Aliquots of 1 μl of the RT reaction products were subjected to PCR amplification in 20 μl with 0.4 U of the high-fidelity Phusion DNA polymerase (Finnzymes) in the presence of HF buffer (Finnzymes), 3% dimethyl sulfoxide, 0.2 mM each dNTP, and 0.5 μM each primer. Reactions consisted of an initial denaturation of 30 s at 98°C followed by 30 cycles of 10 s at 98°C, 30 s at 55°C, and a period at 72°C depending on the length of the expected product (15 s per kbp), with a final incubation of 10 min at 72°C. In most cases, 1 μl of the PCR products was subjected to a second nested PCR under the same conditions but with a new pair of primers.
The first eggplant tRNA ligase cDNA fragment was amplified using the poly(dT) primer I in the RT reaction and primers II and III in the PCR. Primers II and III were designed according to nucleotide sequences conserved in tRNA ligases from other higher plant species. Primer sequences and some description of their use are in Table S1 and Fig. S1 in the supplemental material. After cloning and sequencing of the first eggplant tRNA ligase cDNA fragment, homologous primers were designed for subsequent amplifications. A cDNA corresponding to the 3′ end of the eggplant tRNA ligase mRNA was amplified using primer I in the RT reaction, primers II and IV in the first PCR, and primers V and VI in the second nested PCR. Next, a new cDNA fragment toward the 5′ end of the mRNA was amplified by following a version of a protocol for rapid amplification of cDNA ends (RACE) in which an initial cDNA was obtained by RT with primer VII phosphorylated at the 5′ end. The reaction product was then treated with RNase H (Fermentas) and circularized with T4 RNA ligase (Fermentas). The resulting circular cDNA was used as a template for two successive PCRs with nested primers (VIII and IX in the first reaction and X and XI in the second). A further 5′ cDNA fragment was obtained through a heminested PCR amplification using the RT product previously obtained with primer VII. Primers XII and XIII were used in the first PCR and XII and XIV in the second, heminested reaction. Primer XII was designed from conserved sequences of plant tRNA ligases. A final cDNA corresponding to the 5′ end of the mRNA was amplified by circular RACE using 5′-phosphorylated primer XV in the RT reaction, primers XVI and XVII in the first PCR, and primers XVIII and XIX in the second nested amplification. All the cDNAs were cloned and sequenced.
Expression and purification of a recombinant version of eggplant tRNA ligase.
A cDNA corresponding to the full open reading frame of eggplant tRNA ligase was finally amplified from the eggplant RNA preparation using primer XX in the RT reaction, primers XXI and XXII in the first PCR, and primers XXIII and XXIV in the second nested PCR. The cDNA product was inserted between the NcoI and XhoI sites of plasmid pET-23d(+) (Novagen). The resulting plasmid was used as a template to generate plasmid pESmtRnl by two rounds of PCR to delete sequences coding for extra amino acids at the amino terminus (primers XXV and XXVI) and between the carboxyl terminus of the native eggplant protein and the His6 tag of the recombinant protein (primers XXVII and XXVIII).
A recombinant version of eggplant tRNA ligase, including a carboxy-terminal His6 tail (Fig. 1), was expressed in Escherichia coli Rosetta 2(DE3)pLysS (Novagen) transformed with pESmtRnl. A 250-ml culture at 0.6 U of optical density (OD) at 600 nm was induced with 400 μM isopropyl β-d-1-thiogalactopyranoside for 3 h at 28°C. Cells were harvested by centrifugation, washed, resuspended in 5 ml of H2O containing a cocktail of proteinase inhibitors (Complete; Roche), and frozen. Once the cell suspension was thawed, 0.5 ml of 1 M Tris-HCl (pH 7.5), 1 ml of 10% Nonidet P-40, 25 μl of 1 M MgCl2, 7 μl of 2-mercaptoethanol, and 125 U of Benzonase (Novagen) were added to the cell suspension and the mixture incubated with shaking for 45 min at 4°C. After addition of NaCl (0.3 g), the solution was brought to 10 ml with water and incubated at 4°C for 15 min more. The extract was clarified by centrifugation at 100,000 × g for 30 min and the supernatant brought to 20 mM imidazole. The recombinant protein was purified by chromatography using a 1-ml Ni-Sepharose column (Histrap HP; GE Healthcare) with an ÄKTA Prime Plus liquid chromatography system operated at 4°C with a flow rate of 1 ml/min. The column was equilibrated with 10 ml of buffer (50 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 1% Nonidet P-40, 10 mM 2-mercaptoethanol, 20 mM imidazole) and the extract loaded. The column was washed with 20 ml of buffer and the protein eluted with a 1:1 mix of equilibration buffer and 1 M imidazole-HCl (pH 8.0). The recombinant eggplant tRNA ligase was analyzed in the eluted fractions by electrophoresis in parallel denaturing (0.05% sodium dodecyl sulfate [SDS]) 12.5% polyacrylamide gels. One gel was stained with Coomassie blue and the other subjected to Western blot analysis with an anti-His6 antibody (Clontech).
Fig 1.
Amino acid sequence of eggplant tRNA ligase including the carboxy-terminal His6 tag (gray background) of the expressed recombinant version. The chloroplast transit peptide predicted by the ChloroP algorithm is highlighted. Motifs conserved in eukaryotic tRNA ligases (Lig, Lig I, Lig IIIa, Lig IV, PNK, and CPD) are boxed.
Viroid RNA transcription and purification.
