Abstract
The epidemiology of Staphylococcus epidermidis in U.S. hospitals remains limited. This study aimed to address the genetic backgrounds of linezolid-susceptible and -resistant S. epidermidis strains (isolated in 2010), including cfr-carrying strains. In addition, the antimicrobial susceptibility profiles and linezolid resistance mechanisms among clonal lineages were assessed. A total of 71 S. epidermidis isolates were selected, and linezolid-resistant strains were screened for cfr and mutations in 23S rRNA, L3, and L4. All isolates were subjected to multilocus sequence typing (MLST), and the results were analyzed by eBURST. Overall, 27 sequence types (STs) were detected, and ST5 (21.1%) and ST2 (16.9%) predominated. The majority (62/71; 87.3%) of STs belonged to clonal complex 2 (CC2), which was mostly comprised of subclusters CC2-II (41/62; 66.1%) and CC2-I (21/62; 33.9%). Other STs were grouped within CC23 or CC32 or were singletons. CC2-I strains were more likely to display a methicillin (95.2% versus 33.3 to 70.7%), a linezolid (47.6% versus 0.0 to 7.3%), or a multidrug (81.0% versus 33.3 to 36.6%) resistance phenotype. Among linezolid-resistant isolates, cfr was noted only within CC2 strains, and it was detected equally in the CC2-I (3/10; 30.0%) and CC2-II (1/3; 33.3%) subclusters. 23S rRNA mutations (G2576 [seven strains] and C2534 [one strain]) were observed only among CC2-I (8/10; 80.0%) isolates. Strains showing a G2576 alteration also had M156 (7/7; 100.0%) and/or H146 (6/7; 85.7%) L3 modifications. This study provides an overview of the S. epidermidis clonal distribution and reports higher resistance rates among CC2-I strains. The results show that cfr may be acquired and expressed by both CC2 main subclusters, while 23S rRNA mutations appeared more often within CC2-I strains. Interestingly, these 23S rRNA mutants also had L3 alterations, which may act synergistically or in a compensatory manner to minimize the fitness cost while providing survival advantages under selective pressure.
INTRODUCTION
Staphylococcus epidermidis is ubiquitous in the human skin and mucosal microflora and causes infections in immunocompetent patients when the integrity of the skin barrier is disturbed (23). However, the vast majority of S. epidermidis infections among hospitalized patients have been associated with indwelling medical devices, such as intravascular and intrathecal catheter systems, pacemaker electrodes, urinary tract catheters, and other polymer and metal implants, which are used as vehicles for entering the host (33). It has been demonstrated that the organism possesses great capability for genetic recombination and gene acquisition, including resistance determinants (20). In fact, the rates of methicillin (oxacillin) resistance among S. epidermidis strains currently exceed 70% in many institutions worldwide (1, 7). However, although antimicrobial resistance can compromise therapy, treatment failure has been primarily associated with the species' ability to form biofilms on medical devices, which is a common feature of many nosocomial pathogens (29).
Strains of S. epidermidis resistant to antimicrobial agents other than oxacillin (β-lactams) have also been reported. In large surveillance studies, the linezolid resistance rates are still low; however, there seems to be a trend toward increased rates (7). S. epidermidis appears to be prone to accumulate linezolid resistance mechanisms, such as mutations in the 23S rRNA and in the ribosomal proteins L3 and L4 (7, 10, 13, 18, 19). In addition to alterations in the target site, a more recent linezolid resistance mechanism, the cfr gene, has been increasingly reported in the literature (6, 8, 30). cfr encodes a methyltransferase that catalyzes the posttranscriptional methylation of nucleotide A2503 in the 23S rRNA (6). This gene was initially detected in a transferable plasmid in a Staphylococcus sciuri strain from animal sources (24) and was later observed in several other staphylococci from animal and human origins (3, 8, 22, 25, 28, 30).
Pulsed-field gel electrophoresis (PFGE) has been widely used for characterizing S. epidermidis isolates, though the method has mostly been applied to short-term epidemiological investigations (20). More recently, an improved multilocus sequence typing (MLST) scheme was developed for S. epidermidis (27), and the method provided additional information related to the population structure and epidemiology of the species (21). Subsequently, several reports have described the epidemiology of nosocomial S. epidermidis, including linezolid-resistant isolates (9, 10, 13, 31, 32). However, information regarding the genetic background of S. epidermidis isolates responsible for nosocomial infection within U.S. hospitals remains limited. Therefore, this study addresses the genetic background of linezolid-susceptible and -resistant S. epidermidis strains, including cfr-carrying strains, responsible for infections in monitored U.S. hospitals during the 2010 SENTRY Antimicrobial Surveillance Program. In addition, analyses of the antimicrobial susceptibility profiles and linezolid resistance mechanisms within clonal lineages were evaluated.
