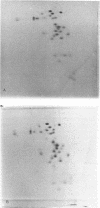
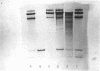
Abstract
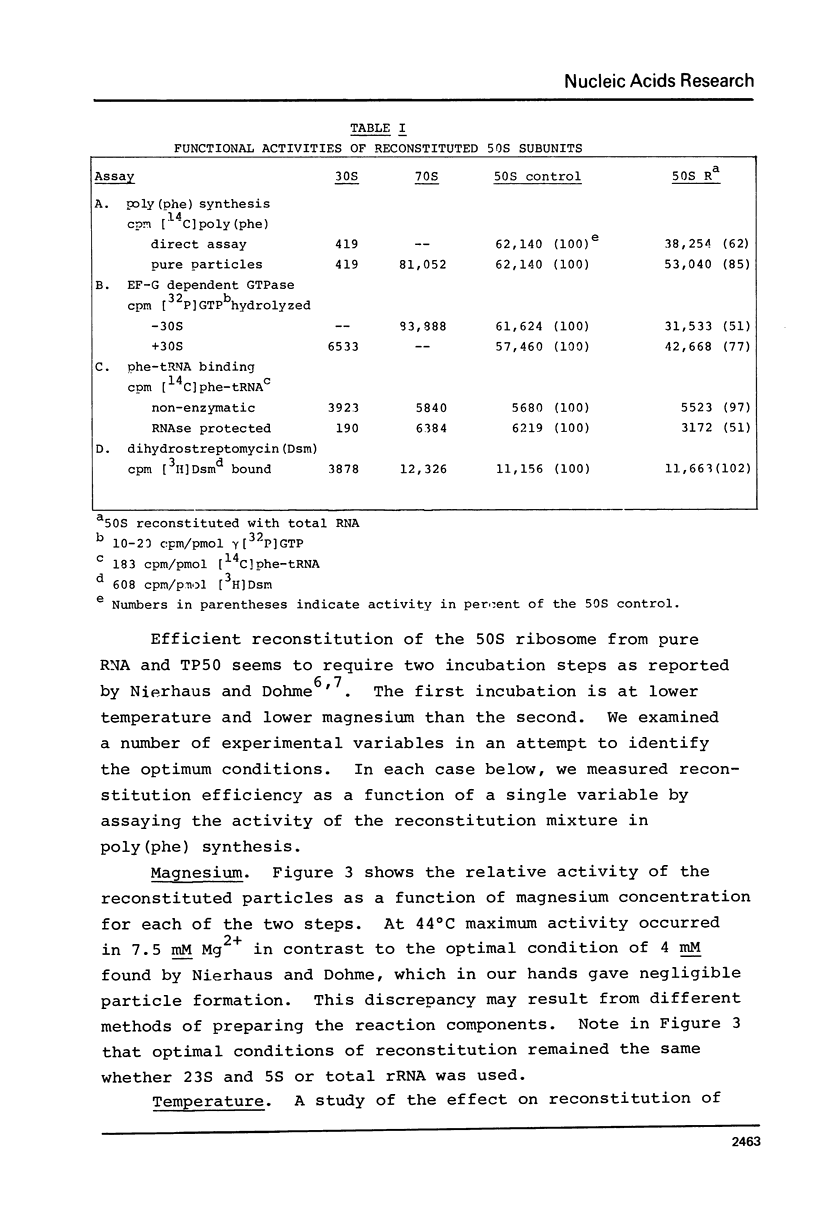
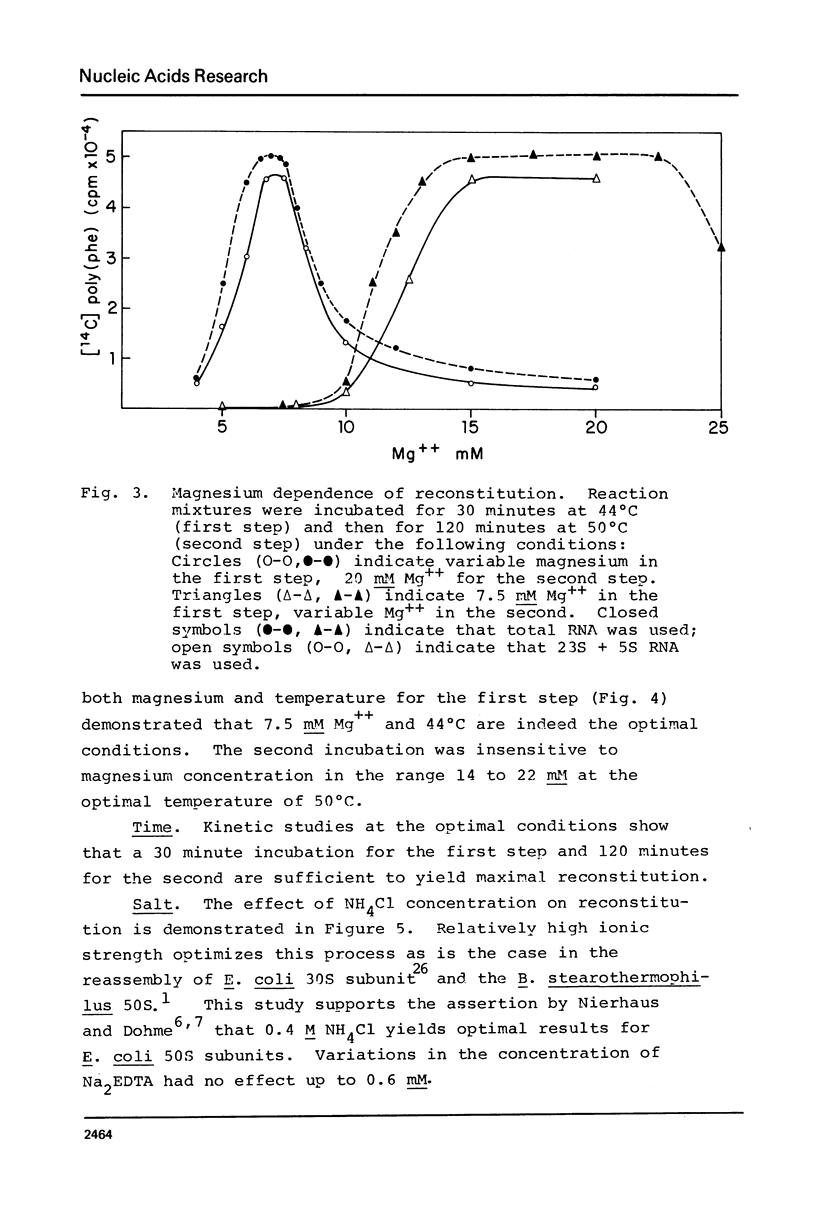
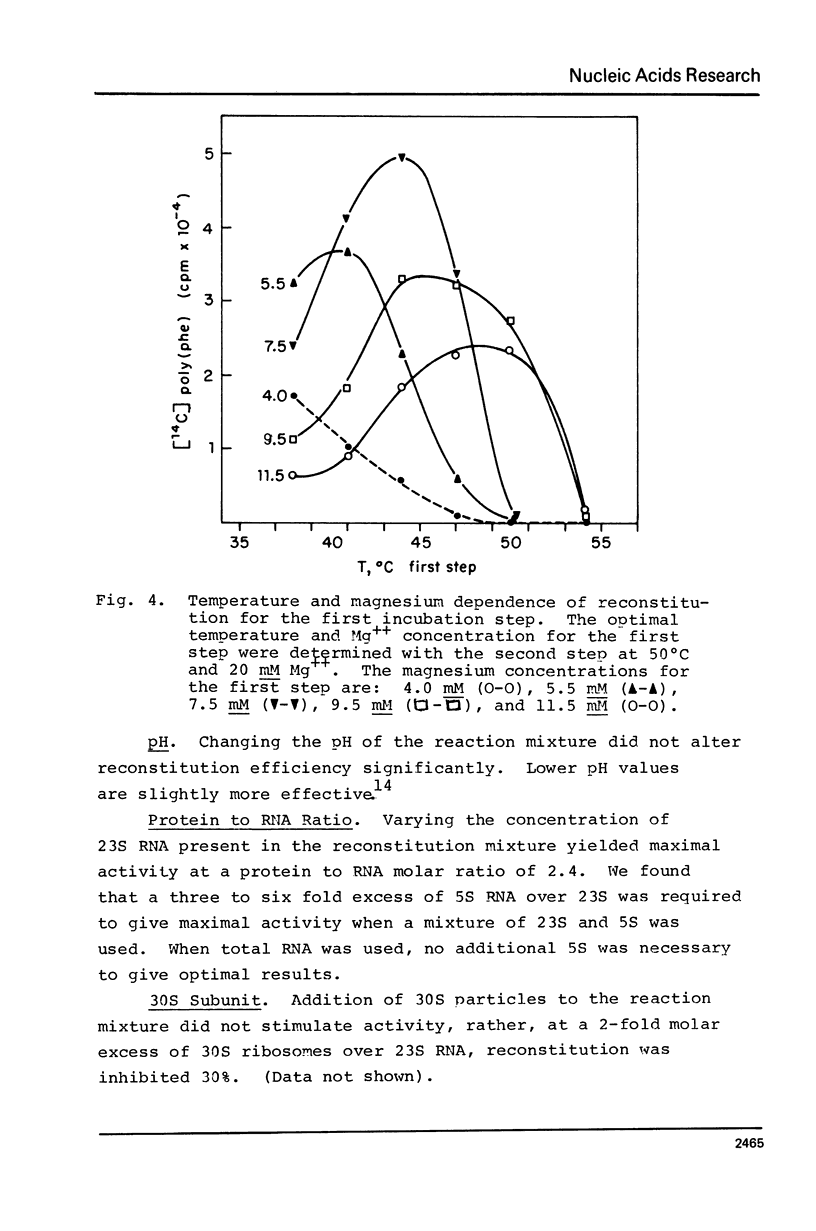
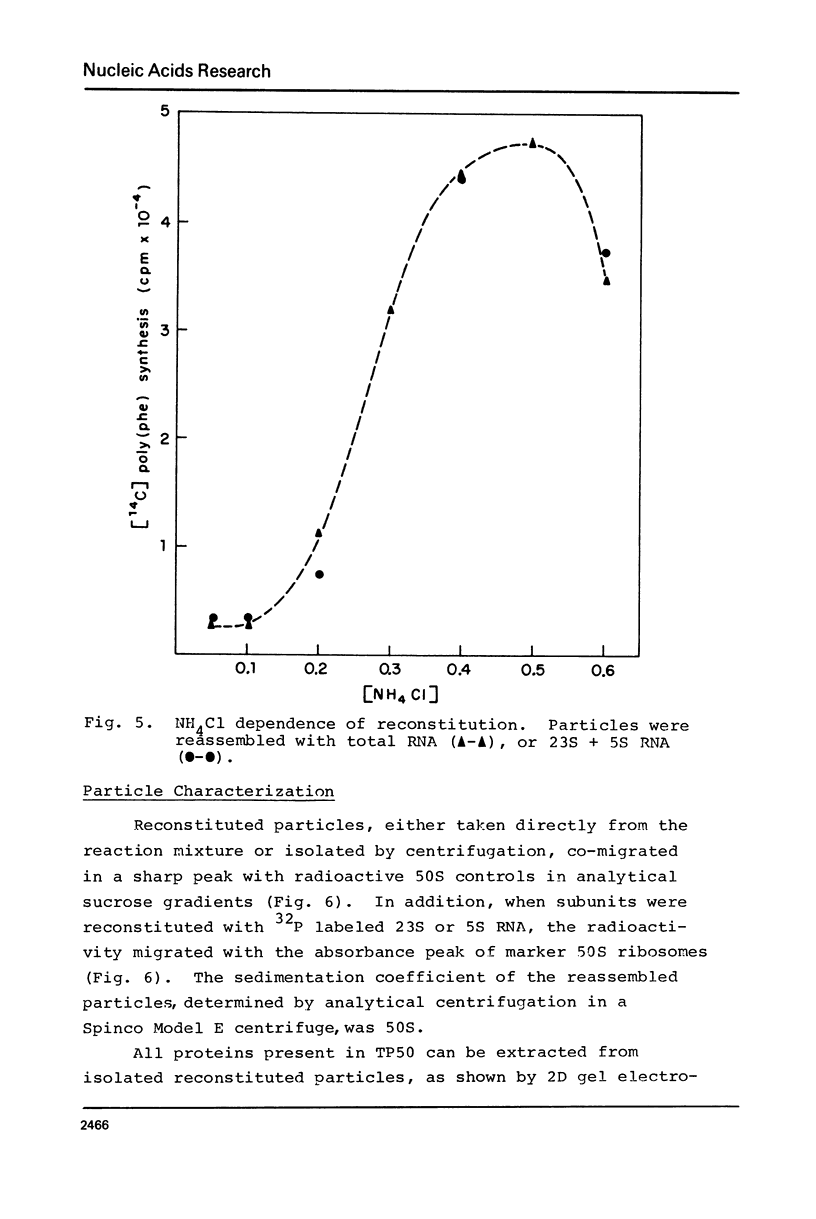
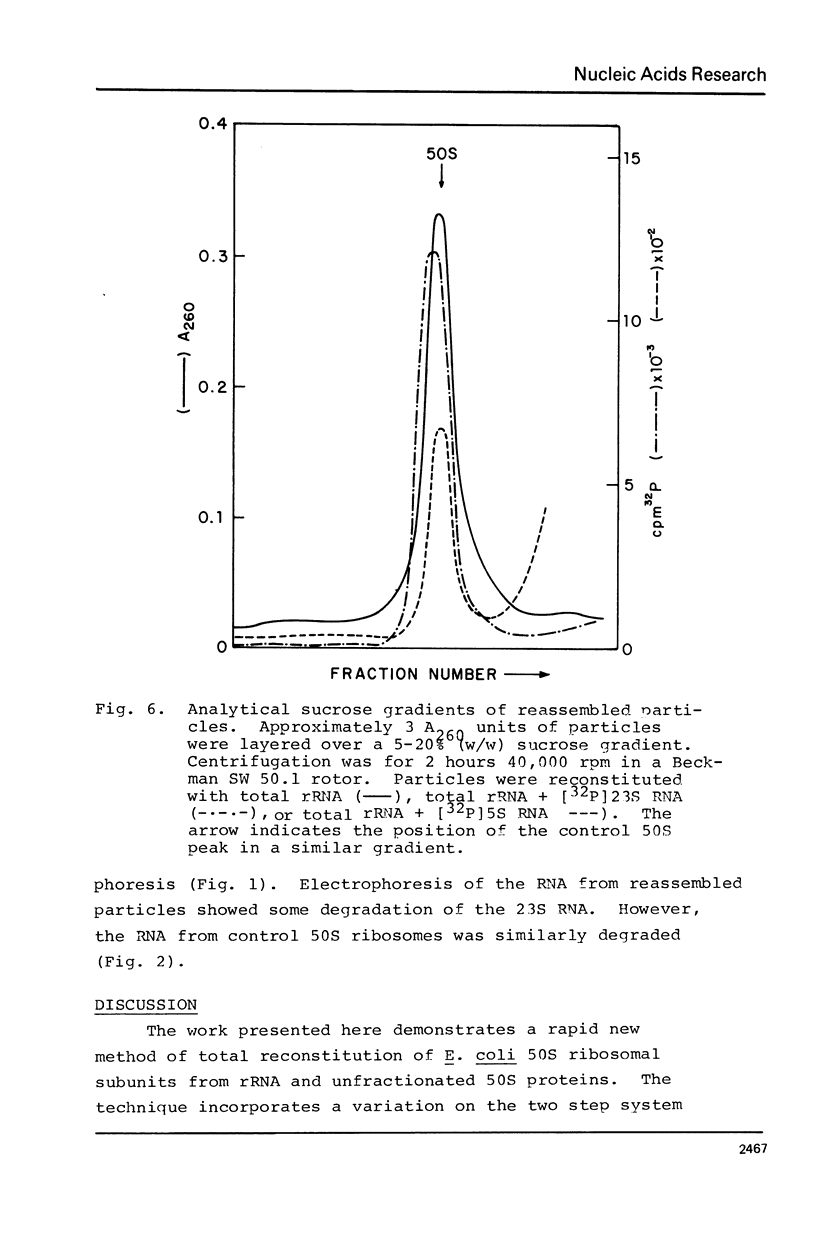
A new, relatively simple technique for the total invitro reconstitution of E. coli 50S ribosomes has been developed. It is a two-step procedure like that previously reported by Nierhaus and Dohme [Proc. Natl. Acad. Sci. 71, 4713 (1974)], but it differs in a number of important aspects. Ribosomal RNA is prepared by direct phenol extraction of 70S particles to minimize nuclease fragmentation. A mixture of 50S proteins is prepared by acetic acid extraction and immediate removal of the acetic acid by thin film dialysis. The resulting protein mixture is soluble and stable. Separate RNA and protein fractions are mixed, incubated first at 44°C in 7.5 mM Mg2+, and then at 50°C in 20 mM Mg2+. The resulting 50S particles comigrate with native 50S particles in analytical gradients. They range from 50 to 100% active in five different functional assays. This is a fairly stringent test of the effectiveness of reconstitution since 50S particles derived from highly active vacant couples were used as a control.