Dimeric (+) and (−) transcripts of ASBVd (GenBank accession no. X52041, from position 154 to 153), PLMVd (AJ005303 with the point mutation C260U, from 199 to 198), CChMVd (AJ878085, from 295 to 294), and ELVd (AJ536613, from 210 to 209) were produced by in vitro runoff transcription with T3 or T7 RNA polymerases (Epicentre) of conveniently linearized plasmid templates. Transcription products were separated by denaturing PAGE in 5% polyacrylamide gels including 8 M urea in TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA). Gels were stained with ethidium bromide, and the products corresponding to the full-length monomeric linear viroid RNAs (resulting from hammerhead-mediated self-cleavage and consequently containing 5′-hydroxyl and 2′,3′-cyclic phosphodiester termini) were eluted by diffusion, precipitated, and quantified spectrophotometrically. RNAs corresponding to full-length monomeric linear (+) ELVd RNAs from position U90 to U89, A104 to A103, C177 to U176, U246 to U245, and G264 to A263 were obtained in the same way but using linearized plasmid templates containing the corresponding monomeric ELVd cDNAs flanked by a modified version of the hammerhead ribozyme from tobacco ringspot virus satellite (sTRSV) RNA (16) and a modified version of the hepatitis delta virus (−) strand ribozyme (43). To produce 32P-labeled (−) ELVd RNA probes, 0.5 mM UTP was replaced by 40 μCi of [α-32P]UTP (800 Ci/mmol) during the transcription reaction.
Viroid RNA circularization assay.
The standard viroid RNA circularization reactions were done in a 20-μl volume in buffer consisting of 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 4 mM MgCl2, 5 mM DTT, and 1 mM ATP, including 25 ng of RNA substrate and 4 μl of the purified protein; reaction mixtures were incubated for 30 min at 30°C. Reactions were stopped by adding 20 μg (1 μl) of proteinase K (New England BioLabs) and 2 μl of buffer consisting of 100 mM Tris-HCl (pH 8.0), 50 mM EDTA, 10 mM 2-mercaptoethanol, and 0.5% SDS and incubating the reaction mixtures successively for 15 min at 42°C and 15 min at 55°C. Reaction products were mixed with 1 volume of loading buffer (98% formamide, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.0025% xylene cyanol, 0.0025% bromophenol blue), denatured by heating for 2 min at 95°C, and separated by denaturing PAGE. ELVd (+) RNAs were detected by Northern blot hybridization using a 32P-labeled ELVd (−) RNA probe as previously described (33). In the circularization assay of the (+) and (−) monomeric linear self-cleavage forms of all viroids (ASBVd, PLMVd, CChMVd, and ELVd), 1 mM ATP was replaced by 5 μCi of [γ-32P]ATP (3,000 Ci/mmol) and the polyacrylamide gels used to separate the reaction products fixed for 30 min in 10% acetic acid–20% methanol, dried under vacuum, and imaged by autoradiography.
VIGS of tRNA ligase from Nicotiana benthamiana Domin.
Plasmid pTRV2-tRnl was generated, which contains an insertion corresponding to position 983 to 1282 of eggplant tRNA ligase cDNA (see Fig. S1 in the supplemental material) cloned in the sense orientation between the EcoRI and XhoI sites of the virus-induced gene silencing (VIGS) vector pTRV2 (GenBank accession no. AF406991) (30). Control plasmid pTRV2-GFP, designed to silence Aequorea victoria green fluorescent protein (GFP), contains an insertion corresponding to position 219 to 480 of the cDNA from GFP variant mgfp5 (GenBank accession no. U87973) (42) also cloned between the EcoRI and XhoI sites of pTRV2 in the sense orientation. Plasmids pCdELVd+ and pCdCCCVd+ contain dimeric cDNAs of ELVd (from position 210 to 209) and coconut cadang-cadang viroid (CCCVd) (J02050 with the point mutation C31T, from position 54 to 53) replacing the GFP coding sequence in pCAMBIA-1302 (AF234298) under the control of the 35S promoter from cauliflower mosaic virus and nos terminator from Agrobacterium tumefaciens. A. tumefaciens C58C1 cells transformed with plasmids pTRV1 (AF406990) (30), pTRV2-tRnl, pTRV2-GFP, pCdELVd+, and pCdCCCVd+ were grown to approximately 0.5 U of OD at 600 nm, recovered by centrifugation, resuspended at 0.5 U of OD in 10 mM MES-NaOH (pH 5.6), 10 mM MgCl2, and 150 μM acetosyringone, and induced for 2 h at 28°C. Transgenic N. benthamiana plants constitutively expressing the GFP variant mgfp5 (line 16c) (42) were agroinoculated at the four-leaf stage with a 1:1 mix of the A. tumefaciens C58C1 cultures transformed with plasmids pTRV1 and pTRV2-tRnl or pTRV1 and pTRV2-GFP. Three to 4 weeks after agroinoculation, GFP silencing was confirmed in the plants inoculated with the A. tumefaciens mix containing pTRV2-GFP by direct observation using a UV lamp. Plants were then infiltrated with A. tumefaciens cultures transformed with pCdELVd+ or pCdCCCVd+. These cultures were also grown and induced at an OD (600 nm) of 0.5 but infiltrated at an OD of 0.05. At different time points, infiltrated tissues were harvested, RNAs purified, and ELVd and CCCVd RNAs analyzed by denaturing PAGE and Northern blot hybridization.
RESULTS
Cloning and expression of eggplant tRNA ligase and ligation of ELVd RNA.
