Abstract
The treatment of infections caused by antibiotic-resistant bacteria is one of the great challenges faced by clinicians in the 21st century. Antibiotic resistance genes are often transferred between bacteria by mobile genetic vectors called plasmids. It is commonly believed that removal of antibiotic pressure will reduce the numbers of antibiotic-resistant bacteria due to the perception that carriage of resistance imposes a fitness cost on the bacterium. This study investigated the ability of the plasmid pCT, a globally distributed plasmid that carries an extended-spectrum-β-lactamase (ESBL) resistance gene (blaCTX-M-14), to persist and disseminate in the absence of antibiotic pressure. We investigated key attributes in plasmid success, including conjugation frequencies, bacterial-host growth rates, ability to cause infection, and impact on the fitness of host strains. We also determined the contribution of the blaCTX-M-14 gene itself to the biology of the plasmid and host bacterium. Carriage of pCT was found to impose no detectable fitness cost on various bacterial hosts. An absence of antibiotic pressure and inactivation of the antibiotic resistance gene also had no effect on plasmid persistence, conjugation frequency, or bacterial-host biology. In conclusion, plasmids such as pCT have evolved to impose little impact on host strains. Therefore, the persistence of antibiotic resistance genes and their vectors is to be expected in the absence of antibiotic selective pressure regardless of antibiotic stewardship. Other means to reduce plasmid stability are needed to prevent the persistence of these vectors and the antibiotic resistance genes they carry.
INTRODUCTION
Modern medicine relies on the action of antibiotics to treat and provide prophylactic cover against bacterial infections. Unfortunately, our ability to treat infections has been compromised by the acquisition of resistance to antibiotics by all major pathogenic bacteria. The recent emergence and rapid dissemination of genes that confer resistance to third-generation cephalosporins and last-resort carbapenems (e.g., CTX-M and NDM-1 β-lactamases, respectively) has been mediated by highly transmissible elements called plasmids (17, 25, 27). The successful spread of such plasmids is often attributed to selective pressure resulting from extensive use of antibiotics in clinical and veterinary medicine (8). Experiments in the 1970s showed that the removal of antibiotic pressure resulted in a corresponding loss of plasmids from bacterial cells (2). These findings suggested that cessation of antibiotic use would result in loss of the corresponding resistance genes/vectors from bacterial populations, and many public health policies are predicated on a reduction of antibiotic use. However, the extent to which antibiotic resistance genes are carried and spread in the absence of antimicrobial selective pressure is not clear. A systematic review of antimicrobial-cycling studies concluded there was insufficient evidence to correlate a fall in resistance with cessation of antibiotic use, and a recent prospective study of the carriage of resistance genes in animals found high levels of resistance to tetracycline and tylosin in an environment where no antibiotics had been used for 2.5 years, covering multiple generations of livestock (5, 21). Lenski et al. showed that after a period of coevolution, expression of a tetracycline resistance gene by plasmid pACYC184 actually conferred a fitness benefit on the host bacterium, indicating a role for a resistance gene in plasmid success (18). Recent advances in DNA-sequencing capability have revealed the ubiquitous nature of plasmids, which are resident within almost all sequenced bacteria. This complement of plasmids includes significant numbers with no antibiotic resistance genes (15). We explored the factors that underpin the spread and persistence of antibiotic resistance and examined the paradigm that plasmids carrying antibiotic resistance genes will be lost in the absence of antibiotic selective pressure. To do this, we investigated the fitness costs associated with carriage of a fully sequenced, globally successful, natural IncK plasmid (pCT) found in bacteria isolated from humans and animals (6, 9, 24). Furthermore, we also investigated the role of the only antibiotic resistance gene (blaCTX-M-14) on the plasmid in its ability to persist and disseminate.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmid pCT (GenBank accession number F868832.1) was chosen as an archetype for our study, as it encodes a clinically relevant extended-spectrum β-lactamase (ESBL) (6). It carries a single antibiotic resistance gene, blaCTX-M-14. Cefotaxime (8 μg/ml) was added to all media for selection and maintenance of pCT. Except for Escherichia coli SW102 (grown at 32°C or 42°C), all bacterial strains were cultured at 37°C with aeration in Luria-Bertani (LB) broth or on LB agar containing appropriate antibiotics. For growth of a mutant lacking blaCTX-M-14 (pCT2, blaCTX-M-14::aph), kanamycin (50 μg/ml) was added to the culture media. Plasmid DNA was harvested using a Qiagen large-construct kit (Qiagen, United Kingdom). Plasmid pCT was transformed into a recombineering E. coli strain (SW102) (20, 23) and selected on LB agar containing cefotaxime (8 μg/ml) to create strain SW102 pCT.