MATERIALS AND METHODS
Bacterial strains.
A total of 71 S. epidermidis clinical isolates collected during the 2010 sampling year of the SENTRY Program were included in this study. At least one representative methicillin-susceptible strain and one methicillin-resistant strain were randomly selected, when available, from each medical institution contributing S. epidermidis isolates. Additional strains were selected for each methicillin susceptibility group (up to 10% of the total surveillance strains submitted) from those medical sites contributing more than 20 strains during the study period. When multiple strains from a single site were included, isolates exhibiting different antimicrobial susceptibility profiles were chosen.
Antimicrobial susceptibility profile.
Susceptibility testing was performed by broth microdilution methods, according to the Clinical and Laboratory Standards Institute (CLSI) recommendations (4). MIC interpretations were applied as described in CLSI document M100-S22 (5). Staphylococcus aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 were concurrently tested for quality assurance purposes. In addition, the inoculum density was monitored by colony counts to ensure an adequate number of cells for each testing event. Isolates exhibiting a resistance phenotype to at least four different classes of antimicrobial agents were considered multidrug resistant (MDR).
Screening for linezolid resistance mechanisms.
All linezolid-resistant strains were screened for the presence of cfr, as well as mutations in the 23S rRNA and ribosomal proteins (L3 and L4), by PCR and sequencing as previously described (19). Amplicons were sequenced on both strands. The ribosomal proteins obtained were compared to those from wild-type S. epidermidis ATCC 12228 using the Lasergene software package (DNAStar, Madison, WI). In addition, seven linezolid-susceptible (all with MIC values of 0.5 μg/ml) strains recovered from the same hospital sites as the linezolid-resistant isolates and belonging to the same sequence type (ST) were also screened for mutations in ribosomal proteins for comparison purposes.
Molecular typing.
All selected strains were subjected to pulsed-field gel electrophoresis (PFGE). SmaI-digested genomic DNA was resolved in CHEF-DR II (Bio-Rad, Richmond, CA), and the profiles obtained were analyzed using GelCompar II software (Applied Math, Kortrijk, Belgium) (20). MLST was performed in all strains according to the methods published for S. epidermidis (27). STs were determined using the S. epidermidis MLST databases and the eBURST method to infer the evolutionary relatedness of STs (http://www.mlst.net/).
RESULTS
Organisms and antimicrobial susceptibility characteristics.
The S. epidermidis clinical isolates included in this study originated from 27 medical centers in 25 cities (19 states and 7 U.S. Census regions), and the majority (49/71; 69.0%) of strains were recovered from blood, followed by other, less prevalent (<13% each) specimen types (Table 1). Overall, 52 of 71 (73.2%) strains were methicillin resistant, with varying susceptibility to other antimicrobial agents, as follows: ciprofloxacin (29.6%), clindamycin (59.1%), erythromycin (31.0%), gentamicin (59.2%), tetracycline (88.7%), trimethoprim-sulfamethoxazole (52.1%), and vancomycin (100.0%). A total of 35 (49.3%) S. epidermidis clinical isolates demonstrated an MDR phenotype, defined as phenotypic resistance to at least four classes of antimicrobial agents described in Table 1. Only 13 (18.3% within this study) strains displaying elevated MIC results for linezolid (16 to >128 μg/ml) were detected during the 2010 sampling year and included in this investigation. All linezolid-resistant S. epidermidis strains were methicillin-resistant.
Table 1.