Using primers derived from conserved sequences in the tRNA ligases from higher plants and an RNA preparation from eggplant, we amplified an 841-bp cDNA by RT-PCR that was cloned and sequenced. When translated in silico, the amino acid sequence of one of the three possible reading frames showed high similarities with the sequences of tRNA ligases from wheat (Triticum aestivum L.), Arabidopsis thaliana L., rice (Oryza sativa L.), and grapevine (Vitis vinifera L.), indicating that this cDNA most likely represented a fragment of the tRNA ligase orthologue from eggplant. The sequence of this cDNA served for designing primers to amplify new cDNAs corresponding to the remaining mRNA portions of the eggplant tRNA ligase. The sequence of this mRNA (GenBank accession no. JX025157; see Fig. S1 in the supplemental material) predicted an amino acid sequence with three conserved catalytic motifs (RNA ligase, polynucleotide kinase, and cyclic phosphodiesterase) characteristic of eukaryotic tRNA ligases (Fig. 1). Further analysis of this sequence with the ChloroP algorithm predicted an amino-terminal transit peptide to the chloroplast (Fig. 1). Previous results from transient-expression assays showed that A. thaliana and rice tRNA ligases contain functional amino-terminal transit peptides directing these proteins to the chloroplast (14). These authors proposed a mechanism of alternative translation initiation to produce in plants two different tRNA ligase isoforms, with or without the transit peptide, targeted to the chloroplasts or nuclei, respectively.
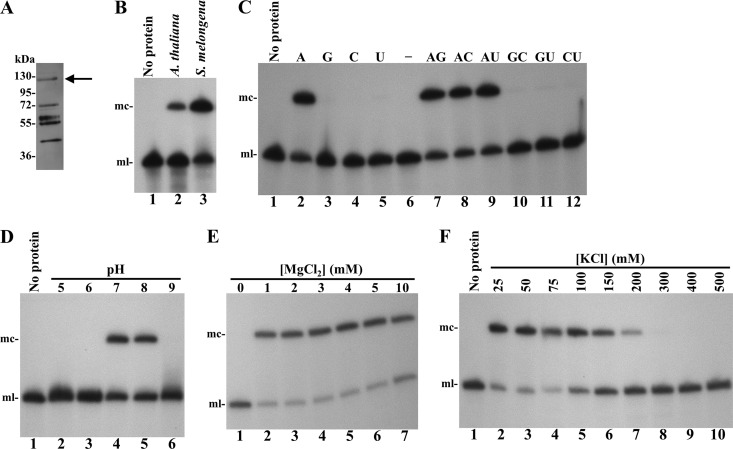
The sequence corresponding to the complete open reading frame of eggplant tRNA ligase, including the predicted transit peptide, was cloned in a vector for expressing in E. coli a recombinant version of this protein tagged at its carboxyl terminus with a His6 tail (Fig. 1). The expressed protein was purified under native conditions by affinity chromatography using a Ni-Sepharose column (Fig. 2A) and tested for its ability to ligate the monomeric linear ELVd RNA of (+) polarity with 5′-hydroxyl and 2′,3′-cyclic phosphodiester termini resulting from in vitro self-cleavage of a dimeric precursor transcript. Analysis of the reaction products, separated by denaturing PAGE and revealed by Northern blot hybridization, showed that the recombinant eggplant tRNA ligase catalyzed efficiently the circularization of the ELVd (+) RNA, similarly to a control preparation of recombinant A. thaliana tRNA ligase (14, 21) (Fig. 2B).
Fig 2.
Characterization of the ELVd RNA circularization activity of recombinant eggplant tRNA ligase. (A) Western blot analysis of an aliquot eluting from the Ni-Sepharose column. Proteins in aliquot no. 3 (elution peak) were separated by SDS-PAGE and analyzed by Western blotting using an anti-His6 antibody. The arrow points the band likely corresponding to the full-length recombinant version of eggplant tRNA ligase. The positions and molecular masses of protein standards are indicated on the left. (B to F) ELVd circularization assays with recombinant eggplant tRNA ligase. Reaction products were separated by denaturing PAGE and ELVd (+) RNAs revealed by Northern blot hybridization. (B) ELVd circularization activity of recombinant A. thaliana (lane 2) and eggplant (lane 3) tRNA ligases. (C) ELVd circularization assay of eggplant tRNA ligase in the presence of ATP, GTP, CTP, and UTP (lanes 2 to 5), with no NTP (lane 6), or in the presence of all possible combinations of two different NTPs (lanes 7 to 12). (D) ELVd circularization assay of eggplant tRNA ligase with buffers at pHs 5, 6, 7, 8, and 9 (lanes 2 to 6). (E) ELVd circularization assay of eggplant tRNA ligase in the absence (lane 1) or in the presence of increasing concentrations of MgCl2 (lanes 2 to 7). (F) ELVd circularization assay of eggplant tRNA ligase in the presence of increasing concentrations of KCl (lanes 2 to 10). In panels B, C, D, and F, lane 1 contains a negative ligation control with no protein added. The positions of monomeric circular (mc) and linear (ml) ELVd forms are indicated on the left of panels B to F.
Next, we aimed at studying the optimal conditions for ELVd circularization in our in vitro reaction system using the recombinant eggplant tRNA ligase. For this purpose, an aliquot of the tRNA ligase preparation was dialyzed and assayed for ELVd circularization under different reaction conditions. Assays in the absence of any NTP or in the presence of 1 mM each NTP or pair combinations showed a strict dependence on ATP (Fig. 2C). The optimal pH determined for eggplant tRNA ligase was quite narrow, since activity was only detected at pHs 7 and 8 (Fig. 2D). The reaction also showed stringent dependence on MgCl2, since no circularization occurred in the absence of this salt (Fig. 2E; compare lane 1 with lanes 2 to 7). Finally, the reaction was inhibited at KCl concentrations above 200 mM (Fig. 2F). From this assay, a standard ligation buffer consisting of 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 4 mM MgCl2, 5 mM dithiothreitol, and 1 mM ATP was used in subsequent experiments.