The strains used in this study were E. coli SW102 [λcI857ind1 (cro-bioA)<>tetRA] ΔgalK, E. coli J-53 (NCTC 50167; K-12 pro meth rpoB), E. coli 3950 (an original host strain of pCT from which pCT had been previously removed), and Salmonella enterica serovar Typhimurium SL1344 (NCTC 13347; hisG46 rpsL).
Inactivation of blaCTX-M-14 on pCT.
To inactivate blaCTX-M-14, pCT was transformed into E. coli SW102. Primers to amplify the kanamycin resistance gene aph from pKD4 (7) had 20 bp of homology to the template DNA and 40 bp of sequence homology to blaCTX-M-14 (5′-TTTATGCGCAGACGAGTGCGGTGCAGCAAAAGCTGGCGGCGTGTAGGCTGGAGCTGCTTC-3′ and 5′CGGCCAGATCACCGCAATATCATTGGTGGTGCCGTAGTCGGGGAATTAGCCATGGTCCAT-3′). Inactivation of blaCTX-M-14 was done via a modification of the method of Datsenko and Wanner (7), which adapts the method for inactivation of chromosomal genes to target plasmid locations (unpublished data). The resulting mutant pCT, in which blaCTX-M-14 had been inactivated, was named pCT2. Strains containing pCT2 were examined for the production of β-lactamases using nitrocefin. PCR was also used to amplify the region across the deletion/insertion site using primers CTX-M-Group 9 F (5′-ATGGTGACAAAGAGAGTGCAAC-3′) and CTX-M-Group 9 R (5′-TTACAGCCCTTCGGCGATG-3′), and amplimers were sequenced to confirm the recombination. The susceptibilities of strains containing pCT and pCT2 (blaCTX-M-14::aph) to antibiotics were determined on three separate occasions by the standardized agar doubling-dilution method following the recommendations of the British Society for Antimicrobial Chemotherapy (3).
Impact of carriage of pCT or pCT2 on host bacteria.
The impact of carriage of pCT or pCT2 on the growth of the host bacterium was determined by monitoring the optical density of bacterial cultures at a wavelength of 600 nm in LB broth in a Fluostar Optima (BMG Labtech, United Kingdom) as previously described (26). In addition, serial dilution and viable counting were used to verify absorbance changes, and cells carrying each plasmid were examined microscopically to check that no gross morphological changes had occurred. The abilities of host bacterial strains carrying pCT or pCT2 to cause infection were measured by determining the rate of killing in the Caenorhabditis elegans model. Bristol N2 C. elegans was cultured using standard methods at 20°C (13). Bacterial killing assays were conducted as previously described (1, 4), with 60 larval stage 4 (L-4) animals used on three separate occasions for each bacterial strain. A Kaplan-Meier estimate was used to determine the probability of C. elegans survival over time. Survival curves were then generated and compared using the log rank test and chi-square analysis to establish whether any difference between two curves was statistically significant.
Effect of antibiotic resistance on plasmid persistence and transfer.
Plasmid persistence was measured by determining the viable counts every 2 h for 12 h. At the 12-, 24-, and 48-h time points, 100 μl of culture was used to inoculate 100 ml of fresh prewarmed (37°C) LB broth, and further viable counts were taken at 24 and 48 h. The plates containing between 20 and 200 colonies were replica plated using sterile velveteen squares onto agar plates containing either 8 μg/ml of cefotaxime (to identify colonies containing pCT) or 50 μg/ml kanamycin (to identify colonies containing pCT2 [blaCTX-M-14::aph]). Colonies were also replica plated onto antibiotic-free agar. Colonies growing on the antibiotic-free plate but not on the antibiotic-containing plates indicated the proportion of bacteria that had lost the plasmid and the percent loss of plasmids from isogenic cultures over time.