Epidemiology, antimicrobial susceptibility testing results, and demographic information related to S. epidermidis isolates included in this study
| Isolate | State | City | Molecular typing |
Antimicrobial susceptibilityb |
MDRc | Specimend | Age (yr)e | Sexf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CCa | MLST | OXA | LZD | CIP | CLI | TET | ERY | GEN | VAN | SXT | |||||||
| 38307 | TX | Houston | 23 | ST23 | R | S | R | R | R | R | I | S | R | Y | WD | 86 | F |
| 7241 | KY | Louisville | 23 | ST23 | R | S | R | R | S | R | I | S | S | Y | BC | 35 | M |
| 11279 | MO | Kansas City | 32 | ST32 | S | S | S | S | S | R | S | S | S | N | UC | 6 | M |
| 18201 | MI | Detroit | 32 | ST358 | S | S | S | S | S | S | S | S | S | N | WD | 11 | F |
| 2758 | AR | Little Rock | 193 | ST193 | S | S | S | S | S | R | S | S | S | N | PF | 16 | M |
| 35920 | PA | Hershey | 249 | ST249 | S | S | S | S | S | R | S | S | S | N | BC | 68 | M |
| 18001 | LA | New Orleans | 347 | ST347 | S | S | S | S | S | S | S | S | S | N | BC | 90 | M |
| 22541 | MI | Lansing | 348 | ST348 | R | S | S | R | S | R | S | S | R | Y | SK | 22 | M |
| 18461 | MI | Detroit | 360 | ST360 | S | S | S | S | S | R | S | S | R | N | BC | 80 | F |
| 18676 | TX | Galveston | 2–I | ST16 | R | S | R | I | S | S | S | S | S | N | BC | 91 | F |
| 49671 | WI | Milwaukee | 2–I | ST16 | R | S | S | S | S | R | S | S | S | N | BC | 76 | M |
| 16644 | TN | Memphis | 2–I | ST16 | R | S | S | S | S | R | S | S | S | N | BC | 49 | F |
| 38449 | AZ | Tempe | 2–I | ST186 | R | R | R | R | S | I | I | S | S | Y | BC | 73 | M |
| 25438 | MA | Boston | 2–I | ST2 | R | R | R | I | S | I | R | S | R | Y | BC | 61 | M |
| 37506 | TX | Houston | 2–I | ST2 | R | R | R | I | S | I | R | S | R | Y | BC | 29 | F |
| 25437 | MA | Boston | 2–I | ST2 | R | R | R | I | S | R | R | S | R | Y | BC | 26 | M |
| 34444 | TX | Houston | 2–I | ST2 | R | R | R | I | S | R | R | S | R | Y | BC | 66 | M |
| 39867 | TX | Houston | 2–I | ST2 | R | R | R | I | S | R | I | S | R | Y | SN | 67 | M |
| 18006 | LA | New Orleans | 2–I | ST2 | R | R | R | S | S | S | R | S | R | Y | BC | 50 | M |
| 38531 | PA | Hershey | 2–I | ST2 | R | R | R | S | S | S | S | S | R | Y | SK | 42 | M |
| 13036 | KY | Lexington | 2–I | ST2 | R | S | R | R | S | R | S | S | R | Y | BC | 60 | M |
| 29257 | CT | Farmington | 2–I | ST2 | R | S | R | R | S | R | R | S | R | Y | BC | 50 | M |
| 35921 | PA | Hershey | 2–I | ST2 | R | S | R | R | S | R | R | S | R | Y | SK | 2 | M |
| 16669 | NY | New York | 2–I | ST2 | R | S | R | R | S | R | R | S | R | Y | WD | 84 | M |
| 5849 | KY | Lexington | 2–I | ST2 | S | S | R | R | S | R | R | S | R | Y | BC | 56 | F |
| 12676 | OH | Akron | 2–I | ST22 | R | R | R | R | S | I | I | S | R | Y | BC | 61 | F |
| 2907 | KY | Lexington | 2–I | ST22 | R | R | R | R | S | I | R | S | R | Y | BC | 55 | F |
| 9611 | NJ | New Brunswick | 2–I | ST22 | R | S | R | S | S | S | R | S | R | Y | BJ | 77 | M |
| 10560 | NY | New York | 2–I | ST346 | R | S | S | S | S | R | S | S | S | N | BC | 52 | M |
| 30077 | MN | St. Paul | 2–I | ST361 | R | S | R | S | S | S | R | S | R | Y | SK | 63 | M |
| 2763 | AR | Little Rock | 2–II–5 | ST130 | R | S | S | R | S | R | S | S | S | N | BC | 21 | M |
| 28062 | MO | Kansas City | 2–II–5 | ST136 | R | S | S | S | S | R | S | S | S | N | BC | 6* | F |
| 36018 | MA | Burlington | 2–II–5 | ST359 | R | S | R | S | S | S | S | S | S | N | BC | 55 | M |
| 4042 | MO | Kansas City | 2–II–5 | ST5 | R | R | R | R | S | R | S | S | S | Y | BC | 49 | F |
| 34374 | MI | Detroit | 2–II–5 | ST5 | R | R | R | S | S | I | R | S | R | Y | BC | 50 | M |
| 22291 | TN | Memphis | 2–II–5 | ST5 | R | R | R | S | S | I | R | S | R | Y | PL | 53 | M |
