Abstract
Herpes simplex virus 1 (HSV-1) invades the nervous system and causes pathological changes. In this study, we defined the remodeling of F-actin and its possible mechanisms during HSV-1 infection of neuronal cells. HSV-1 infection enhanced the formation of F-actin-based structures in the early stage of infection, which was followed by a continuous decrease in F-actin during the later stages of infection. The disruption of F-actin dynamics by chemical inhibitors significantly reduced the efficiency of viral infection and intracellular HSV-1 replication. The active form of the actin-depolymerizing factor cofilin 1 was found to increase at an early stage of infection and then to continuously decrease in a manner that corresponded to the remodeling pattern of F-actin, suggesting that cofilin 1 may be involved in the biphasic F-actin dynamics induced by HSV-1 infection. Knockdown of cofilin 1 impaired HSV-1-induced F-actin assembly during early infection and inhibited viral entry; however, overexpression of cofilin 1 did not affect F-actin assembly or viral entry during early infection but decreased intracellular viral reproduction efficiently. Our results, for the first time, demonstrated the biphasic F-actin dynamics in HSV-1 neuronal infection and confirmed the association of F-actin with the changes in the expression and activity of cofilin 1. These results may provide insight into the mechanism by which HSV-1 productively infects neuronal cells and causes pathogenesis.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) and 2 (HSV-2) are enveloped DNA viruses that belong to the family Herpesviridae and cause lifelong latent infections (5). The stress-induced reactivation of HSV-1 results in productive virus replication that causes perioral lesions of the skin or mucosa or lesions on the cornea, and HSV-2 is primarily associated with genital and newborn infections. HSV-1 can spread from epithelial cells to neurons and causes pathological changes in the central nervous system (CNS) (36). Among these changes, herpes simplex encephalitis (HSE) is considered to be the most common sporadic but fatal encephalitis, over 90% of the cases of which are caused by HSV-1; if left untreated, HSE results in the death of 70% of affected patients (20, 43). The neuronal infection can result in marked neurite damage, neuronal-cell death, and a disruption of cytoskeleton dynamics, which may contribute to virus-induced neurodegenerative processes (11, 46). As an essential facet of virus-cell interactions, HSV interacts with the host cytoskeleton at various stages of the viral life cycle (12, 28, 35, 39). Nevertheless, the detailed roles of F-actin and the related mechanisms, especially those in productive neuronal infection of HSV, remain to be determined.
F-actin consists of two parallel strands of ATP-bound globular actin (G-actin) monomers, with each asymmetric strand possessing a fast-growing barbed end and a slower-growing pointed end. F-actin can be further assembled into a wide range of higher-order cellular structures, including sheet-like protrusive structures and finger-like protrusions (39). The assembly and disassembly of actin structures are highly regulated by various factors (19, 34, 42). The regulation of actin structures plays a critical role in neuronal morphogenesis and migration, and dysfunction of the essential molecular switches (e.g., Rho GTPases) leads to neurological diseases, such as nonsyndromic X-linked mental retardation and William's syndrome (26). The actin-depolymerizing factor (ADF)/cofilin family and gelsolin (GSN) were previously reported to be essential regulators of actin dynamics (3, 38), and ADF/cofilin proteins are considered to be primarily responsible for remodeling the actin cytoskeleton (38). The ADF/cofilin proteins serve to disassemble F-actin by cooperatively binding to older ADP-actin filaments and promoting phosphate (Pi) dissociation from actin subunits, resulting in increased subunit dissociation from the pointed ends and the severing of filaments (42). In mammalian cells, the ADF/cofilin family consists of three highly similar paralogs: cofilin 1 (nonmuscle cofilin), cofilin 2 (muscle cofilin), and ADF (destrin), of which cofilin 1 is the most ubiquitous and has been studied much more widely for its essential role in development (6, 41). Given that F-actin may affect several steps of HSV-1 infection and that F-actin may be involved in all facets of neuronal-cell activities (37), the HSV-induced changes in F-actin dynamics may contribute to the pathogenesis of neurological diseases (e.g., the cognitive deficits exhibited in HSE patients). Thus, it is of considerable importance to investigate the remodeling pattern involved in and the mechanisms that are induced by HSV-1 infection. Several questions currently remain unanswered, including the following: (i) how the actin cytoskeleton of neuronal cells responds to HSV-1 infection during the viral life cycle and (ii) the identity of the host factor(s) involved in the actin regulation processes following HSV-1 infection and how this factor(s) functions.
In this study, we demonstrate that HSV-1 infection induces biphasic dynamics of F-actin to facilitate efficient infection and replication in neuronal cells. Specifically, we found that HSV-1 infection enhances F-actin assembly at an early stage of infection to facilitate viral transport, whereas the total cellular F-actin tends to decrease in favor of viral reproduction at later stages of infection. Additionally, we confirmed that cofilin 1 regulation may be responsible for mediating HSV-1-induced F-actin remodeling in both assembly and disassembly, which occur during the early and late infection stages, respectively. This study demonstrates for the first time the biphasic dynamics of F-actin in response to HSV-1 infection of neuronal cells and the possible roles of cofilin 1 in mediating HSV-1-induced actin cytoskeletal remodeling.
MATERIALS AND METHODS
Cells and virus.
The neuroblastoma cell line SK-N-SH (ATCC HTB-11), purchased from the Cellbank of the Chinese Academy of Sciences, was propagated in Eagle's minimal essential medium (MEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 1.0 mM sodium pyruvate (Sigma), 0.1 mM nonessential amino acids (Invitrogen), and 1.5 g/liter sodium bicarbonate (Sigma). African green monkey kidney cells (Vero; ATCC CCL81), provided by the Wuhan Institute of Virology at the Chinese Academy of Sciences, were cultured in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% FBS (Invitrogen), 0.22% sodium bicarbonate (Sigma), and 50 mg/liter gentamicin (Invitrogen). The cells were cultured at 37°C in a humid atmosphere with 5% CO2. The HSV-1 strain F (ATCC VR733), obtained from Hong Kong University, was propagated in Vero cells and stored at −80°C until use.
Antibodies, reagents, and plasmids.
The primary antibodies used included a mouse monoclonal antibody (MAb) against the HSV-1 and HSV-2 ICP5 major capsid protein (3B6; Abcam), a mouse MAb against HSV-1 and HSV-2 gB (10B7; Abcam), an anti-cofilin rabbit antibody (Cell Signaling Technology), an anti-phosphocofilin (Ser3) rabbit MAb (77G2; Cell Signaling Technology), an Alexa Fluor 647-conjugated anti-beta-tubulin rabbit MAb (9F3; Cell Signaling Technology), an anti-β-actin rabbit antibody (Cell Signaling Technology), and an anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) rabbit MAb (14C10; Cell Signaling Technology). The secondary antibodies used included Alexa Fluor 488-conjugated goat anti-mouse IgG(H+L) (Invitrogen), Alexa Fluor 633-conjugated goat anti-mouse IgG(H+L) (Invitrogen), horseradish peroxidase (HRP)-conjugated anti-mouse IgG (GE Healthcare), and HRP-conjugated anti-rabbit IgG (GE Healthcare). Fluorescein isothiocyanate (FITC)-phalloidin (Sigma-Aldrich) and tetramethylrhodamine B isothiocyanate (TRITC)-phalloidin (Sigma-Aldrich) were used to label F-actin. DAPI (4′,6′-diamidino-2-phenylindole) (Biotium) and propidium iodide (PI) (Sigma) were used to dye the nuclei. The F-actin inhibitors used were cytochalasin D (CytoD) (Gibco), latrunculin A (Lat A) (Invitrogen), and jasplakinolide (Jas) (Invitrogen).
To construct pEGFP-N1-cofilin1, the human cofilin 1 coding sequence (CDS) without the TGA stop codon was cloned from the total cDNA of SK-N-SH cells with the forward primer 5′-TCC AAG CTT ATG GCC TCC GGT GTG GCT-3′ and the reverse primer 5′-TAG GGA TCC AAA GGC TTG CCC TCC AGG GA-3′. The PCR fragment was then digested with HindIII (TaKaRa) and BamH I (TaKaRa) and inserted into the multiple cloning site of the pEGFP-N1 plasmid (Clontech) under the control of the cytomegalovirus (CMV) promoter. The recombinant pCofilin1-EGFP-N1 vector was successfully constructed, as confirmed by DNA-sequencing analysis.
Laser scanning confocal immunofluorescence microscopy.
For the F-actin and microtubule observations, cells were fixed in 4% paraformaldehyde (PFA)–phosphate-buffered saline (PBS) for 15 min, permeabilized with 0.1% Triton X-100–PBS for 5 min, blocked with 5% bovine serum albumin (BSA)-PBS for 1 h, and stained with Alexa Fluor 647-conjugated antibody against beta-tubulin (1:1,000) in 5% BSA-PBS for 1 h. The cells were stained with 5 μM TRITC-phalloidin–PBS for 40 min and 1 mg/ml DAPI-PBS for 15 min to label the F-actin and nuclei, respectively. The cells were gently washed 3 times in PBS after each step in the staining process, and the entire staining process was conducted at room temperature. Fluorescence images were then recorded using a confocal laser scanning microscope (LSM) (LSM 510; Zeiss) under a 63× or 100× oil immersion objective (Carl Zeiss).