Circularization of different viroid RNAs by eggplant tRNA ligase.
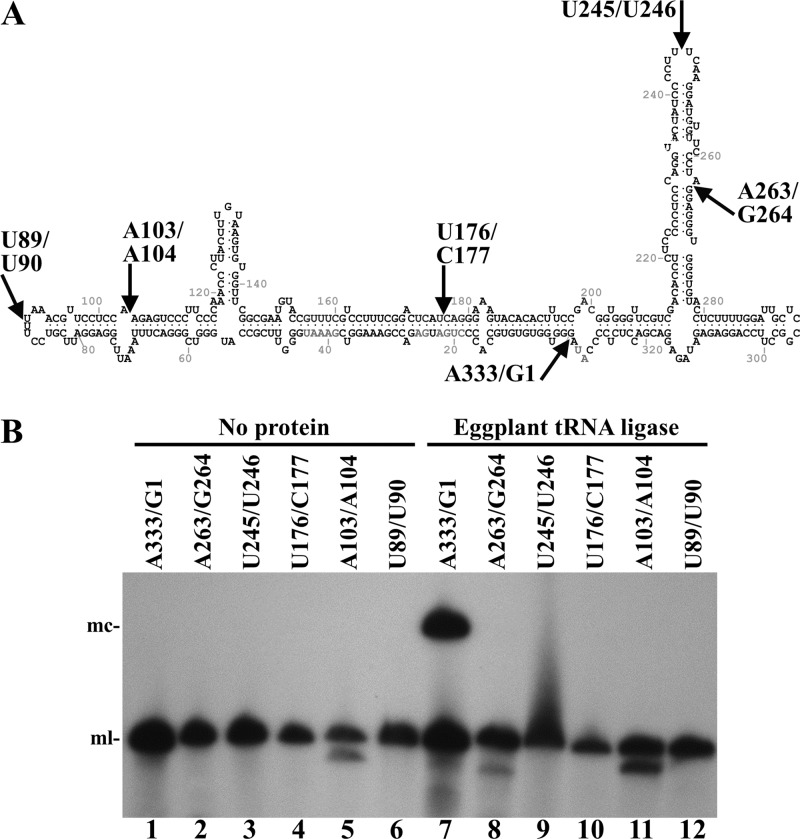
To gain support for the hypothesis that the chloroplastic isoform of plant tRNA ligase mediates RNA circularization during the replication of viroids of the family Avsunviroidae, we assayed ligation using different forms of the monomeric linear ELVd (+) RNA opened at distinct positions in the circular molecule. These substrates were obtained by self-cleavage of dimeric precursors transcript containing 5′ and 3′ engineered ribozymes and resulted in monomeric-length RNAs with 5′-hydroxyl and 2′,3′-cyclic phosphodiester termini. Together with the bona fide monomeric linear (+) ELVd replication intermediate opened between positions A333 and G1, we tested five additional RNAs opened between positions U89 and U90, A103 and A104, U176 and C177, U245 and U246, and A263 and G264, mapping at double-stranded regions, bulges, and loops in the ELVd (+) minimum free-energy conformation (Fig. 3A). Interestingly, eggplant tRNA ligase only circularized the physiological monomeric linear (+) ELVd (A333-G1) (Fig. 3B). This result indicates a high specificity of the eggplant enzyme for the genuine ELVd replication intermediate that may reflect the situation existing in vivo.
Fig 3.
Circularization by eggplant tRNA ligase of different monomeric linear ELVd (+) RNAs opened at different sites. (A) ELVd folded in the predicted minimum free-energy conformation. Arrows indicate positions in which the different linear ELVd substrates subjected to ligation were opened. (B) ELVd circularization assay with recombinant eggplant tRNA ligase. Reaction products were separated by denaturing PAGE and ELVd (+) RNAs revealed by Northern blot hybridization. Lanes 1 to 6, controls with no protein added. Lanes 7 to 12, eggplant tRNA ligase added. Substrate RNAs were opened between positions A333 and G1 (lanes 1 and 7), A263 and G264 (lanes 2 and 8), U245 and U246 (lanes 3 and 9), U176 and C177 (lanes 4 and 10), A103 and A104 (lanes 5 and 11), and U89 and U90 (lanes 6 and 12). The positions of monomeric circular (mc) and linear (ml) ELVd forms are indicated on the left.
Next, we examined the ability of eggplant tRNA ligase to mediate circularization of the physiological monomeric (+) and (−) linear RNAs (resulting from self-cleavage of dimeric transcripts by the embedded hammerhead ribozymes) from the other viroid species within the family Avsunviroidae. The analysis also included the monomeric linear (−) and (+) ELVd RNAs, the latter as a control for comparative purposes. In these ligation assays, we replaced ATP with [γ-32P]ATP. The rationale for this approach is that in the reaction catalyzed by the tRNA ligase, the γ phosphoryl group of an ATP molecule is first transferred to the 5′-hydroxyl terminus of the substrate to produce a 5′-phosphomonoester terminus, with this phosphoryl group finally forming the 3′,5′-phosphodiester linkage resulting from ligation. The linkage also includes a 2′-phosphomonoester resulting from opening the 2′,3′-cyclic phosphodiester termini. The important point is that under these assay conditions, both the monomeric linear and circular forms of all viroid RNAs may become labeled and be easily identified and compared.