Full text
PDF



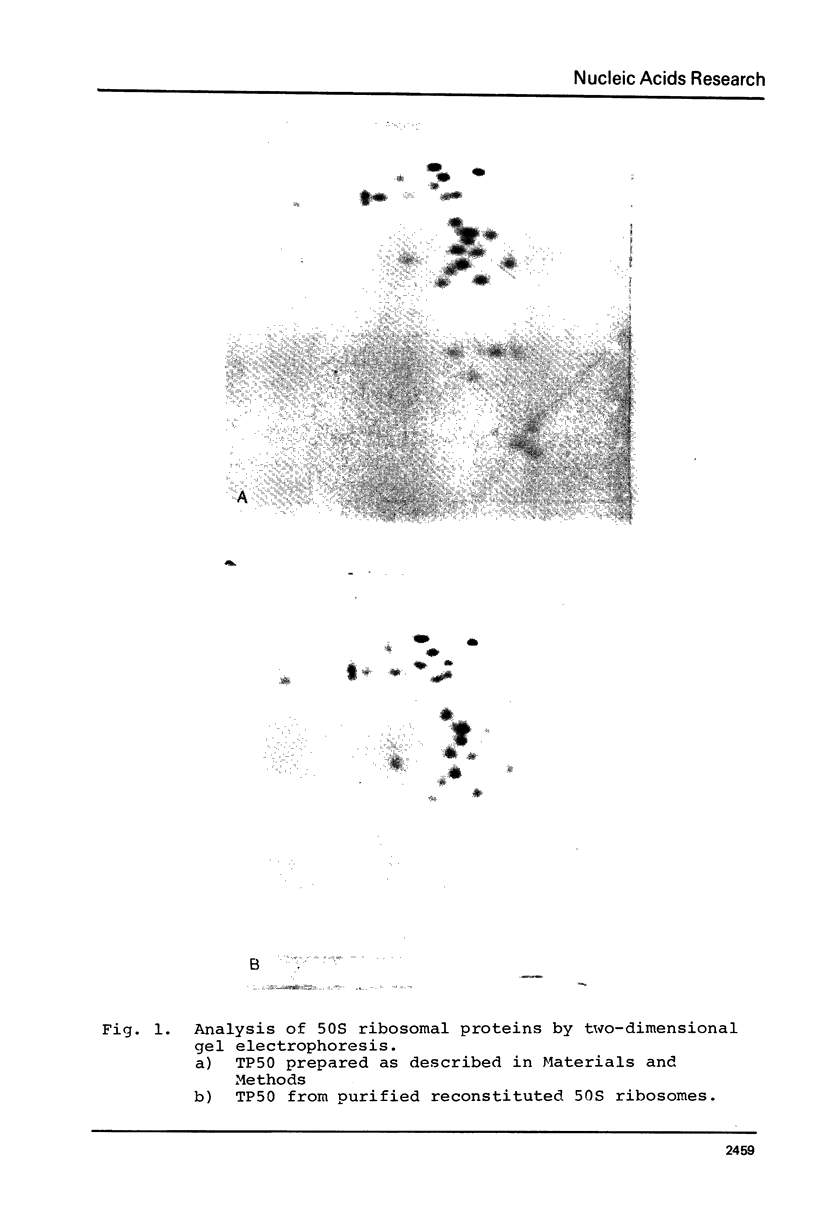

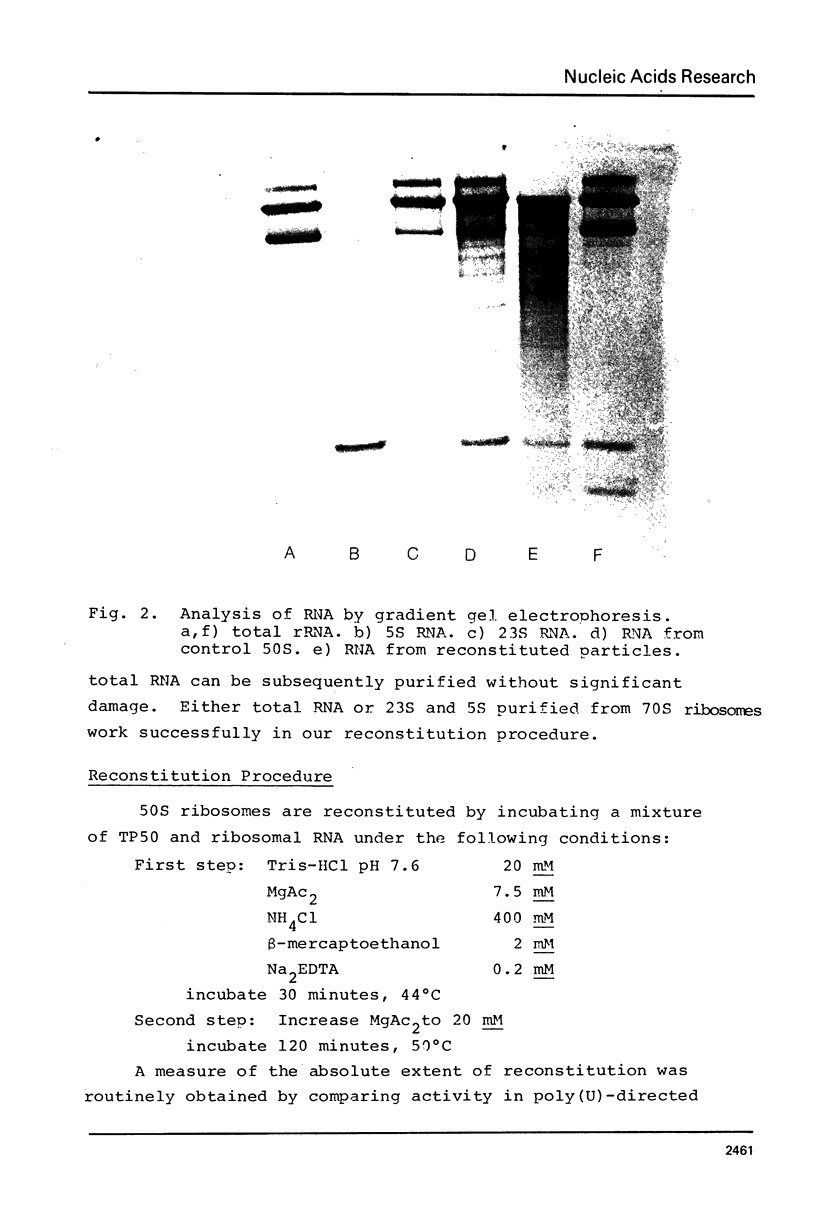




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai N., Kaziro Y. Mechanism of the ribosome-dependent uncoupled GTPase reaction catalyzed by polypeptide chain elongation factor G. J Biochem. 1975 Feb;77(2):439–447. doi: 10.1093/oxfordjournals.jbchem.a130743. [DOI] [PubMed] [Google Scholar]
- Chang F. N., Flaks J. G. Binding of dihydrostreptomycin to Escherichia coli ribosomes: characteristics and equilibrium of the reaction. Antimicrob Agents Chemother. 1972 Oct;2(4):294–307. doi: 10.1128/aac.2.4.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Maeba P. Y. Physical reconstitution of 23 S RNA-50 S protein complexes from Escherichia coli. Can J Biochem. 1973 Feb;51(2):129–139. doi: 10.1139/o73-018. [DOI] [PubMed] [Google Scholar]
- Cohlberg J. A., Nomura M. Reconstitution of Bacillus stearothermophilus 50 S ribosomal subunits from purified molecular components. J Biol Chem. 1976 Jan 10;251(1):209–221. [PubMed] [Google Scholar]
- Dohme F., Nierhaus K. H. Total reconstitution and assembly of 50 S subunits from Escherichia coli Ribosomes in vitro. J Mol Biol. 1976 Nov 15;107(4):585–599. doi: 10.1016/s0022-2836(76)80085-x. [DOI] [PubMed] [Google Scholar]
- Eikenberry E. F., Bickle T. A., Traut R. R., Price C. A. Separation of large quantities of ribosomal subunits by zonal ultracentrifugation. Eur J Biochem. 1970 Jan;12(1):113–116. doi: 10.1111/j.1432-1033.1970.tb00827.x. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A., Doberer H. G. Structure and function of 5S RNA: the role of the 3' terminus in 5S RNA function. Mol Gen Genet. 1972;114(2):89–94. doi: 10.1007/BF00332779. [DOI] [PubMed] [Google Scholar]