To determine whether inactivation of the β-lactamase gene on pCT impacted the ability of the plasmid to persist in vitro, pairwise competition assays were also used. Cultures at a 1:1 ratio were incubated for 24 h at 37°C, and viable counts were determined at the start of the experiment and at 2-h intervals for 12 h and then at 24 and 48 h. In addition, the mixed culture was used to inoculate fresh broth, and viable counts at hours 12 and 24 were determined over time. Agar plates with 20 to 200 colonies were replica plated as described above. The competition index to measure fitness costs imposed by carriage of each plasmid per generation was defined as 1 + ([log10 A] − [log10 B]/number of generations), where A is the fraction of the total population carrying one plasmid at the end of the assay and B is the fraction of the total population carrying the same plasmid at the beginning of the assay (22). This experiment was carried out on three separate occasions.
Another component of plasmid “fitness” examined was the ability of plasmids to transfer to new recipient bacteria. To determine whether inactivation of the β-lactamase gene on pCT affected transfer, the pCT and pCT2 conjugation frequencies were measured from the donor strain E. coli DH5α to the recipient strains E. coli J53-2, E. coli 3950, and Salmonella Typhimurium SL1344. Conjugation was measured on a filter and LB broth after 3 h of mating, as previously described (12). Conjugation frequencies were measured a minimum of three times and calculated using the following formula (19): conjugation frequency = median number of transconjugants/(median number of recipients × initial viable-count ratio of donor to recipient cells). Unpaired Student's t tests were used to determine whether the values differed significantly (a P value of less than 0.05).
RESULTS
Inactivation of the antibiotic resistance gene blaCTX-M-14 on pCT.
To define any role that an antibiotic resistance gene may have in plasmid and bacterial-host fitness, the gene must be inactivated. Conventional gene inactivation methods were ineffective because of a low efficiency of cointroduction of target plasmids and integration cassettes and the requirement for cohabitation of multiple plasmids encoding the necessary recombination machinery. The maintenance and selection of multiple replicons is also challenging due to a limited number of selective markers available. Therefore, we developed a novel protocol for insertional inactivation of specific genes located on plasmids (unpublished data). Plasmid analysis and DNA sequencing confirmed the inactivation of the blaCTX-M-14 gene of pCT and the removal of 606 bp of DNA, including the region encoding the CTX-M-14 active site, and replacement with the aph gene conferring kanamycin resistance. This gave plasmid pCT2 (blaCTX-M-14::aph). To prevent any further recombination events, pCT2 was extracted from SW102 and transformed into E. coli DH5α. The MIC of cefotaxime for E. coli DH5α/pCT2 (blaCTX-M-14::aph) was 0.03 μg/ml compared with 16 μg/ml for E. coli DH5α/pCT, and no β-lactamase activity was detectable with nitrocefin for strains carrying pCT2 (blaCTX-M-14::aph).
The addition of pCT and pCT2 has no impact on bacterial-host growth or virulence.
pCT and pCT2 were transferred by conjugation into three hosts: E. coli J53-2, E. coli 3950, and S. enterica serovar Typhimurium SL1344. There were no significant differences in the growth rates of each bacterial host with or without wild-type pCT or pCT2 (blaCTX-M-14::aph) (Table 1). These data show the aph gene had no effect on the host growth rate. There was also no difference in the ability of the host strain with or without pCT or pCT2 to cause infection over 10 days in the C. elegans virulence model (Fig. 1), with no statistically significant difference in the rates of killing between S. Typhimurium SL1344 carrying pCT or pCT2 (P = 0.2059). SL1344 was able to kill 50% of the population within 4 days regardless of plasmid carriage (all uninfected worms survived for the whole duration of the experiment). The abilities of S. Typhimurium SL1344 carrying either plasmid to adhere to and invade human intestinal cells also were not different (data not shown).
Table 1.