Analysis of the products of the ligation reactions by denaturing PAGE and autoradiography showed that both the monomeric linear and circular RNAs of all viroids became labeled for both polarities (Fig. 4). Comparison of the intensities of the different bands with those arising from the ELVd (+) control indicated that ligation was quite efficient in all cases (Fig. 4; compare lanes 1 to 6 and 8 with lane 7). Even if some differences could be appreciated depending on the particular viroid and the polarity of the strand subjected to ligation (Fig. 4), this result is consistent with the chloroplastic isoform of tRNA ligase catalyzing the circularization of both (+) and (−) monomeric linear intermediates during replication of the Avsunviroidae occurring through a symmetric rolling-circle mechanism.
Fig 4.
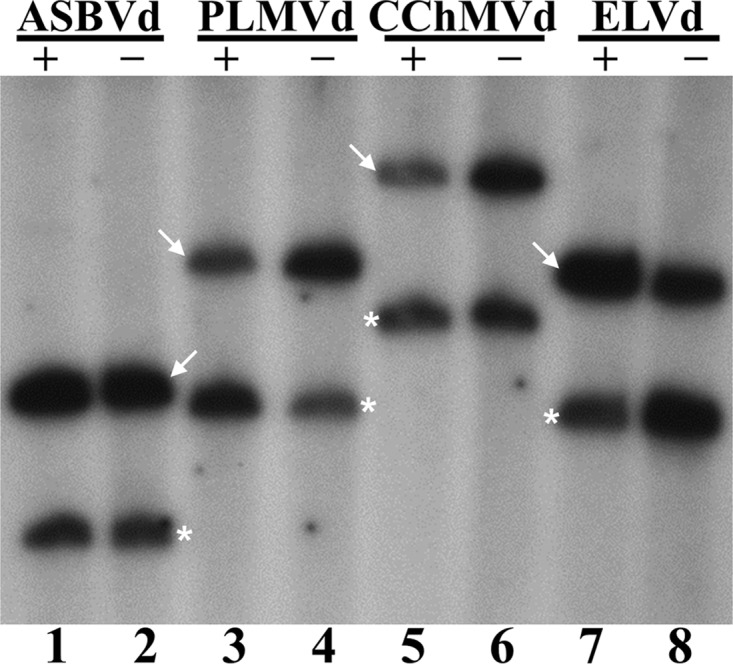
Circularization by the recombinant eggplant tRNA ligase of self-cleavage (+) and (−) monomeric linear ASBVd (lanes 1 and 2), PLMVd (lanes 3 and 4), CChMVd (lanes 5 and 6), and ELVd (lanes 7 and 8) RNAs. Reaction products obtained in the presence of [γ-32P]ATP were separated by denaturing PAGE, and the gel was autoradiographed. The positions of monomeric circular and linear forms of the different viroid RNAs are indicated by an arrow and an asterisk, respectively.
Evidence in vivo for the involvement of eggplant tRNA ligase in ELVd circularization.
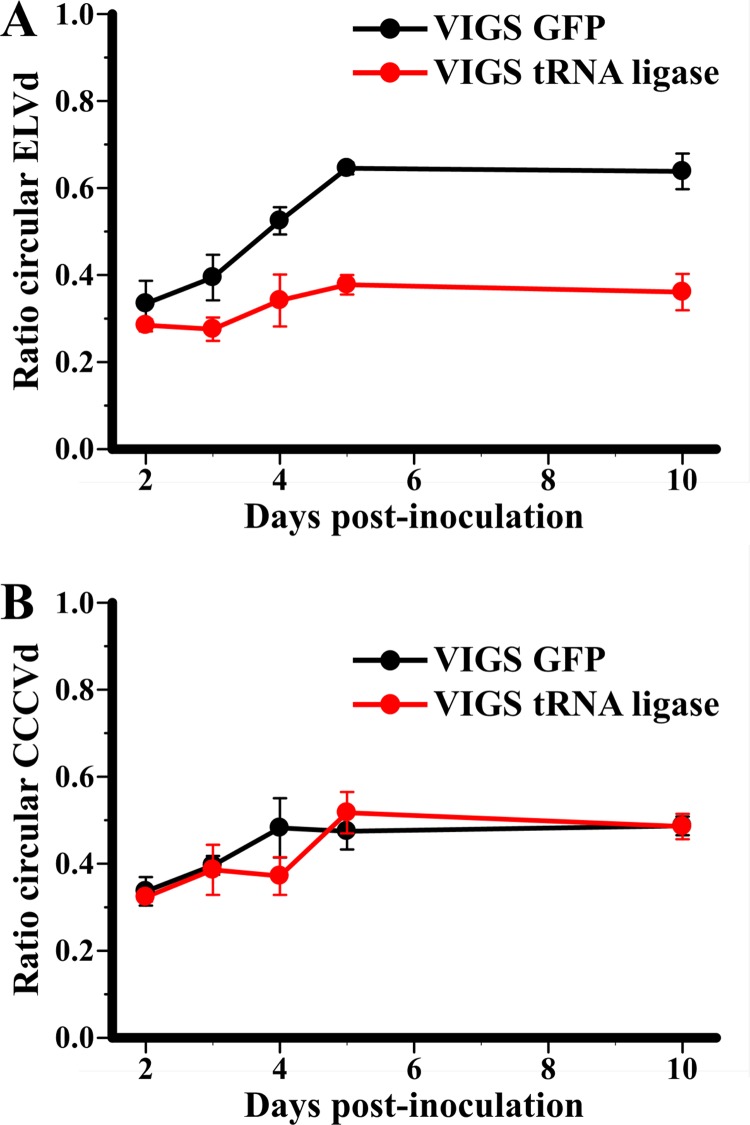
To provide further support for the role of the chloroplastic isoform of tRNA ligase in the replication of the Avsunviroidae, we studied RNA circularization in N. benthamiana plants transiently expressing dimeric ELVd transcripts. Initial attempts to adapt a VIGS strategy to eggplant were unsuccessful, and although N. benthamiana is not a host for ELVd or any other known viroid within the family Avsunviroidae, ELVd dimeric transcripts are processed properly when expressed transiently in this plant. Moreover, an ELVd-derived RNA fused to a GFP mRNA has been shown to traffic into the chloroplast when transiently expressed in N. benthamiana (22). Dimeric transcripts of a viroid replicating in the nucleus (CCCVd, family Pospiviroidae) served as a control in this experiment because when transiently expressed in N. benthamiana, they are also processed into monomeric linear and circular RNAs (J. Marqués and J.-A. Daròs, unpublished results), but through a nuclear pathway involving different enzymes (20, M.-A. Nohales, R. Flores, and J.-A. Daròs, submitted for publication). Moreover, similarly to what occurs with ELVd, N. benthamiana is a nonhost for CCCVd. In these experiments, we used an N. benthamiana transgenic line (16c) constitutively expressing GFP (42) and a vector based on tobacco rattle virus (TRV) (30) for VIGS of the endogenous tRNA ligase or the transgenic GFP, the latter serving as a control and silencing marker. Plants preinoculated with the TRV-derived VIGS vectors were agroinfiltrated with A. tumefaciens cultures transformed with plasmids for expressing dimeric (+) ELVd or CCCVd transcripts. RNA preparations from the agroinfiltrated areas, harvested at several days postinfiltration, were analyzed by denaturing PAGE followed by Northern blot hybridization (see Fig. S2 in the supplemental material). Time course analysis of ELVd processing showed that the ratio of monomeric circular to total monomeric (circular plus linear) ELVd (+) RNA was significantly lower in tissues preinoculated with the VIGS vector to silence the tRNA ligase than in tissues preinoculated with the control VIGS vector to silence GFP (Fig. 5A). This difference was not observed following transient expression of dimeric CCCVd (+) RNA in the same plants preinoculated with the same VIGS vectors (Fig. 5B). This result supports the hypothesis that ELVd circularization catalyzed by tRNA ligase occurs not only in vitro but also in vivo.