| 2969 | IA | Des Moines | 2–II–5 | ST5 | R | S | R | R | R | R | S | S | R | Y | BC | 50 | M |
| 30328 | KY | Lexington | 2–II–5 | ST5 | R | S | R | S | R | R | S | S | R | Y | PF | 39 | F |
| 17996 | LA | New Orleans | 2–II–5 | ST5 | R | S | R | S | S | R | S | S | S | N | BC | 40 | M |
| 34420 | MI | Detroit | 2–II–5 | ST5 | R | S | R | S | S | R | S | S | S | N | BC | 87 | M |
| 17994 | LA | New Orleans | 2–II–5 | ST5 | R | S | R | S | S | R | I | S | R | Y | UT | 80 | F |
| 30079 | MN | St. Paul | 2–II–5 | ST5 | R | S | R | S | S | S | R | S | S | N | BC | 69 | M |
| 42022 | PA | Hershey | 2–II–5 | ST5 | R | S | R | S | S | S | S | S | S | N | BC | 62 | M |
| 497 | NE | Omaha | 2–II–5 | ST5 | R | S | R | S | S | S | S | S | S | N | CT | 70 | M |
| 30078 | MN | St. Paul | 2–II–5 | ST5 | S | S | R | R | S | R | S | S | S | N | U | 72 | M |
| 16733 | OH | Akron | 2–II–5 | ST5 | S | S | R | S | R | R | S | S | S | N | BC | 56 | M |
| 13638 | NY | New York | 2–II–5 | ST5 | S | S | R | S | S | R | S | S | S | N | BC | 71 | F |
| 29249 | CT | Farmington | 2–II–5 | ST5 | S | S | R | S | S | S | S | S | S | N | BC | 50 | F |
| 25442 | MA | Boston | 2–II–5 | ST59 | R | S | R | I | S | R | I | S | S | N | UT | 80 | M |
| 35037 | NE | Omaha | 2–II–5 | ST59 | R | S | R | S | R | S | R | S | S | Y | BC | 71 | F |
| 34419 | MI | Detroit | 2–II–5 | ST59 | R | S | R | S | R | S | R | S | R | Y | BC | 77 | M |
| 69 | NJ | New Brunswick | 2–II–5 | ST59 | R | S | R | S | S | S | S | S | S | N | BC | 79 | F |
| 4045 | MO | Kansas City | 2–II–5 | ST59 | R | S | S | S | S | R | S | S | S | N | BC | 40 | F |
| 29784 | OH | Akron | 2–II–5 | ST69 | R | S | R | S | S | R | S | S | S | N | BC | 28 | M |
| 28061 | MO | Kansas City | 2–II–5 | ST7 | S | S | R | R | S | R | S | S | R | Y | BC | 3 | M |
| 8749 | WI | Milwaukee | 2–II–5 | ST7 | S | S | R | R | S | R | S | S | R | Y | BC | 52 | F |
| 22310 | TN | Memphis | 2–II–5 | ST7 | S | S | R | S | S | R | S | S | R | N | BC | 52 | M |
| 17995 | LA | New Orleans | 2–II–5 | ST7 | S | S | S | S | S | R | S | S | R | N | UT | 30 | F |
| 6711 | AZ | Tucson | 2–II–5 | ST83 | R | S | R | I | S | S | S | S | S | N | BC | 55 | F |
| 34501 | TX | Houston | 2–II–5 | ST83 | R | S | R | S | S | S | R | S | R | Y | BC | 82 | M |
| 2730 | AR | Little Rock | 2–II–5 | ST83 | R | S | R | R | S | R | R | S | R | Y | WD | 2* | M |
| 13657 | OH | Cincinnati | 2–II–5 | ST83 | R | S | R | S | R | R | S | S | R | Y | BC | 59 | M |
| 48458 | MI | Detroit | 2–II–5 | ST83 | R | S | R | S | S | R | S | S | R | Y | BC | 79 | F |
| 2674 | NE | Omaha | 2–II–5 | ST83 | R | S | R | S | S | S | S | S | R | N | BJ | 48 | M |
| 22300 | TN | Memphis | 2–II–5 | ST83 | S | S | S | S | S | S | R | S | S | N | BC | 37 | M |
| 2820 | NJ | New Brunswick | 2–II–6 | ST345 | S | S | S | R | S | R | S | S | S | N | BC | 54 | F |
| 16675 | NY | New York | 2–II–6 | ST73 | S | S | S | I | R | S | S | S | S | N | WD | 60 | F |
| 11175 | UT | Salt Lake City | 2–II–85 | ST20 | R | S | S | R | S | R | R | S | S | Y | BC | 2* | M |
| 5687 | MO | Kansas City | 2–II–85 | ST20 | R | S | S | S | S | R | I | S | S | N | UC | 1* | M |
| 30259 | MI | Detroit | 2–II–89 | ST327 | S | S | S | S | S | S | S | S | S | N | BC | 80 | M |
| 1374 | NJ | New Brunswick | 2–II–89 | ST88 | R | S | S | S | S | S | S | S | S | N | BC | 80 | F |
Clonal complexes were determined according to the eBURST analysis.
MIC interpretive criteria as published in CLSI M100-S22. S, susceptible; I, intermediate; R, resistant; OXA, oxacillin; LZD, linezolid; CIP, ciprofloxacin; CLI, clindamycin; TET, tetracycline; ERY, erythromycin; GEN, gentamicin; VAN, vancomycin; SXT, trimethoprim-sulfamethoxazole.
Multidrug resistance was defined as an isolate exhibiting a resistance phenotype to at least four classes of antimicrobial agents.