Conjugation frequencies and generation times
| Condition | Recipient | Plasmid | Frequency of transfer | SD | P valuea | Generation time (min) | SD | P valuea |
|---|---|---|---|---|---|---|---|---|
| Filter | J53-2b | pCT | 1.70 × 102 | 1.41 × 10−2 | ||||
| pCT2 | 3.40 × 102 | 2.28 × 10−2 | 0.226 | |||||
| 3950b | pCT | 6.12 × 106 | 1.27 × 10−6 | |||||
| pCT2 | 8.71 × 106 | 6.86 × 10−6 | 0.555 | |||||
| SL1344b | pCT | 5.95 × 106 | 5.39 × 10−5 | |||||
| pCT2 | 2.27 × 105 | 9.34 × 10−6 | 0.033 | |||||
| Broth | J53-2 | None | 63.5 | 4.7 | ||||
| pCT | 7.28 × 105 | 6.14 × 10−5 | 67.1 | 3.8 | ||||
| pCT2 | 2.59 × 104 | 3.29 × 10−4 | 0.309 | 63.3 | 5.8 | 0.95 | ||
| 3950 | None | 64.7 | 2.1 | |||||
| pCT | 1.394 × 105 | 1.52 × 10−4 | 68.7 | 5.5 | ||||
| pCT2 | 3.14 × 104 | 5.08 × 10−3 | 0.177 | 69.2 | 5.1 | 0.39 | ||
| SL1344 | None | 94.1 | 6.0 | |||||
| pCT | 5.25 × 106 | 5.92 × 10−6 | 95.2 | 11.7 | ||||
| pCT2 | 2.50 × 105 | 1.98 × 10−5 | 0.179 | 100.7 | 9.8 | 0.28 |
The P values compare the significances of values between data from strains carrying pCT and pCT2.
Recipient strains included E. coli J-53, a common laboratory plasmid host, E. coli 3950 the original parent of pCT and Salmonella Typhimurium SL1344, a wild type of another species.
Fig 1.
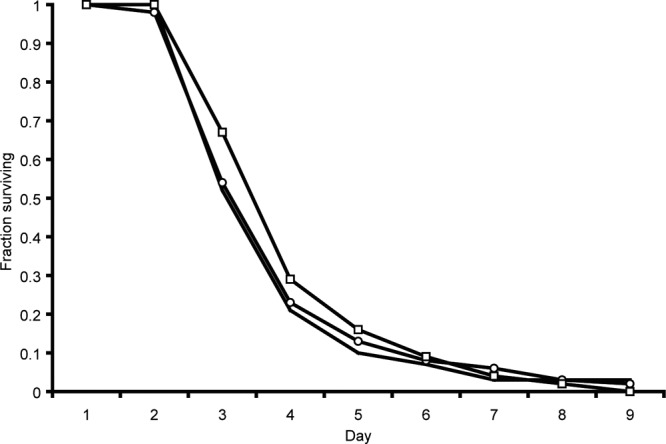
Survival of C. elegans after infection with S. Typhimurium SL1344 without a plasmid. S. Typhimurium SL1344, solid lines; S. Typhimurium SL1344 pCT, lines with circles; S. Typhimurium SL1344 pCT2 (blaCTX-M-14::aph), lines with squares.
Carriage of pCT imposes no detectable fitness cost on the host.
When directly competed against a plasmid-free counterpart, neither plasmid impaired or enhanced host fitness, suggesting little or no fitness burden on the host cell (competition index of E. coli DH5α pCT = 0.99829, P = 0.359, when competed against E. coli DH5α without a plasmid). Furthermore, when host bacteria containing pCT or pCT2 were competed in a 1:1 ratio in vitro, neither plasmid had a significant fitness advantage or disadvantage in the presence of the other (Fig. 2) (competition index of E. coli DH5α pCT2 = 1.001, P = 0.477, when competed against E. coli DH5α pCT).
Fig 2.
