Abstract
The p53 gene is rarely mutated in neuroblastoma, but codon 72 polymorphism that modulates its proapoptotic activity might influence cancer risk and clinical outcome. We investigated whether this polymorphism affects neuroblastoma risk and disease outcome and assessed the biologic effects of the p53-72R and p53-72P isoforms in p53-null cells. Comparison of 288 healthy subjects and 286 neuroblastoma patients revealed that the p53-72 polymorphism had no significant impact on the risk of developing neuroblastoma; however, patients with the Pro/Pro genotype had a shorter survival than those with the Arg/Arg or the Arg/Pro genotypes even in the stage 3 and 4 subgroup without MYCN amplification. By Cox regression analysis, the p53 Pro/Pro genotype seems to be an independent marker of poor prognosis (hazard ratio = 2.74; 95% confidence interval = 1.14–6.55, P = .014) together with clinical stage, MYCN status, and age at diagnosis. In vitro, p53-72P was less effective than p53-72R in inducing apoptosis and inhibiting survival of p53-null LAN-1 cells treated with etoposide, topotecan, or ionizing radiation but not taxol. By contrast, p53-72P was more effective in promoting p21-dependent accelerated senescence, alone or in the presence of etoposide. Thus, the p53-72 Pro/Pro genotype might be a marker of poor outcome independent of MYCN amplification, possibly improving risk stratification. Moreover, the lower apoptosis and the enhanced accelerated senescence by the p53-72P isoform in response to DNA damage suggest that patients with neuroblastoma with the p53-72 Pro/Pro genotype may benefit from therapeutic protocols that do not rely only on cytotoxic drugs that function, in part, through p53 activation.
Introduction
Neuroblastoma (NB) is a childhood solid tumor that accounts for 8% to 10% of all childhood cancers and for ∼15% of all deaths because of pediatric malignancies [1]. NB arises from neuroectodermal precursor cells of the neural crest, and therefore, tumors can develop anywhere in the sympathetic nervous system [1]. Clinically, NB is remarkably heterogeneous: age at diagnosis [2], clinical stage (based on the International Neuroblastoma Staging System [3]), and tumor histology [4] are the most important variables for predicting disease risk and selecting appropriate therapeutic protocols. Children older than 18 months, with tumor at advanced stage (3 or 4) or with unfavorable histologic findings have an adverse outcome despite intensive multimodal treatments such as high-dose myeloablative chemotherapy followed by rescue with autologous bone marrow [5]. Clinical heterogeneity correlates with several genetic abnormalities whose detection in tumor cells has further improved risk stratification [5]. V-myc myelocytomatosis viral-related oncogene NB-derived (MYCN) oncogene amplification [6], hemizygous deletions of chromosomal region 1p36 [7], and unbalanced gain of 17q regions [8] are the most common genomic aberrations in NB. MYCN amplification occurs in ∼20% of cases and represents the most powerful marker of poor outcome [9]. In contrast to other malignancies, only 2% to 3% of NBs have mutations of the p53 gene [10]. p53 is a tumor suppressor gene that encodes for a nuclear phosphoprotein, which, on activation, regulates many biologic processes such as cell cycle checkpoints, apoptosis, and cellular senescence [11]. Impairment of such processes has important implications for clinical behavior and response to therapy of tumors with nonfunctional p53 [12]. p53 also activates the transcription of the murine double minute (MDM2) gene that encodes for the major negative regulator of p53 [13]. In recent years, there has been an increasing interest in identifying and assessing the frequency of gene variants (polymorphisms) as a tool to predict interindividual cancer risk and response to cancer therapies [14].
A polymorphism is defined as a DNA sequence change that occurs in a significant proportion (>1%) of a large population [15]. The most common type of genetic variation is a single nucleotide polymorphism (SNP). For the p53 gene, an SNP has been identified at codon 72 within exon 4 causing an Arg>Pro substitution [16]. The p53-72R isoform seems to be more potent than the p53-72P isoform in inducing apoptosis, and increased mitochondrial localization and reduced affinity of the p53-72R isoform for the p53 inhibitor iASPP are among the proposed mechanisms that may be responsible for such an effect [17,18].