Quantification of HSV-1 trafficking to the nuclei was conducted by a method similar to that in a previous report (18). Briefly, SK-N-SH cells with 40 to 50% confluence were pretreated with different concentrations of cytochalasin D, latrunculin A, or jasplakinolide for 1 h and then infected with HSV-1 at a multiplicity of infection (MOI) of 30 for 4 h in the presence of inhibitors. The cells were then fixed, permeabilized, blocked, stained with primary antibody against HSV-1 (ICP5; 1:1,000), and then stained with Alexa Fluor 488-conjugated or Alexa Fluor 633-conjugated secondary antibody (1:1,000) in 5% BSA-PBS for 1 h. F-actin and nuclei were then stained as described above. Images were acquired by LSM for 5 fields of view per dish to enable the counting of ICP5-positive capsids docked at nuclei. The average number of ICP5-positive capsids per nuclei and the percentage of positive nuclei (nuclei docked with at least 1 ICP5-positive capsid) were calculated to evaluate the viral entry and trafficking efficiencies.
Flow cytometry.
Flow cytometry (FCM) was used to quantify F-actin reorganization. Briefly, cells were washed with PBS and fixed for 5 min in 4% formaldehyde-PBS. After being washed extensively in PBS, the cells were permeabilized with 0.1% Triton X-100–PBS and washed again with PBS. The cells were then stained with 5 μM FITC-phalloidin–PBS for 40 min, and the unbound phalloidin conjugate was removed by several washings with PBS. The fluorescence was then analyzed with a flow cytometer (Becton Dickinson, CA).
Virus titer quantification.
The virus titers were determined by a plaque assay on Vero cell monolayers, as previously reported (44). To determine the effects of the chemical inhibitors on HSV-1 replication, SK-N-SH cells were seeded into 96-well tissue culture plates and cultured overnight in medium. The cells were infected with HSV-1 (MOI = 5) for 4 h, and after the removal of unadsorbed viruses, different inhibitors at the indicated concentrations were added. At 24 h postinfection (p.i.), the cells were lysed by three cycles of freezing (−80°C) and thawing (37°C). The virus titers were then determined by a plaque assay. For the transfection assay, SK-N-SH cells grown in 96-well culture plates to 50 to 60% confluence were transfected with either small interfering RNA (siRNA) or plasmid (as described below). At 24 h after transfection, the cells were infected with HSV-1 (MOI = 5) for 2 h, and the unadsorbed viruses were removed. At 24 h p.i., the cells were lysed, and the virus titers were determined by a plaque assay.
Cytotoxicity assay.
DAPI-PI costaining was conducted to examine the cytotoxicity of the chemical inhibitors in the viral entry and trafficking assay as previously reported (18). Briefly, SK-N-SH cells grown in glass bottom dishes were treated with different inhibitors at the indicated concentrations for 5 h. One hour prior to fixation, the cells were labeled with 10 μg/ml PI-PBS, fixed with 4% paraformaldehyde-PBS for 15 min, and then counterstained with 1 mg/ml DAPI-PBS. Images were then acquired by LSM. The total number of cells and the corresponding PI-positive cells from 5 representative fields were counted and calculated as the percent PI-positive cells. The cells that had been left to air dry for 20 min to induce cell death were used as positive controls.
To examine the cytotoxicity of the chemical inhibitors in the viral growth assay, SK-N-SH cells were seeded into 96-well culture plates, cultured overnight to achieve a monolayer, infected with HSV-1 (MOI = 5) for 4 h, and then incubated with the indicated concentrations of different inhibitors for another 20 h. Cell viability was then determined with a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, as previously described (24). The cytotoxicity resulting from the silencing or transient expression of cofilin 1 was also determined by an MTT assay where SK-N-SK cells were allowed to grow to only 50 to 60% confluence before transfection, and cell viability was assayed at 48 h p.i.
Real-time PCR.
Total RNA was extracted with TRIzol reagent (Invitrogen). The RNA concentrations were measured with a spectrophotometer (Beckman) at wavelengths of 260 nm/280 nm, and 1 μg of RNA was then reverse transcribed with the PrimeScript RT reagent Kit (TaKaRa). A real-time PCR assay was performed using a Bio-Rad CFX96 real-time PCR system with a total volume of 10 μl that contained 0.5 μl cDNA, 5 μl SsoFast EvaGreen Supermix (Bio-Rad), and 250 nM each primer. After the initial denaturation at 95°C for 30 s, the amplification reaction was carried out with 40 cycles, each consisting of denaturation at 95°C for 5 s and annealing/extension at 58°C for 10 s. The results were analyzed with CFX manager software (Bio-Rad). The primer pairs used were Cofilin1 F (5′-GGT GCT CTT CTG CCT GAG TG-3′) and Cofilin1 R (5′-TCT TGA CAA AGG TGG CGT AG-3′), ADF F (5′-TGT GTC CAC ATA ATC CAC G-3′) and ADF R (5′-AGA TGC CAG GTC ACT CTT C-3′), GSN F (5′-AGT GCC TTT TGG AAC TGT C-3′) and GSN R (5′-GCA GGG CTA TTT TTG GAT AC-3′), Rac1 F (5′-CAC GCT GTA TTC TCG CCA GT-3′) and Rac1 R (5′-GGA CAC ACG CCT CCT GTA GT-3′), CDC42 F (5′-CAT TGC TTT TAG TAT GAT GCC G-3′) and CDC42 R (5′-TGG AGC CTC CAG AAC CGA AGA A-3′), RhoA F (5′-GGT CTT CAG CTA CCC GCC TTC GTC-3′) and RhoA R (5′-ACC TCT GGG AAC TGG TCC TTG CT-3′), GAPDH F (5′-CCC ACT CCT CCA CCT TTG AC-3′) and GAPDH R (5′-TCT TCC TCT TGT GCT CTT GC-3′), and UL27 F (5′-GCC TTC TTC GCC TTT CGC-3′) and UL27 R (5′-CGC TCG TGC CCT TCT TCT T-3′).
For HSV-1 DNA detection, SK-N-SH cells grown in 24-well culture plates were treated with different concentrations of CytoD, Lat A, or Jas for 1 h or were transfected with either 1 μg siRNA or 1.5 μg plasmid (as described below) for 24 h. Then, the cells were infected with HSV-1 (MOI = 30) for another 1 h and washed 3 times with PBS, and the internalized viral DNA was isolated using a UNIQ-10 Viral DNA Kit (Sangon, China). A real-time PCR assay was used for the quantification of the obtained viral DNA by detecting the viral UL47 gene with primers UL47F (5′-ACG ATG ATG ATG AGG TTC CC-3′) and UL47R (5′-CAG CTC CTC TAG GAA CAG CG-3′). The PCR amplification product of UL47 was purified using a Gel Extraction Kit II (U-gene, China), diluted serially, and used as a standard for quantitative analysis. The initial copy number of UL47 DNA in each group was calculated using the following formula: CT = −K logX0 + b, where CT is the cycle threshold, and K, X0, and b refer to the slope rate, initial copy number, and constant, respectively.
Western blotting.
SK-N-SH cells grown in 75-cm2 culture flasks (Corning) were infected with HSV-1 (MOI = 5) for 0, 2, 6, 12, and 20 h. After three washes with PBS, the cells were harvested and lysed in RIPA buffer (Beyotime, Jiangsu, China) for 30 min on ice, and the protein concentrations were measured with an enhanced bicinchoninic acid (BCA) protein assay kit (Beyotime, Jiangsu, China). The cell lysates were mixed with 5× SDS-PAGE buffer (Beyotime, Jiangsu, China) and boiled for 5 min. Twenty micrograms of each sample was subjected to SDS-12% to 15% PAGE and Western blotting as previously reported (24). To detect the protein levels after cofilin 1 transfection, SK-N-SH cells grown in a 75-cm2 culture flask were first transfected with pCofilin1-EGFP-N1 or pEGFP-N1 and then infected with HSV-1 (MOI = 5) 24 h posttransfection. The cells were harvested 24 h p.i., lysed, and analyzed for protein expression as described above.
Transfection.