- Fahnestock S. R. Identification of homologues of a functionally important 50 S ribosomal protein in different bacterial species. Arch Biochem Biophys. 1977 Apr 30;180(2):555–561. doi: 10.1016/0003-9861(77)90073-x. [DOI] [PubMed] [Google Scholar]
- Hapke B., Noll H. Structural dynamics of bacterial ribosomes. IV. Classification of ribosomes by subunit interaction. J Mol Biol. 1976 Jul 25;105(1):97–109. doi: 10.1016/0022-2836(76)90196-0. [DOI] [PubMed] [Google Scholar]
- Hardy S. J., Kurland C. G., Voynow P., Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry. 1969 Jul;8(7):2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- Howard G. A., Traut R. R. Separation and radioautography of microgram quantities of ribosomal proteins by two-dimensional polyacrylamide gel electrophoresis. FEBS Lett. 1973 Jan 15;29(2):177–180. doi: 10.1016/0014-5793(73)80555-1. [DOI] [PubMed] [Google Scholar]
- Huang K. H., Fairclough R. H., Cantor C. R. Singlet energy transfer studies of the arrangement of proteins in the 30 S Escherichia coli ribosome. J Mol Biol. 1975 Oct 5;97(4):443–470. doi: 10.1016/s0022-2836(75)80053-2. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt E., Wittmann H. G. Ribosomal proteins. VII. Two-dimensional polyacrylamide gel electrophoresis for fingerprinting of ribosomal proteins. Anal Biochem. 1970 Aug;36(2):401–412. doi: 10.1016/0003-2697(70)90376-3. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Maruta H., Tsuchiya T., Mizuno D. In vitro reassembly of functionally active 50 s ribosomal particles from ribosomal proteins and RNA's of Escherichia coli. J Mol Biol. 1971 Oct 14;61(1):123–134. doi: 10.1016/0022-2836(71)90210-5. [DOI] [PubMed] [Google Scholar]