In this study, we compared the frequency of the p53 codon 72 Arg/Arg, Arg/Pro, and Pro/Pro genotypes in 288 control subjects and 286 newly diagnosed NBs and correlated these frequencies with clinical-biologic variables such as age at diagnosis, primary tumor site, clinical stage, and MYCN amplification. We report here that patients with the Pro/Pro genotype, including those with normal MYCN status and advanced disease stages, seem to have a shorter 5-year survival than those with the Arg/Pro or Arg/Arg genotype.
The more aggressive disease of patients with NB with the Pro/Pro genotype correlated with lower apoptosis and enhanced survival of cytotoxic drug or ionizing radiation (IR)-treated p53-null LAN-1 cells expressing the p53-72P compared with the p53-72R isoform. By contrast, expression of the p53-72P isoform, alone or in the presence of a low concentration of etoposide, induced an increase in senescent cells.
Together, these findings suggest that, although relatively rare, the p53 codon 72 Pro/Pro genotype might identify a subgroup of patients with NB with aggressive disease independently of the status of other markers predictive of poor outcome. Moreover, patients with this genotype may respond less efficiently to treatments that induce DNA damage and increased p53 expression/activity and may benefit from therapeutic protocols that include cytotoxic drugs with p53-independent mechanisms of action.
Materials and Methods
Subjects
Two hundred eighty-six patients with NB (258 Italian patients, most of whom were previously included to assess the frequency of the MDM2 SNP-309 polymorphism [19] and 28 British patients) were selected for the analysis of the p53 codon 72 genotype. Their clinical and biologic characteristics are listed in Table W1. Tumor DNA was analyzed in each patient, whereas peripheral blood DNA was also genotyped in 40 cases to confirm that polymorphism frequency did not reflect somatic mutation. Selection criteria were lack of previous treatments and availability of adequate amount of DNA. Tumor DNA was used because peripheral blood lymphocytes are not always readily attainable from patients with NB. Institutional written informed consent was obtained from parents or legal guardians of patients with NB. The study underwent ethical review and approval according to local institutional guidelines.
Controls
The p53 codon 72 genotype was also evaluated in peripheral blood DNA of 288 healthy individuals from anonymous blood donors randomly selected during a 5-year period (2000–2005) at several Northern Italy Blood Banks and from control subjects stored at the “Oncology Institute of Veneto,” Padova, Italy. Their age ranged from 25 to 60 years (median, 45 years) and the male-to-female ratio was approximately 46% to 54%.
NB Cell Lines DNA
DNA from NB-69, NBL-W, NBL-S, BE1N, TR14, NGP, NB1691, and LS NB cell lines was from Dr Tweddle's laboratory, whereas DNA from RN-GA, SH-EP, SK-, GI-CAN, and SK-NBE2 cell lines was from Dr Raschellà's laboratory. DNA from LAN-5, HTLA-230, GI-CAN, SH-SY5Y, IMR32, LAN-1, and SK-N-AS cell lines was obtained using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) (Table W2).
p53 Codon 72 Genotyping
The p53 DNA segment for codon 72 genotype was amplified by polymerase chin reaction (PCR) using 150 ng of DNA and a pair of forward (5′-TTGCCGTCCCAAGCAATGGATGA-3′) and reverse (5′-TCTGGGAAGGGACAGAAGATGAC-3′) primers that generated a 199-bp DNA product. DNA amplification was done as follows: 95°C for 1 minute, 60°C for 1 minute, and 72°C for 2 minutes for 40 cycles. PCR products were separated by electrophoresis in 1% agarose gel with ethidium bromide, extracted using a PCR purification kit (Roche Diagnostics GmbH, Mannheim, Germany), and each fragment was sequenced from both ends on an ABI PRISM 377 DNA Sequencer using the ABI PRISM Big Dye Terminator (PE Biosystem, Foster City, CA). Thirty tumor and control DNA were independently resequenced to confirm the genotype's accuracy.
Statistical Analysis
The frequencies of p53 polymorphism at codon 72 were cross-tabulated in patients with NB versus healthy controls using a two-sided Fisher exact test. The frequencies of p53 polymorphism at codon 72 in patients with NB versus known prognostic factors were also cross-tabulated using the Fisher exact test to evaluate the significance of the association.
Clinical-pathologic variables were categorized as follows: age at diagnosis (<18 vs ≥18 months) [2], primary site (adrenal vs nonadrenal), clinical stage according to the International Neuroblastoma Staging System [3] (stage 1, 2, and 4 S vs 3 and 4), and MYCN status (single copy vs amplified).