The transfection of SK-N-SH cells with the pEGFP-N1-Cofilin1 or pEGFP-N1 plasmid was performed with the Lipofectamine LTX and Plus reagents (Invitrogen). For the 24-well transfection, 1.5 μg of the pCofilin1-EGFP-N1 or pEGFP-N1 plasmid was diluted in 100 μl Opti-MEM I reduced-serum medium (Invitrogen) and mixed thoroughly, and then 1.5 μl Plus reagent was added. After 5 min of incubation, 3 μl Lipofectamine LTX was added to the DNA dilution and incubated for another 30 min. The transfection mixture was then added to each well containing SK-N-SH cells (50 to 60% confluence). A transfection mixture without any plasmid was also added as the mock-transfected control. At 6 h after transfection, the mixture was replaced with growth medium. At 24 h posttransfection, the cells were infected with HSV-1 for further studies.
siRNA transfection was conducted using the Lipofectamine RNAiMax reagents (Invitrogen). The cofilin 1 siRNA duplex consisted of oligonucleotides with the sequences 5′-GGA TCA AGC ATG AAT TGC AAG CAA A-3′ and 5′-CCT ACG CCA CCT TTG TCA AGA TGC T-3′. The scrambled siRNA duplex of oligonucleotides 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and 5′-ACG UGA CAC GUU CGG AGA ATT-3′, which does not target any gene product, was used as a negative control. All siRNAs were obtained from Shanghai GenePharma Co., Ltd. For the 24-well transfection, 1 μg of RNAi duplex was diluted in 50 μl Opti-MEM I reduced-serum medium and then mixed with another 50 μl Opti-MEM I reduced-serum medium containing 1 μl RNAiMax and incubated for 20 min at room temperature. The transfection mixture was then added to each well containing SK-N-SH cells (30 to 50% confluence). The mixture was replaced with growth medium 6 h after transfection. At 24 h posttransfection, the cells were infected with HSV-1 for further studies.
RESULTS
HSV-1 infection induces biphasic F-actin dynamics in neuronal cells.
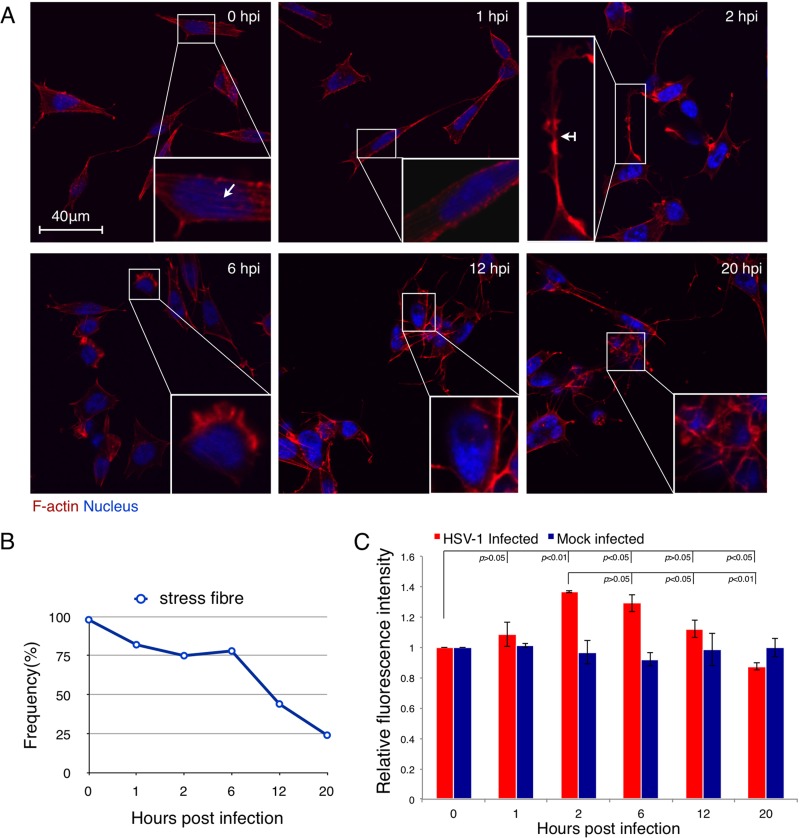
SK-N-SH neuroblastoma cells were infected with HSV-1 at an MOI of 5, and the F-actin structures were monitored under LSM at different time points. One noteworthy change following infection involved the intracellular stress fiber breakdown (Fig. 1A). This change was semiquantitatively analyzed and is shown in Fig. 1B. In the absence of HSV-1 infection, 98% of cells displayed obvious stress fiber structures. After infection, the appearance frequencies of stress fiber were 82% (n = 62), 75% (n = 71), 78% (n = 81), 44% (n = 81), and 24% (n = 49) of all cells at 1, 2, 6, 12, and 20 h p.i., respectively. A marked stress fiber breakdown occurred after 6 h p.i. These changes in the stress fibers were more obvious at higher MOIs (MOI = 30), at which nearly all of the cells displayed stress fiber breakdown as early as 4 h p.i. (see Fig. S1A in the supplemental material).
Fig 1.
HSV-1 infection induces biphasic dynamics of F-actin in SK-N-SH neuroblastoma cells. (A) F-actin reorganization of SK-N-SH cells after HSV-1 infection. Cells were infected with HSV-1 (MOI = 5) for 0, 1, 2, 6, 12, or 20 h and then fixed, permeabilized, and stained with TRITC-phalloidin and DAPI. Images were then recorded with a confocal LSM. Typical cells are enlarged in the insets, where the arrow indicates a stress fiber and the arrow with a tail indicates dendritic filopodia. Cells at 2 h p.i. showed enhanced outgrowth of neuritic extensions, while cells at 6 h p.i., 12 h p.i., and 20 h p.i. showed marked stress fiber breakdown. The results are representative of 3 separate experiments. (B) Quantification of changes in the F-actin-based stress fibers at different time points postinfection. Cells with F-actin stress fibers were designated positive cells; each value represents the mean of 50 to 80 cells from at least 5 fields of 1 representative experiment. (C) Quantification of F-actin reorganization in SK-N-SH cells after HSV-1 infection. Cells were infected with HSV-1 (MOI = 5) for 0, 1, 2, 6, 12, or 20 h and then fixed, permeabilized, and stained with FITC-phalloidin. The fluorescence intensity was analyzed by flow cytometry. Each value represents the mean ± standard deviation (SD) of 2 separate experiments.
An enhanced outgrowth of neuritic extensions, specifically, the growth of dendritic filopodia and lamellipodia, was observed in early infection (Fig. 1A; see Fig. S1B in the supplemental material, white arrow with a tail). In comparison with the uninfected cells at the same culture time (see Fig. S1B-I in the supplemental material), the infected cells showed longer filopodia with more and expanded branches (see Fig. S1B-II in the supplemental material) or extended lamellipodia (see Fig. S1B-III in the supplemental material), although the uninfected cells also displayed outgrowths of these two structures during cell culture. This enhancement was more obvious at higher MOIs (see Fig. S1A in the supplemental material). At later stages of infection, however, the growth of these neuritic extensions was attenuated, especially at 20 h p.i., when adjacent cells were fused with each other and distinct cell boundaries were lost (Fig. 1A). Also, the attenuation of the growth of neuritic extensions was more obvious at higher MOIs, which happened as early as 4 h p.i. (see Fig. S1A in the supplemental material).
We further quantified the total cellular F-actin in either HSV-1-infected cells or mock-infected cells using FCM. As shown in Fig. 1C, the total F-actin of infected cells increased at an early stage of infection (2 h p.i.) and then decreased from 6 h to 20 h p.i. In contrast, the uninfected cells did not display distinct alteration in the F-actin fluorescence intensity during the duration of the culture period. The cell viabilities at 2, 6, 12, and 20 h p.i., as determined by MTT assay, were 102.6%, 103.6%, 94.5%, and 82.1% of that at 0 h p.i., respectively (see Fig. S2 in the supplemental material). Thus, although the decrease of F-actin at 20 h p.i. may partly result from decreased cell viability at that time, the decline of F-actin from 2 h p.i. to 12 h p.i. still confirmed the disassembly tendency of total F-actin during this period. Together, it can be concluded that HSV-1 can promote the overall formation of F-actin-based structures in neuronal cells at early stages of infection but decreases the overall F-actin assembly at the late stages of infection, which indicates that HSV-1 infection can induce the biphasic dynamics of F-actin in neuronal cells.
F-actin dynamics are crucial for efficient HSV-1 infection.