Fig 5.
Effect of tRNA ligase silencing on circularization of ELVd (A) and CCCVd (B) monomeric RNAs. Dimeric (+) ELVd and CCCVd transcripts were infiltrated in N. benthamiana 16c plants preinoculated with a VIGS vector to silence the GFP transgene or the endogenous tRNA ligase. RNAs were purified at different days from the agroinfiltrated areas of three different plants infected with each VIGS vector. Monomeric circular and linear (+) ELVd and CCCVd RNAs were revealed by Northern blot hybridization and quantified by phosphorimetry. The plots represent the average ratio of monomeric circular to total monomeric (circular plus linear (+) ELVd (A) and CCCVd (B) RNAs at different days postinfiltration in GFP-silenced plants (black symbols) and in tRNA ligase-silenced plants (red symbols). Error bars indicate the standard deviations of the triplicate measurements.
DISCUSSION
RNA transcription during viroid replication through a rolling-circle mechanism produces oligomeric viroid strands (4). In members of the family Pospiviroidae, only the (+) oligomeric RNAs are processed to monomers (3). This processing has been proposed to occur through a conserved mechanism in which the capping loops of two hairpin I motifs (formed by sequences in the upper CCR strand and flanking inverted repeats) of two contiguous units in the oligomeric RNA intermediate interact (kissing loops) and promote the adoption of a palindromic quasi-double-stranded structure that would be the substrate of a host type III RNase (20). The resulting monomeric linear viroid intermediate with 5′-phosphomonoester and 3′-hydroxyl termini (21) would be ultimately circularized by the host DNA ligase 1 redirected to act as an RNA ligase (Nohales et al., submitted).
In members of the family Avsunviroidae, oligomeric viroid RNAs of both (+) and (−) polarities self-cleave to monomers through the embedded hammerhead ribozymes (9, 15, 23, 26, 35). Host RNA chaperones may facilitate this reaction in vivo (8). Here we have shown that the chloroplastic isoform of eggplant tRNA ligase most likely mediates circularization of both (+) and (−) monomeric linear RNAs during replication of ELVd. This proposal is supported by several observations. First, a recombinant version of this enzyme catalyzes efficient in vitro circularization of the monomeric linear ELVd (+) RNA, resulting from the hammerhead ribozyme self-cleavage of a head-to-tail dimeric transcript (mimicking the replication intermediates in vivo), but not of five other monomeric-length linear ELVd (+) RNAs with their ends mapping at different sites along the molecule, despite having the same 5′-hydroxyl and 2′,3′-cyclic phosphodiester terminal groups (Fig. 3). Second, the recombinant eggplant tRNA ligase efficiently catalyzes in vitro circularization of the ELVd (−) monomeric RNA replication intermediate resulting from self-cleavage (Fig. 4). And third, the ratio of monomeric circular to total monomeric ELVd (+) RNA decreased when dimeric (+) ELVd transcripts were transiently expressed in N. benthamiana plants preinoculated with a VIGS vector to induce silencing of endogenous tRNA ligase, in contrast to the situation observed with CCCVd, a member of the family Pospiviroidae, with a different processing pathway (Fig. 5).