WD, wound; UT, urinary tract; UC, urethral catheter; SK, skin; SN, sinus; BJ, joint/bone; CT, catheter; PF peritoneal fluid; PL, pleural fluid.
An asterisk indicates age in months.
F, female; M, male.
Linezolid resistance mechanisms.
Among linezolid-resistant strains, four carried cfr, while the remaining isolates possessed modifications at the G2576 or C2534 position in the 23S rRNA (Table 2). The presence of cfr or mutations in the 23S rRNA was always associated with alterations in L3 and/or L4 ribosomal proteins, except for strains 22291 and 34374, which had alterations only in L3 and L4. One S. epidermidis isolate (38449) demonstrated a combination of 23S rRNA alteration (C2534T) and the cfr gene. The most common alterations in the L3 protein were at positions 146, 154, 156, and 157, while an insertion at position 71 of L4 was present in 69.2% (9/13) of linezolid-resistant strains. Among linezolid-susceptible strains selected for additional screening for mutations in the ribosomal proteins, all isolates demonstrated wild-type amino acid sequences (i.e., sequences identical to those of ATCC 12228) (data not shown).
Table 2.
Linezolid resistance mechanisms observed among S. epidermidis strains recovered from USA hospitals and selected for this study.
| Isolate | City | State | Site | Linezolid MIC (μg/ml) | Molecular typing |
Linezolid resistance mechanismb |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| CCa | MLST | cfrc | 23S rRNA | L3 | L4 | |||||
| 4042 | Kansas City | MO | 449 | 16 | 2–II–5 | ST5 | + | WT | A157R | WT |
| 34374 | Detroit | MI | 003 | 16 | 2–II–5 | ST5 | − | WT | H146Q/V154L/A157R | 71G72 |
| 22291 | Memphis | TN | 412 | 16 | 2–II–5 | ST5 | − | WT | H146Q/V154L/A157R | 71G72 |
| 18006 | New Orleans | LA | 448 | 32 | 2–I | ST2 | − | G2576T | H146P/M156T | WT |
| 38531 | Hershey | PA | 453 | 32 | 2–I | ST2 | − | G2576T | V154L/M156T | WT |
| 39867 | Houston | TX | 024 | 64 | 2–I | ST2 | − | G2576T | H146R/M156T | 71G72 |
| 37506 | Houston | TX | 116 | 64 | 2–I | ST2 | − | G2576T | H146R/M156T | 71G72 |
| 34444 | Houston | TX | 116 | 128 | 2–I | ST2 | − | G2576T | H146R/M156R | 71G72 |
| 25438 | Boston | MA | 441 | 128 | 2–I | ST2 | − | G2576T | G137S/H146P/M156R | G69R/71G72 |
| 25437 | Boston | MA | 441 | 128 | 2–I | ST2 | − | G2576T | G137D/H146R/V154L/M156T | 71G72 |
| 2907 | Lexington | KY | 107 | 128 | 2–I | ST22 | + | WT | V154L/A157R | WT |
| 12676 | Akron | OH | 004 | >128 | 2–I | ST22 | + | WT | H146Q/V154L/A157R | 71G72 |
| 38449 | Tempe | AZ | 426 | >128 | 2–I | ST186 | + | C2534T | H146Q/V154L/A157R | 71G72 |
Clonal complexes were determined according to the eBURST analysis.
WT, wild type.
+, present; −, absent.
Genetic background.
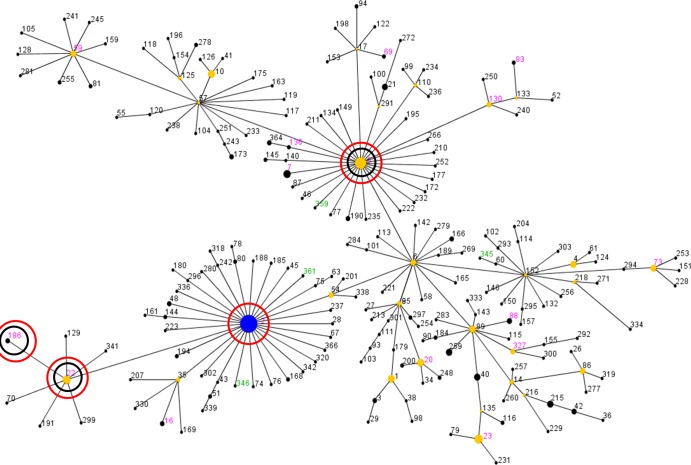
A total of 27 STs were observed; ST5 (21.1%) and ST2 (16.9%) predominated, followed by ST83 (9.9%), ST59 (7.0%), and ST7 (5.6%). Other STs had occurrence rates below 4.5% (Table 1). A total of 19 STs had been previously recorded in the MLST database (as of January 2012), while 8 STs were unique to this study (Fig. 1). The eBURST algorithm clustered these STs into one major clonal complex (clonal complex 2 [CC2]; 62/71; 87.3%), two minor CCs (CC23 and CC32 [2/71; 2.8% each]), and five singletons (CC193, CC249, CC347, CC348, and CC360) (Table 1 and Fig. 1). It has been proposed (20) that CC2 can be segregated into clusters CC2-II (41/62; 66.1%) and CC2-I (21/62; 33.9%) and that cluster CC2-II can be further subdivided into four groups, represented in this study by CC2-II-5 (35/41; 85.4%), CC2-II-6 (2/41; 4.9%), CC2-II-85 (2/41; 4.9%), and CC2-II-89 (2/41; 4.9%) (Table 1). Analysis of SmaI PFGE patterns clustered the S. epidermidis isolates into 41 major types and 62 subtypes. Two PFGE types showed the highest number of strains (8/71; 11.3%) and were associated with CC2-I or CC2-II (data not shown).