Five-year overall survival curves on the basis of the p53 genotype at codon 72 in all patients with NB or in subgroups defined on the basis of established prognostic factors were calculated according to Kaplan and Meier [20], and the statistical significance of the differences was assessed using log-rank test. Cox multiple regression analysis [21] was carried out, including clinical stage, MYCN status, age at diagnosis, and p53 polymorphism as covariates. All statistical tests were two-sided, and P < .05 was considered statistically significant. The analyses were carried out using the software package SPSS 11.0 for Windows (SPSS, Inc, Chicago, IL). The Hardy-Weinberg equilibrium was assessed by χ2 test.
Plasmids
p53-72R-ERTAM and p53-72P-ERTAM were generated as follows: the p53-72R and p53-72P coding sequences were amplified by reverse transcription-polymerase chain reaction from total RNA of ACN and NBL-S cells, respectively. Sense primer (5′GTTAACATGGAGGAGCCGCA-3′) contains a 5′-flapping HpaI site flanked by the ATG, and antisense primer (5′GGATCCGTCTGAGTCAGGC-3′) contains a 5′-flapping BamHI and a mutated p53 stop codon. The ligand-binding domain of the mutated (Gly525Arg) tamoxifen-responsive murine estrogen receptor (ER) was obtained by BamHI/EcoRI digestion of the ΔuORF-C/EBPα-ERTAM vector [22]. Each PCR product and the digested ERTAM fragment were subcloned into the HpaI/EcoRI-digested MigRI vector.
Cells Cultures and Retroviral and Lentiviral Infections
p53-null human NB cell line LAN-1 [23] was cultured in Dulbecco modified Eagle medium with Glutamax (Invitrogen-GIBCO, Carlsbad, CA) supplemented by 10% fetal bovine serum and penicillin and streptomycin (100 µg/ml each) at 37°C, 5% CO2. For retroviral infections, Phoenix cells were transiently transfected with the indicated plasmids. The infectious supernatant was collected 48 hours later and was used to infect (a 48-hour procedure) LAN-1 cells. Twenty-four hours later, infected cells were sorted (EPICS Profile Analyzer; Coulter, Hialeah, FL) for green florescent protein (GFP) expression and maintained in culture as described [24].
For lentiviral infections, 293T cells were cotransfected with p21shRNA pLKO.1 (20 µg) and the packaging plasmids pCMVd8.2 (10 µg) and pCMV-VSVG (15 µg) using the calcium phosphate transfection kit (Invitrogen). Lentiviral stocks were harvested 48 hours later, centrifuged for 5 minutes at 3000 rpm, aliquoted, and stored at -80°C. Lentiviral titers were determined by transduction of 3 x 104 293T cells with serial dilutions of the viral stocks in 24-well plates supplemented with puromycin (2 µg/ml). Then, LAN-1-p53-72R-ER or LAN-1-p53-72P-ER plated at a cell density of 2 x 104 cells per well and transduced with a multiplicity of infection of 10 using one round of cosedimentation and incubation. Lentivirally transduced cells were selected in the presence of puromycin (2 µg/ml).
Protein Analysis
Cell lysates from 5 x 105 cells were suspended in 50 µl of 2x Laemmli lysis buffer (BioRad, Richmond, CA) and boiled for 10 minutes. For Western blot analysis, 20 µl (equivalent to 200,000 cells) was resolved by 10% SDS-PAGE, transferred to nitrocellulose membranes, and blotted with the indicated antibodies: anti-p53 (DO-1, sc-126; Santa Cruz Biotechnology, Santa Cruz, CA), anti-phospho Ser-15 p53 (no. 07-388; Cell Signaling), anti-β-actin (sc-47778; Santa Cruz Biotechnology), anti-p21 (DF10; Calbiochem, San Diego, CA), and antiMDM2 (no. 0P46; Calbiochem). Secondary antibodies were peroxidase labeled, and peroxidase was detected with Enhanced Chemiluminescence Kit (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
Irradiation Treatment
Cells were irradiated with different x-ray doses (20, 40, and 80 cGy) using a Gilardoni x-ray generator (Gilardoni SpA, Mandello del Lario, Lecco, Italy). Irradiation times varied from 15 to 30 seconds depending on the dose. After irradiation, medium was changed, and cells were kept at 37°C in the incubator for 24 hours. Subsequently, cells were plated for colony formation assays or used for flow cytometric measurements carried out after an additional 48 hours (72 hours after irradiation).