Previous data showed efficient ELVd circularization in C. reinhardtii expressing dimeric ELVd (+) transcripts in the chloroplast (33). This result can now be interpreted as a circularization mediated by the chloroplastic tRNA ligase of this green alga. The specificity reported here regarding the ligation site is also in accordance with the circularization of ELVd RNA in C. reinhardtii chloroplasts demanding a quasi-double-stranded structure in the middle of the molecule wherein the ligation site maps at a short loop motif (33).
The recombinant eggplant tRNA ligase also mediates the efficient circularization in vitro of the (+) and (−) self-cleavage monomeric linear RNAs of ASBVd, PLMVd, and CChMVd, the other three components of the family Avsunviroidae (Fig. 4), thus supporting the involvement in replication of the tRNA ligase homologues from their corresponding hosts. In the case of PLMVd, the monomeric linear RNAs resulting from self-cleavage self-ligates in vitro in the presence of Mg2+ through a reaction which is not the reverse of the cleavage by the hammerhead ribozyme, because a 2′,5′-phosphodiester bond is produced (7). The monomeric linear ELVd (+) RNA resulting from self-cleavage also shows the same behavior (D. Molina-Serrano, R. Flores, and J.-A. Daròs, unpublished results). However, despite the detection of some circular PLMVd RNA forms locked through a 2′,5′-phosphodiester linkage in infected peach tissue (6), it is unlikely that this is the main circularization mechanism during replication in the family. First, and similar to what occurs with the hammerhead ribozyme reverse reaction, the efficiency of this reaction is very low compared to that of the tRNA ligase-mediated circularization. And second, reverse transcription analysis of the monomeric circular ASBVd and ELVd (+) RNAs isolated from infected tissues indicates that they are mainly closed through typical 3′,5′-phosphodiester bonds (15, 34). Incidentally, this observation makes also unlikely the involvement of a recently described chloroplastic RNA ligase activity analogous to the bacterial and archaeal 2′-5′ RNA ligase (34).
RNA ligation mediated by plant tRNA ligases produces a 3′,5′-phosphodiester, 2′-phosphomonoester junction (13). However, the 2′-phosphomonoester group seems to be absent in mature ASBVd and ELVd circular RNAs accumulating in infected tissues (15, 34). In contrast, the presence of a 2′-phosphomonoester group was previously reported at the ligation site of two viroid-like satellite RNAs and considered to be a signature of circularization by an RNA ligase (28). RT-PCR amplification of circular PLMVd and CChMVd RNAs accumulating in infected tissues suggests that a minor fraction of the molecules have an anomalous ligation site (2, 11), which may arise from a 2′-phosphomonoester group remaining from a tRNA ligase-mediated circularization or, less likely, from a 2′,5′-phosphodiester linkage. Removal of the 2′-phosphomonoester after viroid circularization may result from the activity of a 2′-phosphotransferase, similar to what occurs during eukaryotic tRNA maturation (1, 46). Interestingly, A. thaliana and rice 2′-phosphotransferases also contain amino-terminal transit peptides and have been shown to target the chloroplast during transient expression (14).
Finally, viroids belonging to the family Avsunviroidae are characterized for their restricted host range, since they only infect the plants in which they were initially discovered and a few phylogenetically closely related species (15, 17). Our finding that the chloroplastic isoform of eggplant tRNA ligase efficiently mediates in vitro circularization of the genuine monomeric RNAs of both polarities of all Avsunviroidae implies that the remarkable host specificity of this family does not result from this replication step in the infectious cycle.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Ministerio de Ciencia e Innovación (MICINN) from Spain through grants BIO2008-01986, BIO2011-26741, and BFU2008-03154. M. A. Nohales and D. Molina-Serrano were the recipients of predoctoral fellowships from the Spanish Ministerio de Educación y Ciencia.
We thank Verónica Aragonés for excellent technical assistance. We thank Aaron J. Plys (Department of Biological Sciences, University of Cyprus, Nicosia) for critical reading of the manuscript.
Footnotes
Published ahead of print 23 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abelson J, Trotta CR, Li H. 1998. tRNA splicing. J. Biol. Chem. 273:12685–12688 [DOI] [PubMed] [Google Scholar]
- 2. Ambrós S, Hernández C, Desvignes JC, Flores R. 1998. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasispecies. J. Virol. 72:7397–7406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Branch AD, Benenfeld BJ, Robertson HD. 1988. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U. S. A. 85:9128–9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Branch AD, Robertson HD. 1984. A replication cycle for viroids and other small infectious RNAs. Science 223:450–455 [DOI] [PubMed] [Google Scholar]
- 5. Canny MD, Jucker FM, Pardi A. 2007. Efficient ligation of the Schistosoma hammerhead ribozyme. Biochemistry 46:3826–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Côté F, Lévesque D, Perreault JP. 2001. Natural 2′,5′-phosphodiester bonds found at the ligation sites of peach latent mosaic viroid. J. Virol. 75:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Côté F, Perreault JP. 1997. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J. Mol. Biol. 273:533–543 [DOI] [PubMed] [Google Scholar]
- 8. Daròs JA, Flores R. 2002. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daròs JA, Marcos JF, Hernández C, Flores R. 1994. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. U. S. A. 91:12813–12817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De la Peña M, Gago S, Flores R. 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22:5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de la Peña M, Navarro B, Flores R. 1999. Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation. Proc. Natl. Acad. Sci. U. S. A. 96:9960–9965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding B. 2009. The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47:105–131 [DOI] [PubMed] [Google Scholar]