Fig 1.
eBURST analysis of S. epidermidis CC2 using all 312 STs available in the MLST database as of January 2012. Each ST is represented by a dot, and lines connect single-locus variants. The blue dot (ST2) represents the putative founder of CC2. The yellow dots represent putative subgroup founders. The pink STs represent those previously recorded in the MLST database, while the green STs represent new STs detected in this study. The red circles indicate STs associated with linezolid-resistant isolates, and the inner black circles indicate STs associated with Cfr-mediated resistance.
Resistance profile and genetic background.
All S. epidermidis strains belonging to CC2-I were methicillin resistant (20/21; 95.2%) except for one strain. Lower methicillin resistance rates were observed among CC2-II (29/41; 70.7%) and other clusters (3/9; 33.3%) (Tables 1 and 3), the latter represented by CC23 (two strains), CC32 (two strains), CC193 (one strain), CC249 (one strain), CC347 (one strain), CC348 (one strain), and CC360 (one strain). Linezolid resistance was noted only within CC2-I (10/21; 47.6%) and CC2-II (3/41; 7.3%) S. epidermidis isolates. Among the linezolid-resistant strains, Cfr-mediated resistance was detected equally among CC2-I (3/10; 30.0%) and CC2-II (1/3; 33.3%) strains, while modifications in the 23S rRNA were present only among CC2-I isolates (8/10; 80.0%) (Tables 1, 2, and 3). Overall, strains exhibiting an alteration at position G2576 of 23S rRNA also demonstrated modifications in the L3 ribosomal protein at position M156 (7/7; 100.0%) and/or H146 (6/7; 85.7%). An MDR phenotype was observed among 81.0% (17/21) of the strains belonging to CC2-I, whereas rates between 33.3% and 36.3% were noted within other clusters (Table 3).
Table 3.
Occurrences of antimicrobial resistance phenotypes according to clonal complexes among S. epidermidis clinical strains
| Resistance phenotype | CCa | Occurrence (no./total) | Percentage |
|---|---|---|---|
| Methicillin resistance | 2–I | 20/21 | 95.2 |
| 2–II | 29/41 | 70.7 | |
| Other | 3/9 | 33.3 | |
| Linezolid resistance | 2–I | 10/21 | 47.6 |
| 2–II | 3/41 | 7.3 | |
| Other | 0/9 | 0.0 | |
| Multidrug resistanceb | 2–I | 17/21 | 81.0 |
| 2–II | 15/41 | 36.6 | |
| Other | 3/9 | 33.3 |
CC2–II includes subclusters 2–II–5 (35 strains), 2–II–6 (two strains), 2–II–85 (two strains), and 2–II–89 (two strains). Other CCs include CC23 (two strains), CC32 (two strains), CC193 (one strain), CC249 (one strain), CC347 (one strain), CC348 (one strain), and CC360 (one strain).
Multidrug resistance represents strains exhibiting a resistance phenotype to at least four different classes of antimicrobial agents.
DISCUSSION
A high degree of genetic diversity (41 PFGE types) was observed within a contemporary collection of S. epidermidis strains. This was also indicated by the elevated number of STs (27 STs among 71 strains) detected. The proportion of STs observed in this study was higher than that previously reported (8 STs among 40 strains) among linezolid-resistant coagulase-negative staphylococcus (CoNS) isolates from the United States (32). However, 25% of the CoNS strains included in this study originated from a single medical site. A larger study, which analyzed the diversity of S. epidermidis strains collected from several countries worldwide, also identified high genotypic diversity (74 STs among 217 strains) (21). These data corroborate the genetic diversity reported here, which was expected due to the strain selection criteria applied and the fact that the isolates had very diverse geographic origins, as well.