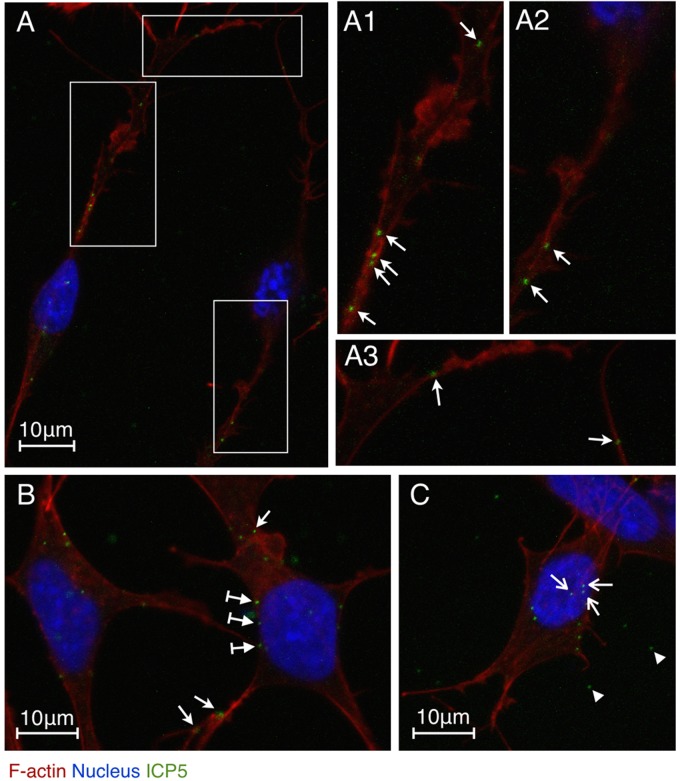
To investigate the effects of enhanced F-actin assembly on viral infection, we detected the localization of HSV-1 virions. HSV-1 internalizes into neuronal cells via the fusion of the viral envelope with the plasma membrane rather than through a pH-dependent endocytic pathway (27, 30); thus, the capsid proteins are appropriate targets to localize viral particles in infected cells. Here, the HSV-1 major capsid protein ICP5 was immunostained as an indication of the localization of HSV-1 particles. HSV-1-infected SK-N-SH cells (MOI = 10) were induced to grow long, branched dendrites, and filopodia docked with viral particles (Fig. 2A1, A2, and A3, arrows) as early as 0.5 h p.i. This finding suggests that HSV-1 might interact with F-actin-rich structures for efficient viral transport to the soma, which is in agreement with an earlier report (11). We also observed an asynchrony of the infection process; whereas some viral particles were localized on filopodia and lamellipodia away from soma (Fig. 2B and C, arrows), other viral particles were attached to the cellular membrane approaching the nucleus (Fig. 2B, arrows with tails) or were already docked at the nucleus (Fig. 2C, arrows). Free viral particles between the intercellular spaces were also observed (Fig. 2C, arrowheads). This asynchronous localization of viruses indicates that the viral particles that were randomly distributed around the cells were approaching the soma and nucleus from various directions and from different distances. Despite this asynchronous localization, the viral capsids were observed mainly in the nucleus region at 4 h p.i. (data not shown), and we conducted a further evaluation of the viral transport efficiency at that time.
Fig 2.
localization of HSV-1 particles in SK-N-SH neuroblastoma cells during early infection. Cells were infected with HSV-1 (MOI = 10) for 0.5 h and then fixed, permeabilized, and stained with an HSV-1 ICP5 primary antibody, an Alexa Fluor 488-conjugated secondary antibody, TRITC-phalloidin, and DAPI. The images were recorded by LSM. (A) Two infected neuronal cells. (A1 and A2) Higher magnifications of the vertical boxed areas showing viral particles attached to actin-rich dendrites (arrows). (A3) Higher magnification of the horizontal boxed area showing viral particles attached to actin-rich filopodia (arrows). (B) Infected cells with viral particles attached to the surface of the cellular membrane (arrows with tails), the dendritic filopodia (bottom two arrows), and the lamellipodia (the top arrow). (C) Infected cell with viral particles docked at the nucleus (arrows). Free viral particles (arrowheads) are also shown.
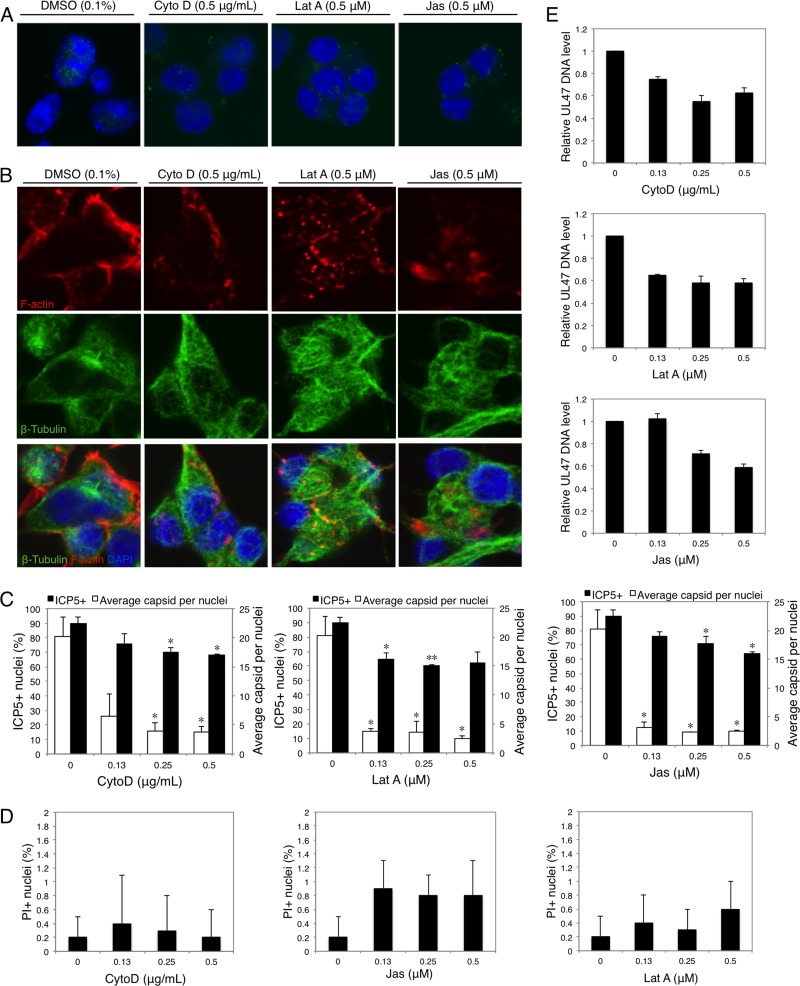
SK-N-SH cells were treated with chemical inhibitors of F-actin dynamics. These inhibitors included the following: CytoD, which induces the depolymerization of existing actin filaments; Lat A, which targets monomeric G-actin and prevents actin polymerization; and Jas, which induces the polymerization and stabilization of actin filaments, depleting the cellular pool of free actin monomers available for de novo polymerization (18). At 4 h p.i., the cells were stained for HSV-1 ICP5, and the percentage of positive nuclei (nuclei docked with at least 1 ICP5-positive dot) and the average number of ICP5-positive dots per nucleus were determined to evaluate the viral trafficking efficiency. In the presence of CytoD, Lat A, and Jas, the percentages of ICP5-positive nuclei were all reduced, although the inhibitory ratios were distinct and dependent on different inhibitors and concentrations (Fig. 3A and C). When the average number of capsids docked at positive nuclei was calculated, the inhibitory effects were more obvious: cells treated with the inhibitors were docked with significantly fewer capsids than untreated cells (Fig. 3C). This transport inhibition was not a consequence of reduced cell viability caused by inhibitors, as demonstrated by a PI-DAPI costaining assay (Fig. 3D; see Fig. S3 in the supplemental material). Treatments with each of these chemical inhibitors resulted in the disruption of F-actin dynamics (Fig. 3B), while the effects on microtubule networks were not obvious in HSV-1-infected and uninfected cells (Fig. 3B; see Fig. S4 and S5 in the supplemental material).
Fig 3.
Disruption of F-actin dynamics reduces the early transport of HSV-1. (A) Nuclear docking with ICP5-positive capsids in the presence or absence of CytoD, Lat A, or Jas. Cells were pretreated with CytoD, Lat A, or Jas at different concentrations for 1 h and then infected with HSV-1 (MOI = 30) for 4 h in the presence of inhibitors. The cells were then fixed and stained for LSM observation. (B) Structures of F-actin and microtubules in the presence of CytoD (0.5 μg/ml), Lat A (0.5 μM), or Jas (0.5 μM). (C) Quantification of viral transport efficiency in treated or untreated (0) cells. The average number of ICP5-positive capsids per nucleus and the percentage of positive nuclei (nuclei docked with at least 1 ICP5-positive capsid) were determined to evaluate the viral entry and trafficking efficiencies. Each value represents the mean and SD of 2 separate experiments, and at least 100 cells from 5 representative fields were counted in each experiment (*, P < 0.05, and **, P < 0.01 compared with the control). (D) The cytotoxicity of chemical inhibitors was determined by DAPI-PI costaining. The total number of cells and the corresponding number of PI-positive cells were determined and calculated as percent PI-positive cells. Each value represents the mean and SD of 3 separate experiments, and at least 100 cells from 5 representative fields were counted in each experiment. (E) Real-time PCR assay of internalized HSV-1 DNA. Cells were pretreated with CytoD, Lat A, or Jas at different concentrations for 1 h and then infected with HSV-1 (MOI = 30) for 1 h in the presence of inhibitors. Total intracellular viral DNA was isolated, and the quantities of UL47 DNA were assayed. The results are representative of 2 separate experiments. Each value represents the mean and SD of triplicate wells.