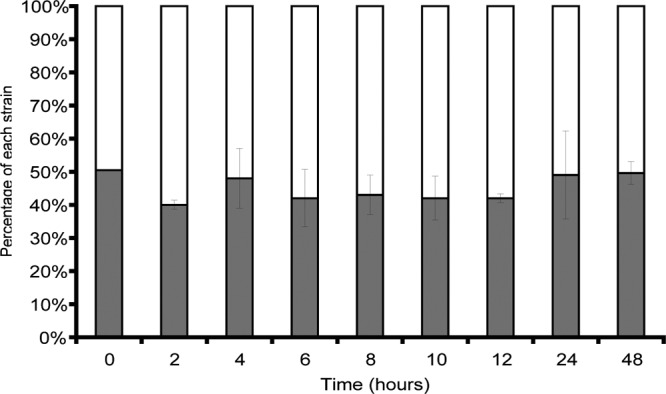
Pairwise in vitro competition showing the proportion of each plasmid present as a percentage at each time point. The white proportion of each bar represents E. coli DH5α pCT; the solid portions represent E. coli DH5α pCT2. The error bars indicate 1 standard deviation for each sample.
pCT is persistent in the absence of antibiotic pressure, and the blaCTX-M-14 gene has no impact on persistence or plasmid transfer rates.
To determine the effect of the antibiotic resistance gene on plasmid transfer, the conjugation frequencies of pCT and pCT2 from the donor E. coli strain DH5α to three recipient host strains were measured on a solid surface with the use of a filter and in liquid media. Both plasmids were able to conjugate from E. coli DH5α to all the tested recipients, and there was no difference between the transfer rates of pCT and pCT2 to any host. Conjugation into E. coli J-53 occurred at a higher frequency than into other strains (Table 1).
pCT and pCT2 also persisted in vitro for ∼70 generations in all daughter cells of the four bacterial host strains in the absence of antibiotic pressure. These data indicate that the lack of blaCTX-M-14 (pCT2) had no effect on plasmid maintenance in the absence of selective antibiotic pressure under these experimental conditions.
DISCUSSION
Here, we have shown that a natural, globally distributed ESBL-carrying plasmid (pCT) is able to successfully persist and spread in the absence of antibiotic selective pressure and that the plasmid confers little or no fitness burden on the bacterial host strains tested. We have also shown that disruption of the only antibiotic resistance gene on plasmid pCT had no significant effect on any of the basic bacterial-host and plasmid biological factors tested and did not impair plasmid fitness when directly competed with the wild-type plasmid in vitro. These data indicate that the antibiotic resistance gene may not be the sole driver responsible for the evolutionary success or worldwide spread of this epidemic plasmid. The use of antimicrobials both clinically and in veterinary medicine has been considered to have led to the increased prevalence of antibiotic-resistant bacteria and resistance genes worldwide (8, 10, 25). While the antibiotic resistance gene on pCT ensures plasmid persistence in the presence of many commonly used β-lactams, the extent to which antibiotic selective pressure has led to the persistence of plasmids such as pCT once the resistance gene has been stably incorporated within the plasmid genome is not known. Previous literature has assumed that the presence of antibiotic resistance genes has driven the evolution and spread of naturally occurring plasmids such as pCT; however, our study challenges this dogma and suggests that natural plasmids are well adapted to spread and persist in the absence of antimicrobial selection, which is likely to be an intermittent pressure.
These results predict that both plasmids and the antibiotic resistance genes they carry will persist once established in bacterial populations. Persistence occurs regardless of antibiotic administration or stewardship, although antibiotic pressure provokes an amplification of resistance plasmids from a wider reservoir. It may be that the persistence of these antibiotic resistance vectors in the absence of selective pressure is the reason why the various strategies to remove resistant strains, such as by antibiotic “cycling,” have been largely ineffective (16). We found no cost associated with carriage of blaCTX-M-14 in this study; either the gene has no intrinsic fitness cost or pCT carries a factor(s) that ameliorates the impact on the cell, as has previously been described for blaCMY-2 in Salmonella (14). Highly persistent and transmissible plasmids have an advantage in the face of host strain evolution, where plasmids with a limited transfer ability may be lost if their host is displaced (11). Our data are based on laboratory models, with all the associated caveats and limitations; however, our data suggest that future treatment regimes and antibiotic stewardship policies should consider the persistent nature of both antibiotic resistance genes and the vectors that carry them. A better understanding of the key components of successful vectors is vital to devising strategies to prevent and minimize plasmid spread.
ACKNOWLEDGMENT
The work of J.L.C. was funded in part by the Veterinary Laboratories Agency through a Defra-funded project.