The sequence types ST5 (21.1%) and ST2 (16.9%) predominated in this investigation. ST2 has usually been the most prevalent ST observed in previous epidemiological studies among S. epidermidis strains worldwide (non-United States) (9, 12, 20, 21, 31). Similar findings (45% and 10% of ST2 and ST5, respectively) were observed by Wong et al. (32), who described the genetic backgrounds of linezolid-resistant S. epidermidis isolates from U.S. hospitals. Conversely, the study presented here detected an overall greater proportion of ST5 strains. However, further analysis of linezolid-resistant strains showed that this resistant subset was mostly comprised of ST2 strains (7/13; 53.8%) and a smaller percentage of ST5 isolates (3/13; 23.1%), corroborating the data previously reported (32).
Although less frequent, S. epidermidis strains associated with CC2-I were more likely to display a methicillin resistance phenotype (95.2% versus 33.3 to 70.7% in other clusters). A linezolid resistance (47.6% versus 0.0 to 7.3%) and an MDR (81.0% versus 33.3 to 36.6%) phenotype were also more commonly observed in CC2-I. Previous studies demonstrated that S. epidermidis isolates have high rates of recombination events and acquisition of foreign DNA, including resistance determinants (20). Therefore, it is tempting to speculate that among different populations of S. epidermidis, those associated with CC2-I may be more prone to acquire, maintain, and express resistance genes. In fact, it has been demonstrated that ST2 (CC2-I) isolates are associated with health care infections in the United States and Europe and with biofilm production (23, 33). Thus, the ability of this lineage to acquire resistance genes renders the clone highly adapted to the nosocomial environment. Furthermore, cfr was noted only within CC2 strains, and the gene was detected equally in subclusters CC2-I and CC2-II, suggesting that both groups can acquire and express the gene. This may be due to the fact that minimal fitness costs have been associated with cfr (11), although the number of cfr strains included here was limited.
All linezolid-resistant ST2 strains described in this study exhibited a G2576T mutation, along with several alterations in L3 and/or L4. Similar results were observed for isolate 38449 (ST186, a double-locus variant of ST2), which showed an alteration at position C2534 of 23S rRNA and L3/L4 mutations. In contrast, linezolid-resistant ST5 isolates did not show 23S rRNA alterations. It is well known that the accumulation of 23S rRNA modifications is associated with reduced fitness (26). The findings here suggest that 23S rRNA mutations may affect the biological fitness cost of CC2-I isolates to a lesser extent than with other clones. It can also be hypothesized that the L3 changes act synergistically, requiring a lower number of 23S rRNA mutated alleles (16). In addition, certain L3 modifications detected in this study (i.e., at position H146 and/or M156) may compensate for any deleterious effects associated with 23S rRNA mutations. Compensatory mutations within genes responsible for decreased susceptibility or resistance or mutations at distinct sites have been shown to increase fitness (17). Actually, it has been reported that a mutation at position Y137 in L3 (which corresponds to amino acid F147 in S. epidermidis) restores the fitness cost associated with G2576T in Streptococcus pneumoniae (2). These mutations in L3 may be implicated in releasing constraints associated with unfavorable 23S rRNA mutations by altering the conformation, thereby increasing/restoring protein stability (2). It is important to emphasize that selected linezolid-susceptible strains did not exhibit L3/L4 mutations, suggesting that the alterations observed among linezolid-resistant isolates do not represent polymorphisms inherent to these lineages (14–16, 26).
This investigation was designed to select clinical isolates from several geographic regions within the United States to establish the genetic backgrounds of S. epidermidis nosocomial isolates. Therefore, this database does not provide a precise prevalence rate for each clonal lineage, since these rates may have been biased by the applied selection criteria. The fact that only 13 linezolid-resistant strains were included also limits possible associations between resistance mechanisms and lineages of S. epidermidis. However, in summary, this study provides a contemporary overview of the genetic backgrounds of S. epidermidis clinical strains recovered from U.S. hospitals and shows a remarkable presence of antimicrobial resistance among CC2-I strains compared with other clusters. In addition, the data presented suggest that 23S rRNA and L3 mutations may act synergistically or in a compensatory manner to minimize the fitness cost in CC2-I S. epidermidis strains, mainly ST2 strains, while providing survival advantages under selective pressure.
ACKNOWLEDGMENTS
We thank the following staff members at JMI Laboratories (North Liberty, IA) for technical support and manuscript assistance: S. Benning, M. Castanheira, S. Farrell, G. J. Moet, and P. R. Rhomberg. We also thank Ronald N. Jones for carefully reviewing the manuscript.