To further confirm the effects of disrupted F-actin dynamics on HSV-1 early infection, the internalized viral DNA at 1 h p.i. was quantified using real-time PCR. As shown in Fig. 3E, treatment with CytoD, Lat A, or Jas all decreased the level of internalized HSV-1 DNA following infection, indicating their inhibitory effects on HSV-1 early infection. Together, these results suggest that the early infection process of HSV-1 in neuronal cells requires the F-actin dynamics, and HSV-1 may achieve efficient infection by enhancing F-actin assembly at this stage.
Manipulating F-actin dynamics affects intracellular HSV-1 replication.
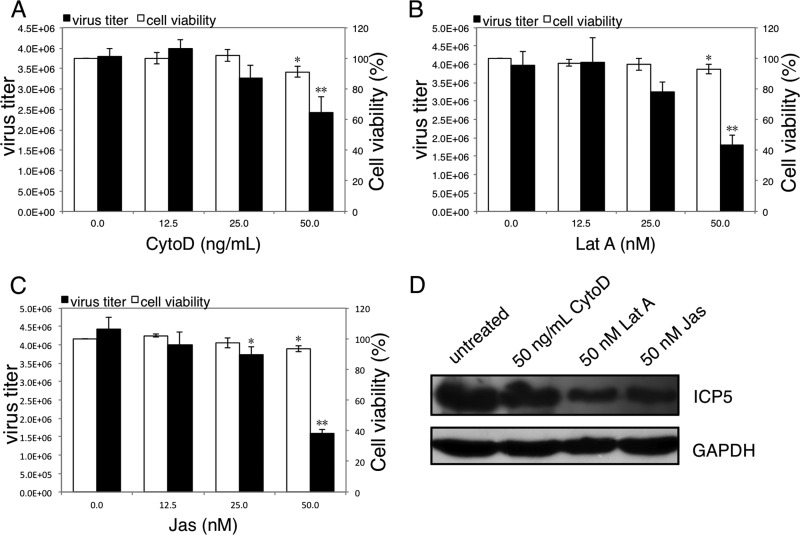
To investigate the correlation between the F-actin dynamics and intracellular HSV-1 reproduction, SK-N-SH cells were infected with HSV-1 (MOI = 5) for 4 h to allow complete viral infection and then treated with the chemical inhibitors CytoD, Lat A, and Jas for another 20 h. In these assays, lower concentrations of CytoD, Lat A, or Jas were used because of the prolonged incubation period compared to the viral entry assay. In comparison with untreated cells, the total progeny virus titers at 24 h p.i. were significantly reduced in the presence of 50 ng/ml CytoD, 50 nM Lat A, or 50 nM Jas, with inhibitory rates of 36%, 55%, and 64%, respectively (Fig. 4A, B, and C). An MTT assay revealed that the cell viabilities after a 20-h incubation in the presence of 50 ng/ml CytoD, 50 nM Lat A, or 50 nM Jas in infected cells were 91%, 93%, and 94% of the viral control, respectively.
Fig 4.
Disruption of F-actin dynamics inhibits the intracellular reproduction of HSV-1. (A, B, and C) Cytotoxicity of CytoD, Lat A, and Jas on HSV-1-infected cells and their effects on production of progeny viruses. SK-N-SH cells were infected with HSV-1 (MOI = 5) for 4 h. After the removal of unadsorbed viral particles, CytoD (A), Lat A (B), or Jas (C) was added at the indicated concentrations, and at 24 h p.i., the total progeny viruses were quantified by a plaque assay. Meanwhile, cells infected with HSV-1 (MOI = 5) for 4 h were incubated with chemical inhibitors at different concentrations for another 20 h, and the cell viability was determined by the MTT assay. Each value represents the mean ± SD of 3 separate experiments (*, P < 0.05, and **, P < 0.01 compared with the control). (D) Effects of CytoD, Lat A, and Jas on viral protein synthesis. SK-N-SH cells were infected with HSV-1 (MOI = 5) for 4 h, and then 50 ng/ml CytoD, 50 nM Lat A, or 50 nM Jas was added and incubated for another 20 h. Cells were harvested and lysed in RIPA buffer. Each sample (20 μg) was then subjected to Western blot analysis for HSV-1 ICP5 detection. The results are representative of three separate experiments.
Meanwhile, cells infected with HSV-1 (MOI = 5) for 4 h were incubated with 50 ng/ml CytoD, 50 nM Lat A, or 50 nM Jas for another 20 h, and the expression levels of viral ICP5 protein were detected using Western blotting. As shown in Fig. 4D, compared to the untreated control, the chemical inhibitors all decreased the synthesis of ICP5 protein. Overall, these results demonstrated that intracellular HSV-1 reproduction in neuronal cells requires F-actin dynamics, whereas the disruption of F-actin dynamics can reduce viral replication.
Cofilin 1 activity shows biphasic modulation by HSV-1 infection.
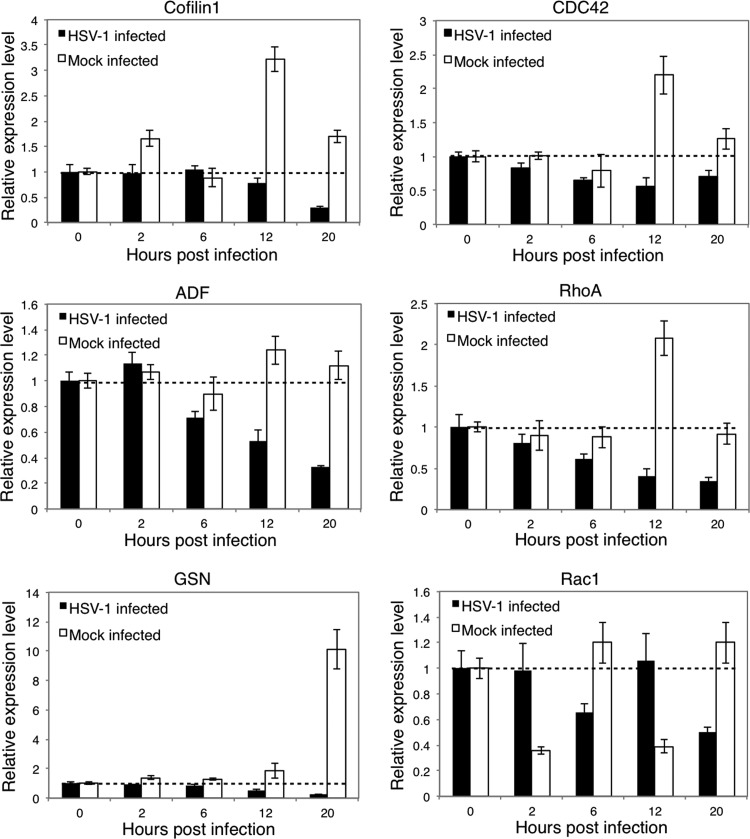
We further analyzed the expression of several essential F-actin regulators, including cofilin 1, ADF, and GSN, and their upstream Rho GTPases, including CDC42, RhoA, and Rac1. As shown in Fig. 5, cofilin 1, ADF, and GSN all displayed gradually reduced mRNA expression following HSV-1 infection, with decreased relative expression levels of 29%, 33%, and 23%, respectively, at 20 h p.i. compared with those at 0 h p.i. In contrast, the mRNA expression of these regulators in uninfected cells did not show obvious alterations; one exception was the expression of GSN, which gradually increased during cell culture. Similarly, both CDC42 and RhoA showed continuously reduced expression levels after infection, with decreased relative expression levels of 71% and 35%, respectively, at 20 h p.i. compared with those at 0 h p.i., indicating that they may be involved in regulating the downstream proteins cofilin 1, ADF, and GSN. In contrast, Rac1 did not show a continuous decrease in expression during the culture period, but it did display a converse tendency of expression compared with uninfected cells at each time point tested. This finding demonstrates the expression modulation caused by HSV-1 infection. By comparing the expression tendencies of these genes in uninfected cells, it was observed that CDC42 and RhoA might primarily mediate the regulation of cofilin 1; this phenomenon was more obvious at 12 h p.i., when cofilin 1, CDC42, and RhoA all displayed an expression peak.
Fig 5.
Expression of F-actin regulators after HSV-1 infection. SK-N-SH cells were infected with HSV-1 (MOI = 5) for 0, 2, 6, 12, or 20 h, and the total RNA was extracted and reverse transcribed. A real-time PCR assay was performed to determine the mRNA levels of cofilin 1, ADF, GSN, CDC42, RhoA, and Rac1. The results are representative of 3 separate experiments from 1 culture. Each value represents the mean ± SD of triplicate wells.