Footnotes
Published ahead of print 18 June 2012
REFERENCES
- 1. Aballay A, Ausubel FM. 2001. Programmed cell death mediated by ced-3 and ced-4 protects Caenorhabditis elegans from Salmonella Typhimurium-mediated killing. Proc. Natl. Acad. Sci. U. S. A. 98:2735–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson JD. 1974. The Effect of R-factor carriage on the survival of Escherichia coli in the human intestine. J. Med. Microbiol. 7:85–90 [DOI] [PubMed] [Google Scholar]
- 3. Andrews JM. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48:5–16 [DOI] [PubMed] [Google Scholar]
- 4. Bailey AM, et al. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown EM, Nathwani D. 2005. Antibiotic cycling or rotation: a systematic review of the evidence of efficacy. J. Antimicrob. Chemother. 55:6–9 [DOI] [PubMed] [Google Scholar]
- 6. Cottell JL, et al. 2011. Complete sequence and molecular epidemiology of an IncK epidemic plasmid encoding blaCTX-M-14. Emerg. Infect. Dis. 17:645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74:417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dhanji H, et al. 2012. Dissemination of pCT-like IncK plasmids harbouring CTX-M-14 ESBL among clinical Escherichia coli in the United Kingdom. Antimicrob. Agents Chemother. 56:3376–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dhanji H, et al. 2011. Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum β-lactamase from UK river water. J. Antimicrob. Chemother. 66:512–516 [DOI] [PubMed] [Google Scholar]
- 11. Gibreel TM, et al. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J. Antimicrob. Chemother. 67:346–356 [DOI] [PubMed] [Google Scholar]
- 12. Hartskeerl R, Zuidweg E, Van Geffen M, Hoekstra W. 1985. The IncI plasmids R144, R64 and ColIb belong to one exclusion group. J. Gen. Microbiol. 131:1305–1311 [DOI] [PubMed] [Google Scholar]
- 13. Hope IA. 1999. C. elegans: a practical approach, 1st ed Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 14. Hossain A, Reisbig MD, Hanson ND. 2004. Plasmid-encoded functions compensate for the biological cost of AmpC overexpression in a clinical isolate of Salmonella typhimurium. J. Antimicrob. Chemother. 53:964–970 [DOI] [PubMed] [Google Scholar]
- 15. Iredell J, Ellem J, Zong Z, Partridge S. 2011. The ecology of major resistance plasmids in E. coli, poster P560. 21st Eur. Congr. Clin. Microbiol. Infect. Dis [Google Scholar]
- 16. Kollef MH. 2006. Is antibiotic cycling the answer to preventing the emergence of bacterial resistance in the intensive care unit? Clini. Infect. Dis. 43:S82–S88 [DOI] [PubMed] [Google Scholar]
- 17. Kumarasamy KK, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lenski RE, Simpson SC, Nguyen TT. 1994. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176:3140–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorian V. 1996. Antibiotics in laboratory medicine, 4th ed Williams and Wilkins, New York, NY [Google Scholar]
- 20. Murphy KC. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pakpour S, Jabaji S, Chenier MR. 20121 Frequency of antibiotic resistance in a swine facility 2.5 years after a ban on antibiotics. Microb. Ecol. 63:41–50 [DOI] [PubMed] [Google Scholar]
- 22. Pope CF, Gillespie SH, Moore JE, McHugh TD. 2010. Approaches to measure the fitness of Burkholderia cepacia complex isolates. J. Med. Microbiol. 59:679–686 [DOI] [PubMed] [Google Scholar]
- 23. Sharan SK, Thomason LC, Kuznetsov SG, Court DL. 2009. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4:206–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stokes MO, et al. 2012. Detection and characterization of pCT-like plasmid vectors for blaCTX-M-14 in Escherichia coli isolates from humans, turkeys and cattle in England and Wales. J. Antimicrob. Chemother. 67:1639–1644 [DOI] [PubMed] [Google Scholar]
- 25. Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 26. Webber MA, Randall LP, Cooles S, Woodward MJ, Piddock LJV. 2008. Triclosan resistance in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 62:83–91 [DOI] [PubMed] [Google Scholar]
- 27. Woodford N, et al. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499 and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53:4472–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]