- 13. Englert M, Beier H. 2005. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 33:388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Englert M, et al. 2007. Plant pre-tRNA splicing enzymes are targeted to multiple cellular compartments. Biochimie 89:1351–1365 [DOI] [PubMed] [Google Scholar]
- 15. Fadda Z, Daròs JA, Fagoaga C, Flores R, Duran-Vila N. 2003. Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). J. Virol. 77:6528–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feldstein PA, Hu Y, Owens RA. 1998. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc. Natl. Acad. Sci. U. S. A. 95:6560–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flores R, Daròs JA, Hernández C. 2000. The Avsunviroidae family: viroids containing hammerhead ribozymes. Adv. Virus Res. 55:271–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flores R, Hernández C, Martínez de Alba AE, Daròs JA, Di Serio F. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117–139 [DOI] [PubMed] [Google Scholar]
- 19. Flores R, Owens RA. 2008. Viroids, p 332–342 In Mahy BWJ, Van Regenmortel MHV. (ed), Encyclopedia of virology. Elsevier, Oxford, United Kingdom [Google Scholar]
- 20. Gas ME, Hernández C, Flores R, Daròs JA. 2007. Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog. 3:1813–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gas ME, Molina-Serrano D, Hernández C, Flores R, Daròs JA. 2008. Monomeric linear RNA of citrus exocortis viroid resulting from processing in vivo has 5′-phosphomonoester and 3′-hydroxyl termini: implications for the RNase and RNA ligase involved in replication. J. Virol. 82:10321–10325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gómez G, Pallás V. 2010. Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS One 5:e12269 doi:10.1371/journal.pone.0012269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hernández C, Flores R. 1992. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc. Natl. Acad. Sci. U. S. A. 89:3711–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hertel KJ, Herschlag D, Uhlenbeck OC. 1994. A kinetic and thermodynamic framework for the hammerhead ribozyme reaction. Biochemistry 33:3374–3385 [DOI] [PubMed] [Google Scholar]
- 25. Hutchins CJ, et al. 1985. Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol. 4:293–304 [DOI] [PubMed] [Google Scholar]
- 26. Hutchins CJ, Rathjen PD, Forster AC, Symons RH. 1986. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14:3627–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khvorova A, Lescoute A, Westhof E, Jayasena SD. 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 10:708–712 [DOI] [PubMed] [Google Scholar]
- 28. Kiberstis PA, Haseloff J, Zimmern D. 1985. 2′ phosphomonoester, 3′-5′ phosphodiester bond at a unique site in a circular viral RNA. EMBO J. 4:817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Konarska M, Filipowicz W, Domdey H, Gross HJ. 1981. Formation of a 2′-phosphomonoester, 3′,5′-phosphodiester linkage by a novel RNA ligase in wheat germ. Nature 293:112–116 [DOI] [PubMed] [Google Scholar]
- 30. Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. 2002. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30:415–429 [DOI] [PubMed] [Google Scholar]
- 31. Makino S, Sawasaki T, Endo Y, Takai K. 2005. Purification and sequence determination of an RNA ligase from wheat embryos. Nucleic Acids Symp. Ser. 2005:319–320 [DOI] [PubMed] [Google Scholar]
- 32. Marcos JF, Flores R. 1993. The 5′ end generated in the in vitro self-cleavage reaction of avocado sunblotch viroid RNAs is present in naturally occurring linear viroid molecules. J. Gen. Virol. 74:907–910 [DOI] [PubMed] [Google Scholar]
- 33. Martínez F, Marqués J, Salvador ML, Daròs JA. 2009. Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. J. Gen. Virol. 90:3057–3065 [DOI] [PubMed] [Google Scholar]
- 34. Molina-Serrano D, Marqués J, Nohales MA, Flores R, Daròs JA. 2012. A chloroplastic RNA ligase activity analogous to the bacterial and archaeal 2–5′ RNA ligase. RNA Biol. 9:326–333 [DOI] [PubMed] [Google Scholar]
- 35. Navarro B, Flores R. 1997. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc. Natl. Acad. Sci. U. S. A. 94:11262–11267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Navarro JA, Daròs JA, Flores R. 1999. Complexes containing both polarity strands of avocado sunblotch viroid: identification in chloroplasts and characterization. Virology 253:77–85 [DOI] [PubMed] [Google Scholar]
- 37. Navarro JA, Vera A, Flores R. 2000. A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology 268:218–225 [DOI] [PubMed] [Google Scholar]
- 38. Nelson JA, Shepotinovskaya I, Uhlenbeck OC. 2005. Hammerheads derived from sTRSV show enhanced cleavage and ligation rate constants. Biochemistry 44:14577–14585 [DOI] [PubMed] [Google Scholar]
- 39. Pick L, Furneaux H, Hurwitz J. 1986. Purification of wheat germ RNA ligase. II. Mechanism of action of wheat germ RNA ligase. J. Biol. Chem. 261:6694–6704 [PubMed] [Google Scholar]
- 40. Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. 1986. Autolytic processing of dimeric plant-virus satellite RNA. Science 231:1577–1580 [DOI] [PubMed] [Google Scholar]
- 41. Rodio ME, et al. 2007. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 19:3610–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruiz MT, Voinnet O, Baulcombe DC. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schürer H, Lang K, Schuster J, Mörl M. 2002. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 30:e56 doi:10.1093/nar/gnf055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwartz RC, Greer CL, Gegenheimer P, Abelson J. 1983. Enzymatic mechanism of an RNA ligase from wheat germ. J. Biol. Chem. 258:8374–8383 [PubMed] [Google Scholar]
- 45. Tsagris EM, Martínez de Alba AE, Gozmanova M, Kalantidis K. 2008. Viroids. Cell. Microbiol. 10:2168–2179 [DOI] [PubMed] [Google Scholar]
- 46. Wang LK, Shuman S. 2005. Structure-function analysis of yeast tRNA ligase. RNA 11:966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.