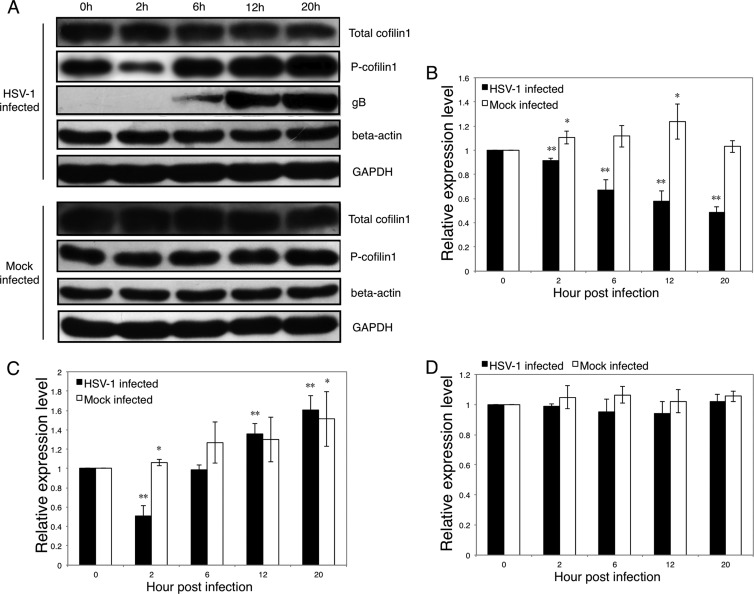
We further investigated the regulation of cofilin 1, which is regulated by the phosphorylation (inactive form) and the dephosphorylation (active form) of the serine residue at position 3 (1, 6, 29). As determined by Western blot analysis, the total cofilin 1 protein level in uninfected cells did not display obvious alterations. However, the total cofilin 1 protein level in infected cells decreased gradually after HSV-1 infection; at 20 h p.i., the cofilin 1 protein had an expression level that was 48% of the expression level at 0 h p.i. (Fig. 6A and B). This finding agrees with the results of the mRNA assay (Fig. 5). During the culture period, the phosphorylated-cofilin 1 (P-cofilin 1) (inactive form) levels increased both in infected and in uninfected cells. At 2 h p.i., the P-cofilin 1 protein level in infected cells decreased to 51% of the baseline level (0 h p.i.), whereas the total cofilin 1 level decreased to 92% of the baseline level (Fig. 6A and C), suggesting that the active form of cofilin 1 was upregulated at an early stage of infection. In contrast, the total cofilin 1 level decreased and the P-cofilin 1 level increased at the later stages of infection, suggesting that the active form of cofilin 1 was downregulated.
Fig 6.
Regulation of cofilin 1 after HSV-1 infection. SK-N-SH cells were infected with HSV-1 (MOI = 5) for 0, 2, 6, 12, and 20 h, and then the cells were harvested and lysed in RIPA buffer. Each sample (20 μg) was subjected to Western blot analysis. (A) Western blot analysis of the total amounts of cofilin 1, P-cofilin 1, HSV-1 gB, beta-actin, and GAPDH. The results are representative of three separate experiments. (B, C, and D) Quantification of the total amounts of cofilin 1, P-cofilin 1, and beta-actin with or without HSV-1 infection. The amount of GAPDH was measured as an internal control. Each value represents the mean ± SD of 3 experiments from 3 separate cultures (*, P < 0.05, and **, P < 0.01 compared with the control).
Cofilin 1 may promote actin assembly or disassembly, based on the ratio of cofilin 1 to actin (2, 8, 41); thus, we evaluated the cellular beta-actin levels after infection. A low cofilin 1/actin ratio is considered to result in actin filament severing, but at a high cofilin 1/actin ratio, cofilin 1 tends to stabilize filamentous actin in a twisted form or nucleates filaments (2, 8). As shown in Fig. 6A and D, no significant changes in beta-actin were observed in either HSV-1-infected or uninfected cells. These results indicate that the ratio of active cofilin 1 to actin may also exhibit biphasic changes during infection. Overall, the biphasic changes in active cofilin 1 after HSV-1 infection coincided with the changes in F-actin (Fig. 1C) and suggest that cofilin 1 may be responsible for mediating the biphasic F-actin dynamics during HSV-1 infection of neuronal cells.
Modulation of cofilin 1 level affects F-actin dynamics, viral infection, and replication.
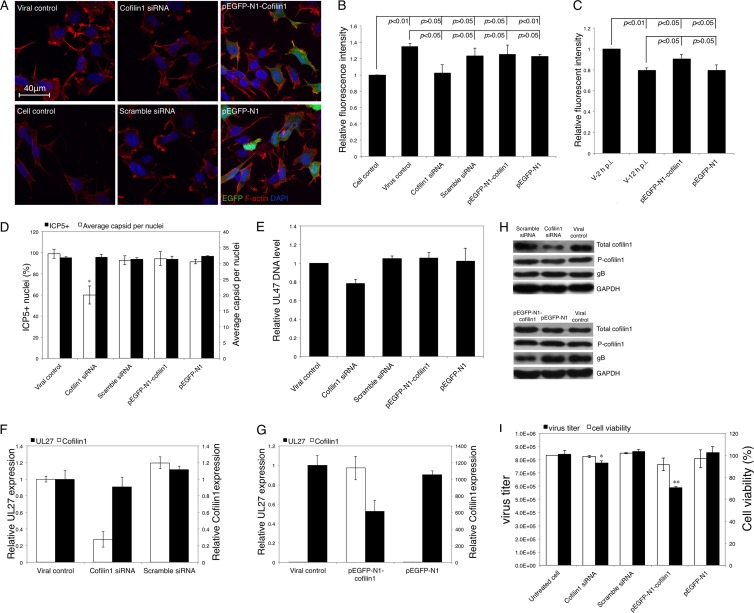
To further confirm the role of cofilin 1 in F-actin dynamics during viral infection and replication, we knocked down or overexpressed cofilin 1 by transfecting cofilin 1-targeting siRNA or pEGFP-N1-Cofilin1, respectively. At 24 h posttransfection, SK-N-SH cells were infected with HSV-1 (MOI = 5), and the virally induced F-actin assembly during the early stage of infection was examined. The downregulation of cofilin 1 decreased the HSV-1-induced formation of F-actin-based structures, especially the outgrowth of lamellipodia, at 2 h p.i. (Fig. 7A). This finding was confirmed by the quantitation of F-actin intensity using FCM (Fig. 7B); the enhancement of F-actin assembly following HSV-1 infection for 2 h was significantly inhibited by cofilin 1 siRNA (P < 0.05). In contrast, both LSM observation and FCM quantification demonstrated that there was no obvious difference between the viral control and the pEGFP-N-Cofilin1-transfected group (Fig. 7A and B). At 12 h p.i., cofilin 1 overexpression restored F-actin decrease induced by HSV-1 infection, although the F-actin fluorescence intensity was still lower than that at 2 h p.i. (Fig. 7C). These results further demonstrate the essential role of cofilin 1 in HSV-1-induced F-actin dynamics; specifically, it was demonstrated that cofilin 1 may promote F-actin assembly during the HSV-1 infection of neuronal cells.
Fig 7.
Effects of cofilin 1 down- or upregulation on F-actin remodeling, viral infection, and reproduction. (A) Effects of the cofilin 1 expression level on F-actin remodeling. SK-N-SH cells were transfected with cofilin 1 siRNA, scrambled siRNA, pEGFP-N1-Cofilin1, or pEGFP-N1. At 24 h posttransfection, the cells were infected with HSV-1 (MOI = 5) for 2 h. The cells were then fixed, permeabilized, stained for F-actin, and observed under LSM. (B) Quantification of F-actin remodeling following cofilin 1 knockdown or overexpression using FCM. SK-N-SH cells were transfected, infected, and harvested as described above and subjected to fluorescence analysis by flow cytometry. Each value represents the mean and SD of 2 separate experiments. (C) Quantification of F-actin remodeling by cofilin 1 overexpression using FCM. SK-N-SH cells were transfected and infected with HSV-1 as described above. At 2 h p.i. or 12 h p.i., cells were harvested and subjected to fluorescence analysis by flow cytometry. (D) Effects of the cofilin 1 expression level on the early transport of HSV-1 particles. SK-N-SH cells were transfected with cofilin 1 siRNA, scrambled siRNA, pEGFP-N1-Cofilin1, or pEGFP-N1. At 24 h posttransfection, the cells were infected with HSV-1 (MOI = 30) for 4 h. The infected cells were then fixed, permeabilized, and stained with HSV-1 ICP5 primary antibody, Alexa Fluor 633-conjugated secondary antibody, and DAPI to detect capsid docking. The average number of ICP5-positive capsids per nucleus and the percentage of positive nuclei (nuclei docked with at least 1 ICP5-positive capsid) were determined to evaluate viral entry and trafficking efficiencies. Each value represents the mean ± SD of 2 separate experiments, and at least 50 cells from 5 representative fields were counted in each separate experiment (*, P < 0.05 compared with the viral control). (E) Real-time PCR assay of internalized HSV-1 DNA. Cells transfected with either siRNA or plasmid for 24 h were infected with HSV-1 (MOI = 30) for 1 h. Then, total intracellular viral DNA was isolated, and the quantities of UL47 DNA were assayed. The results are representative of 2 separate experiments. Each value represents the mean and SD of triplicate wells. (F and G) Effects of cofilin 1 regulation on the expression of cofilin 1 and HSV-1 mRNA. SK-N-SH cells transfected with cofilin 1 siRNA, scrambled siRNA, pEGFP-N1-Cofilin1, or pEGFP-N1 for 24 h were infected with HSV-1 (MOI = 5). At 24 h p.i., the total RNA was extracted and reverse transcribed. A real-time PCR assay was performed to determine the mRNA expression of cofilin 1 and UL27 encoding HSV-1 gB. The results are representative of 3 separate experiments. Each value represents the mean ± SD of triplicate wells. (H) Protein expression after modulation of cofilin 1 expression. SK-N-SH cells were transfected with pEGFP-N1-Cofilin1 or pEGFP-N1 for 24 h and infected with HSV-1 (MOI = 5). At 24 h p.i., the cells were harvested and lysed in RIPA buffer. Twenty micrograms of each sample was then subjected to Western blot analysis. The results are representative of three separate experiments. (I) Effects of the cofilin 1 expression level on the production of progeny viruses. SK-N-SH cells were transfected and infected as described for panel H. At 24 h p.i., the cells were harvested, and the total virus titer was determined by a plaque assay. The cytotoxicities of the cofilin 1 siRNA, scrambled siRNA, pEGFP-N1-Cofilin1, and pEGFP-N1 were examined with the MTT assay 48 h posttransfection without HSV-1 infection. Each value represents the mean ± SD of 3 separate experiments (*, P < 0.05, and **, P < 0.01 compared with the control).