- Moore P. B., Langer J. A., Schoenborn B. P., Engelman D. M. Triangulation of proteins in the 30 S ribosomal subunit of Exherichia coli. J Mol Biol. 1977 May 15;112(2):199–227. doi: 10.1016/s0022-2836(77)80139-3. [DOI] [PubMed] [Google Scholar]
- NIRENBERG M., LEDER P. RNA CODEWORDS AND PROTEIN SYNTHESIS. THE EFFECT OF TRINUCLEOTIDES UPON THE BINDING OF SRNA TO RIBOSOMES. Science. 1964 Sep 25;145(3639):1399–1407. doi: 10.1126/science.145.3639.1399. [DOI] [PubMed] [Google Scholar]
- Nierhaus K. H., Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y., Lipmann F. The interrelationship between guanosine triphosphatase and amino acid polymerization. Arch Biochem Biophys. 1966 Sep 26;116(1):344–351. doi: 10.1016/0003-9861(66)90040-3. [DOI] [PubMed] [Google Scholar]
- Nomura M. Assembly of bacterial ribosomes. Science. 1973 Mar 2;179(4076):864–873. doi: 10.1126/science.179.4076.864. [DOI] [PubMed] [Google Scholar]
- Nomura M., Erdmann V. A. Reconstitution of 50S ribosomal subunits from dissociated molecular components. Nature. 1970 Nov 21;228(5273):744–748. doi: 10.1038/228744a0. [DOI] [PubMed] [Google Scholar]
- Pestka S. Ribonuclease sensitivity of aminoacyl-tRNA: an assay for codon recognition and interaction of aminoacryl-tRNA with 50 S subunits. Methods Enzymol. 1974;30:439–452. doi: 10.1016/0076-6879(74)30044-4. [DOI] [PubMed] [Google Scholar]
- Traub P., Nomura M. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30 s ribosomes studied in vitro. J Mol Biol. 1969 Mar 28;40(3):391–413. doi: 10.1016/0022-2836(69)90161-2. [DOI] [PubMed] [Google Scholar]
- Wishnia A., Boussert A., Graffe M., Dessen P. H., Grunberg-Manago M. Kinetics of the reversible association of ribosomal subunits: stopped-flow studies of the rate law and of the effect of Mg2+. J Mol Biol. 1975 Apr 25;93(4):499–415. doi: 10.1016/0022-2836(75)90242-9. [DOI] [PubMed] [Google Scholar]