Footnotes
Published ahead of print 11 June 2012
REFERENCES
- 1. Biedenbach DJ, Farrell DJ, Mendes RE, Ross JE, Jones RN. 2010. Stability of linezolid activity in an era of mobile oxazolidinone resistance determinants: Results from the 2009 Zyvox Annual Appraisal of Potency and Spectrum program. Diagn. Microbiol. Infect. Dis. 68:459–467 [DOI] [PubMed] [Google Scholar]
- 2. Billal DS, Feng J, Leprohon P, Legare D, Ouellette M. 2011. Whole genome analysis of linezolid resistance in Streptococcus pneumoniae reveals resistance and compensatory mutations. BMC Genomics 12:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonilla H, et al. 2010. Multicity outbreak of linezolid-resistant Staphylococcus epidermidis associated with clonal spread of a cfr-containing strain. Clin. Infect. Dis. 51:796–800 [DOI] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2012. M07-A9. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2012. M100-S22. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 6. Diaz L, et al. 2012. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob. Agents Chemother. 56:3917–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2011. LEADER Program results for 2009: an activity and spectrum analysis of linezolid using 6,414 clinical isolates from the United States (56 medical centers). Antimicrob. Agents Chemother. 55:3684–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gopegui ER, Juan C, Zamorano L, Perez JL, Oliver A. 2012. Transferable multidrug resistance plasmid carrying cfr associated with tet(L), ant(4′)-Ia, and dfrK genes from a clinical methicillin-resistant Staphylococcus aureus ST125 strain. Antimicrob. Agents Chemother. 56:2139–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ibrahem S, Salmenlinna S, Lyytikainen O, Vaara M, Vuopio-Varkila J. 2008. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin. Microbiol. Infect. 14:1020–1027 [DOI] [PubMed] [Google Scholar]
- 10. Kosowska-Shick K, Julian KG, McGhee PL, Appelbaum PC, Whitener CJ. 2010. Molecular and epidemiologic characteristics of linezolid-resistant coagulase-negative staphylococci at a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 68:34–39 [DOI] [PubMed] [Google Scholar]
- 11. LaMarre JM, Locke JB, Shaw KJ, Mankin AS. 2011. Low fitness cost of the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 55:3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li M, Wang X, Gao Q, Lu Y. 2009. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58:456–461 [DOI] [PubMed] [Google Scholar]
- 13. Liakopoulos A, et al. 2010. Dissemination of two international linezolid-resistant Staphylococcus epidermidis clones in Greek hospitals. J. Antimicrob. Chemother. 65:1070–1071 [DOI] [PubMed] [Google Scholar]
- 14. Locke JB, Hilgers M, Shaw KJ. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob. Agents Chemother. 53:5275–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Locke JB, Hilgers M, Shaw KJ. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 53:5265–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Long KS, Vester B. 2012. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 56:603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marcusson LL, Frimodt-Moller N, Hughes D. 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5:e1000541 doi:10.1371/journal.ppat.1000541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mendes RE, et al. 2010. First report of staphylococcal clinical isolates in Mexico with linezolid resistance caused by cfr: evidence of in vivo cfr mobilization. J. Clin. Microbiol. 48:3041–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendes RE, et al. 2010. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J. Antimicrob. Chemother. 65:2329–2335 [DOI] [PubMed] [Google Scholar]
- 20. Miragaia M, et al. 2008. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: proposal for clone definition. J. Clin. Microbiol. 46:118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanchez Garcia M, et al. 2010. Clinical outbreak of linezolid-resistant Staphylococcus aureus in an intensive care unit. JAMA 303:2260–2264 [DOI] [PubMed] [Google Scholar]
- 23. Schoenfelder SM, et al. 2010. Success through diversity: how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int. J. Med. Microbiol. 300:380–386 [DOI] [PubMed] [Google Scholar]
- 24. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seral C, et al. 2011. Nosocomial outbreak of methicillin- and linezolid-resistant Staphylococcus epidermidis associated with catheter-related infections in intensive care unit patients. Int. J. Med. Microbiol. 301:354–358 [DOI] [PubMed] [Google Scholar]
- 26. Shaw KJ, Barbachyn MR. 2011. The oxazolidinones: past, present, and future. Ann. N. Y. Acad. Sci. 1241:48–70 [DOI] [PubMed] [Google Scholar]
- 27. Thomas JC, et al. 2007. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45:616–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toh SM, et al. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vuong C, Otto M. 2002. Staphylococcus epidermidis infections. Microbes Infect. 4:481–489 [DOI] [PubMed] [Google Scholar]
- 30. Wang Y, et al. 2011. Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J. Antimicrob. Chemother. 66:2521–2526 [DOI] [PubMed] [Google Scholar]
- 31. Widerstrom M, et al. 2009. Clonality among multidrug-resistant hospital-associated Staphylococcus epidermidis in northern Europe. Scand. J. Infect. Dis. 41:642–649 [DOI] [PubMed] [Google Scholar]
- 32. Wong A, et al. 2010. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob. Agents Chemother. 54:742–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ziebuhr W, et al. 2006. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int. J. Antimicrob. Agents 28(Suppl. 1):S14–S20 [DOI] [PubMed] [Google Scholar]