Besides F-actin remodeling in the presence of cofilin 1 down- or upregulation, the efficiencies of HSV-1 entry were investigated. As shown in Fig. 7D, although the percentages of ICP5-positive nuclei at 4 h p.i. were not affected by either cofilin 1 siRNA or pEGFP-N1-Cofilin1 transfection, the average number of ICP5-positive dots per nucleus was significantly reduced by cofilin 1 siRNA transfection, suggesting that the downregulation of cofilin 1 inhibits HSV-1 early infection. The inhibitory effect of cofilin 1 downregulation on viral entry was further confirmed by the quantification of internalized HSV-1 DNA using real-time PCR (Fig. 7E). Further, the effect of cofilin 1 on intracellular replication of HSV-1 was investigated, as shown in Fig. 7F; even though the cofilin 1 mRNA level was reduced by siRNA to 27% of the control, only a slight decrease (approximately 10% of the control) in the HSV-1 UL27 mRNA level was observed. In contrast, in the pEGFP-N1-Cofilin1-transfected group, in which cofilin 1 mRNA expression was increased over 1,000-fold, viral UL27 mRNA expression was decreased to 53% of that of the viral control (Fig. 7G). Western blot analysis confirmed the inhibitory effect of cofilin 1 overexpression on viral protein expression, whereas the effects of cofilin 1 knockdown were not obvious (Fig. 7H). In addition, cofilin 1 overexpression largely decreased the progeny virus titer by 30% compared with that of the untreated control without significantly affecting cell viability; however, only an 8% decrease in the virus titer was observed when cofilin 1 was knocked down (Fig. 7I). Taken together, these results demonstrated that the downregulation of cofilin 1 mainly affects HSV-1 early infection, whereas the upregulation of cofilin 1 disturbs intracellular HSV-1 replication.
DISCUSSION
Studies on the productive infection of other cell types with HSV have provided clues for understanding the correlation between HSV and the F-actin cytoskeleton. The primary roles of F-actin in HSV infection, as separately determined in CHO cells, Madin-Darby canine kidney II (MDCK II) cells, HEp-2 cells, and Vero cells, may involve virus entry, short-range movements underneath the cell membrane, the movement of the capsid to the periphery of the nucleus, and the egress and cell-to-cell spread of viruses, respectively (10, 14, 21, 28, 40). Nevertheless, the exact roles of F-actin in the HSV life cycle seem to be cell type dependent and remain controversial (12).
In this study, we have provided the first evidence that HSV-1 infection induces the biphasic F-actin dynamics in neuronal cells. We found that F-actin assembly was enhanced in the early stages of HSV-1 infection. This enhancement was observed as an increase in the outgrowth of F-actin-containing sheet-like extensions and finger-like protrusions. We demonstrated the critical roles of F-actin dynamics in early HSV-1 infection. During early HSV-1 infection, F-actin may participate in “viral surfing” along the outer membrane of dendrites to reach the soma efficiently and may participate in short-range transport in the cell cortex (11, 28, 31). Although the involvement of F-actin in this process is currently not well understood, our results indicate that actin dynamics, especially the enhanced outgrowth of F-actin-based structures, may facilitate early HSV-1 infection in neuronal cells.
HSV-1 infection induced a continuous decrease in cellular F-actin levels during the later stages of infection. However, the chemical disruption of F-actin dynamics inhibited intracellular HSV-1 reproduction. These results suggest that only continuous dynamics of F-actin, either in the early (assembly tending) or in the late (disassembly tending) stage of infection, would benefit HSV-1 infection or replication. Nonetheless, we confirmed that the intracellular reproduction of HSV-1 in neuronal cells relies on F-actin dynamics, and the present results suggest that HSV-1-induced biphasic F-actin dynamics would benefit viral infection and intracellular replication.
The ADF/cofilin proteins interact with actin dimers to promote disassembly, which can be initiated by the activity of GSN, and cofilin 1 is considered to be primarily responsible for remodeling the actin cytoskeleton (6, 38, 41). In this study, we confirmed that these three regulators were downregulated by HSV-1 infection and further focused on cofilin 1 to elucidate its crucial role in F-actin dynamics during infection. The modulation of cofilin 1 by HSV-1 infection was displayed on two levels: at the level of total protein and at the level of the phosphorylated form of the protein (the inactive form). The level of active cofilin 1 correlated well with the biphasic dynamics of F-actin following infection. This finding was confirmed by the results showing that cofilin 1 knockdown reduced virus-induced F-actin assembly during early infection. The basic function of cofilin 1 is to depolymerize and cleave F-actin (15). Nevertheless, recent studies have suggested novel roles for cofilin 1 as a promoter of F-actin assembly, including in lamellipodium extension, protrusion generation, and cell migration (7, 17, 23, 25, 47). It has recently been reported that cofilin might contribute to stimulus-induced F-actin assembly by generating an abundant pool of cytoplasmic actin monomers (25). Our results indicate that cofilin 1 activation and inactivation may be associated with virus-induced F-actin assembly and disassembly at different stages of the HSV-1 life cycle. Thus, it was suggested that cofilin 1 may also serve as a promoter of F-actin assembly during HSV-1 infection of neuronal cells. Further, this relationship suggests that the HSV-1-induced and cofilin 1-mediated biphasic dynamics of F-actin may provide the conditions required for HSV-1 infection or replication during different stages of the viral life cycle. This hypothesis was confirmed by the results showing that cofilin 1 knockdown inhibits viral entry and that its overexpression decreases intracellular reproduction. Nevertheless, it was reported that HIV activates cofilin to promote actin depolymerization in resting CD4 T cells (45), and in our previous work, we showed that cofilin 1 downregulation in MRC-5 cells inhibited actin depolymerization (32); these findings suggest that the virus-induced F-actin dynamics may occur in a cell-type-dependent or virus-type-dependent manner.
Cofilin 1 works primarily in response to upstream regulator RhoGTPases that serve to phosphorylate cofilin 1 (12, 26, 39). Viruses were reported to manipulate the F-actin dynamics by interfering with the RhoGTPase-mediated signal transduction pathways (12, 39). In HSV infection, a phagocytosis-like uptake of HSV-1 into primary corneal fibroblasts and nectin-1-expressing CHO cells involves the growth of F-actin-containing cell extensions mediated by RhoA activation (10). Early HSV-1 infection of epithelial MDCK II cells and keratinocytes also affects Rac1 and CDC42 activity (21, 33). In addition, other factors that are involved in the RhoGTPase signal pathway might be affected after infection, such as the effect of HSV infection on FAK phosphorylation (9). Nevertheless, whether and how the virus-induced modulation of RhoGTPases or other factors relates to alteration of cofilin 1 activity remain unclear. In this study, we extended the observation period (ranging from the early to the late stages of infection) and detected the expression of the three primary RhoGTPases. HSV-1 infection altered the expression patterns of CDC42, RhoA, and Rac1 compared with mock-infected cells. In comparison with Rac1, the regulation of CDC42 and RhoA is more closely correlated with downstream factors, including cofilin 1, suggesting that their roles in cofilin 1 regulation during HSV-1 infection are different. Overall, we can conclude that the HSV-1-induced changes in cofilin 1 activity might be regulated by a RhoGTPase-mediated signal transduction pathway.
The regulation of F-actin is essential for neuronal functions. Protrusions that extend from the surfaces of neurons might be fundamental for either axon extension and guidance or for the formation of axon branches and synaptic structures (13, 16, 48). The modulation of F-actin dynamics or relevant regulators might therefore result in the disruption of cellular functions or in neurological diseases (26). The invasion of the nervous system by HSV-1 results in the acute brain disease HSE and related neurological dysfunctions. As a consequence, affected patients face high mortality rates and severe comorbidity, which may be related to virus-induced cytoskeleton alterations (11, 46). Our results further suggest the possibility that HSV-1 may cause dysfunction of the nervous system by its ability to remodel F-actin dynamics. The involvement of cofilin 1, a factor that may be related to neurodegenerative processes (4, 22), further indicates the correlation between HSV-1 infection and neuronal dysfunction. Thus, our results might also provide a basis for understanding the pathogenesis of HSV-1-induced neurological diseases.
We conclude that HSV-1 infection induces biphasic dynamics of F-actin that might be required for efficient viral infection and replication. The mechanism by which HSV-1 remodels F-actin involves both protein expression and activity regulation of cofilin 1. Here, we confirm the biphasic mode of cofilin 1 and F-actin regulation in neuronal cells, and we speculate that, even in other cell types, the impact of HSV-1, and perhaps other viruses, on cofilin 1 and F-actin involves a dynamic process of regulation that depends on the requirements of the virus at different stages of infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Twelfth Five-Year National Science and Technology Support Program (2012BAI29B06); the State Key Laboratory of Natural and Biomimetic Drugs, Beijing University (K20120208); and the Fundamental Research Funds for the Central Universities (21611394).
Footnotes
Published ahead of print 23 May 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Agnew BJ, Minamide LS, Bamburg JR. 1995. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 270:17582–17587 [DOI] [PubMed] [Google Scholar]
- 2. Andrianantoandro E, Pollard TD. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24:13–23 [DOI] [PubMed] [Google Scholar]
- 3. Bamburg JR. 1999. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230 [DOI] [PubMed] [Google Scholar]
- 4. Bamburg JR, et al. 2010. ADF/cofilin-actin rods in neurodegenerative diseases. Curr. Alzheimer Res. 7:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baringer JR, Swoveland P. 1973. Recovery of herpes-simplex virus from human trigeminal ganglions. N. Engl. J. Med. 288:648–650 [DOI] [PubMed] [Google Scholar]
- 6. Bernstein BW, Bamburg JR. 2010. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. 2000. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148:531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan C, Beltzner CC, Pollard TD. 2009. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr. Biol. 19:537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheshenko N, et al. 2003. Herpes simplex virus triggers activation of calcium-signaling pathways. J. Cell Biol. 163:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clement C, et al. 2006. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J. Cell Biol. 174:1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dixit R, Tiwari V, Shukla D. 2008. Herpes simplex virus type 1 induces filopodia in differentiated P19 neural cells to facilitate viral spread. Neurosci. Lett. 440:113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Favoreel HW, Enquist LW, Feierbach B. 2007. Actin and Rho GTPases in herpesvirus biology. Trends Microbiol. 15:426–433 [DOI] [PubMed] [Google Scholar]
- 13. Fiala JC, Feinberg M, Popov V, Harris KM. 1998. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J. Neurosci. 18:8900–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forest T, Barnard S, Baines JD. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7:429–431 [DOI] [PubMed] [Google Scholar]
- 15. Galkin VE, et al. 2003. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J. Cell Biol. 163:1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gallo G. 2011. The neuronal actin cytoskeleton and the protrusion of lamellipodia and filopodia, p 7–22 In Gallo G, Lanier LM. (ed), Neurobiology of actin: from neurulation to synaptic function. Springer Press, New York, NY [Google Scholar]
- 17. Ghosh M, et al. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304:743–746 [DOI] [PubMed] [Google Scholar]
- 18. Greene W, Gao SJ. 2009. Actin dynamics regulate multiple endosomal steps during Kaposi's sarcoma-associated herpesvirus entry and trafficking in endothelial cells. Plos. Pathog. 5:e1000512 doi:10.1371/journal.ppat.1000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goley ED, Welch MD. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7:713–726 [DOI] [PubMed] [Google Scholar]
- 20. Hjalmarsson A, Blomqvist P, Skoldenberg B. 2007. Herpes simplex encephalitis in Sweden, 1990-2001: incidence, morbidity, and mortality. Clin. Infect. Dis. 45:875–880 [DOI] [PubMed] [Google Scholar]
- 21. Hoppe S, et al. 2006. Early herpes simplex virus type 1 infection is dependent on regulated Rac1/Cdc42 signalling in epithelial MDCKII cells. J. Gen. Virol. 87:3483–3494 [DOI] [PubMed] [Google Scholar]
- 22. Jang DH, et al. 2005. Cofilin expression induces cofilin-actin rod formation and disrupts synaptic structure and function in Aplysia synapses. Proc. Natl. Acad. Sci. U. S. A. 102:16072–16077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jovceva E, Larsen MR, Waterfield MD, Baum B, Timms JF. 2007. Dynamic cofilin phosphorylation in the control of lamellipodial actin homeostasis. J. Cell Sci. 120:1888–1897 [DOI] [PubMed] [Google Scholar]
- 24. Ju HQ, et al. 2011. BJ-B11, a novel Hsp90 inhibitor, induces apoptosis in human chronic myeloid leukemia K562 cells through the mitochondria-dependent pathway. Eur. J. Pharmacol. 666:26–34 [DOI] [PubMed] [Google Scholar]
- 25. Kiuchi T, Ohashi K, Kurita S, Mizuno K. 2007. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 177:465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo LQ. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1:173–180 [DOI] [PubMed] [Google Scholar]
- 27. Lycke EHB, Johansson M, Krotochwil A, Lycke J, Svennerholm B. 1988. Herpes simplex virus infection of the human sensory neuron. An electron microscopy study. Arch. Virol. 101:87–104 [DOI] [PubMed] [Google Scholar]
- 28. Lyman MG, Enquist LW. 2009. Herpesvirus interactions with the host cytoskeleton. J. Virol. 83:2058–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moriyama K, Iida K, Yahara I. 1996. Phosphorylation of Ser-3 of cofilin regulates its essential function on actin. Genes Cells 1:73–86 [DOI] [PubMed] [Google Scholar]
- 30. Nicola AV, Hou J, Major EO, Straus SE. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 79:7609–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oh MJ, Akhtar J, Desai P, Shukla D. 2010. A role for heparan sulfate in viral surfing. Biochem. Biophys. Res. Commun. 391:176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pei Y, et al. 2011. Pentagalloylglucose downregulates cofilin1 and inhibits HSV-1 infection. Antiviral Res. 89:98–108 [DOI] [PubMed] [Google Scholar]
- 33. Petermann P, Haase I, Knebel-Morsdorf D. 2009. Impact of Rac1 and Cdc42 signaling during early herpes simplex virus type 1 infection of keratinocytes. J. Virol. 83:9759–9772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qualmann B, Kessels MM. 2008. Actin nucleation: putting the brakes on Arp2/3. Curr. Biol. 18:R420–R423 [DOI] [PubMed] [Google Scholar]
- 35. Radtke K, Dohner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol. 8:387–400 [DOI] [PubMed] [Google Scholar]
- 36. Roizman BKD, Whitley RJ. 2007. Herpes simplex viruses, p 2502–2601 In Knipe DM, Howley PM. (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 37. Sarmiere PD, Bamburg JR. 2004. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J. Neurobiol. 58:103–117 [DOI] [PubMed] [Google Scholar]
- 38. Southwick FS. 2000. Gelsolin and ADF/cofilin enhance the actin dynamics of motile cells. Proc. Natl. Acad. Sci. U. S. A. 97:6936–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor MP, Koyuncu OO, Enquist LW. 2011. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 9:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Leeuwen H, Elliott G, O'Hare P. 2002. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J. Virol. 76:3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Van Troys M, et al. 2008. Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87:649–667 [DOI] [PubMed] [Google Scholar]
- 42. Wear MM, Schafer DA, Cooper JA. 2000. Actin dynamics: assembly and disassembly of actin networks. Curr. Biol. 10:R891–R895 [DOI] [PubMed] [Google Scholar]
- 43. Whitley RJ, Gnann JW. 2002. Viral encephalitis: familiar infections and emerging pathogens. Lancet 359:507–513 [DOI] [PubMed] [Google Scholar]
- 44. Xiang Y, et al. 2011. In vitro anti-herpes simplex virus activity of 1,2,4,6-tetra-O-galloyl-beta-d-glucose from Phyllanthus emblica L. (Euphorbiaceae). Phytother. Res. 25:975–982 [DOI] [PubMed] [Google Scholar]
- 45. Yoder A, Yu DY, Dong L. 2008. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 134:782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zambrano A, et al. 2008. Neuronal cytoskeletal dynamic modification and neurodegeneration induced by infection with herpes simplex virus type 1. J. Alzheimers Dis. 14:259–269 [DOI] [PubMed] [Google Scholar]
- 47. Zebda N, et al. 2000. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J. Cell Biol. 151:1119–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ziv NE, Smith SJ. 1996. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17:91–102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.