Flow Cytometry Analysis
Cells were harvested, and pellets were washed twice with PBS and diluted in 2x Nicoletti solution (0.1% Na citrate, 0.1% Triton X-100, 50 µg/ml propidium iodide) for 5 minutes before the analyses. Samples were analyzed by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). For each sample, at least 2 x 104 cells were analyzed. Cell cycle distribution and hypodiploid DNA content were calculated by Cell Quest software (BD Biosciences). Statistical significance (P) was calculated by two-tailed Student's t test.
Confocal Immunofluorescence
LAN-1 (parental and p53-ER-derivative) cells were plated on Culture Slides (BD Falcon) and, 36 hours later, treated with 250 nM 4-hydroxy-tamoxifen (4-HT). After 2 hours, cells were washed twice this PBS, fixed with ice-cold methanol for 20 minutes, permeabilized in 0.1% Triton X-100 in PBS (5 minutes), washed twice with PBS, and blocked in 1% bovine serum albumin/PBS for 30 minutes at room temperature.
Slides were stained with an anti-p53 monoclonal antibody (PAb 1801; Calbiochem) diluted 1:80 in 5% milk-Tris-buffered saline and Tween 20, incubated for 1 hour at room temperature. After a 30-minute incubation at room temperature with antirabbit Alexa Fluor 555 conjugate (A-21428; Invitrogen) diluted 1:300 in 5% milk-Tris-buffered saline and Tween 20, slides were washed three times in PBS and rinsed in water. For nuclei staining, Hoechst 33258 was added in the last PBS wash. As control, LAN-1 cells were stained in the absence of the anti-p53 antibody. Cells were visualized by Leica TCS SP2 confocal laser scanner microscope (Leica Microsystems, Wetzlar, Germany) using a 40x oil immersion objective, and images were analyzed with Leica software.
Colony Assay
Parental, p53-72R-ER, and p53-72P-ER-derivative LAN-1 cells were seeded at 3 x 105 cells per well in six-well plates. The following day, cells were treated with etoposide (5, 10 or 20 µM) or taxol (2 µM). After 3 hours, cells were washed and seeded (5 x 103, 1x104, 2x104 cells perplate) in six-well plates with or without 4-HT (250 nM). After 7 days, plates were stained with Crystal violet, and colonies were counted.
SA-β-gal Staining Assay
The staining for SA-β-galactosidase activity was performed as described [25]. Briefly, cells were seeded in 24-well plates and treated with 4-HT alone (250 nM, 48 hours) when 80% confluent or with 4-HT and etoposide (25 nM etoposide for 3 hours and then 250 nM 4-HT for 48 hours) when 50% confluent. At the end of the treatment, cells were washed in PBS and fixed in 2% formaldehyde/0.2% glutaraldehyde. Cells were then washed and incubated at 37°C for 4 hours with fresh SA-β-gal staining solution (1 mg of 5-bromo-4-chloro-3-indolyl-β-D-galactoside per milliliter, 40 mM citric acid/sodium phosphate [pH 6.0], 150 mM NaCl, 2 mM MgCl2, 5 mM potassium ferricyanide).
Real-time PCR Expression Analyses
For real-time quantitative PCR, total RNA was isolated from p53-72R-ER- and p53-72P-ER-LAN-1 cells using the RNeasy Mini kit (Qiagen). RNA was treated for 1 hour at 37°C with DNase-RNase free (Roche Applied Science, Mannheim, Germany), which was then deactivated for 15 minutes at 65°C. Four micrograms of total mRNA was reverse-transcribed using SuperScript III Reverse Transcriptase kit (Invitrogen), and the resulting complementary DNA was used as a PCR template. All reactions were done in triplicate, and total RNA was extracted from two separate experiments.
Primer pairs of analyzed genes (MDM2: FW 5′-caagttactgtgtatcaggcaggg-3′, RV 5′-tctgttgcaatgtgatggaagg-3′; BAX: FW 5′-ggagcggcggtgatggac-3′, RV 5′-ggcccctgtcttcatgatctgc-3′; NOXA: FW 5′cagagctggaagtcgagtgtg-3′, RV 5′-gttcctgagcagaagagtttgg-3′; PERP: FW 5′-cccgtgaagtacacccagacc-3′, RV 5′-gcccacccaaagccgtagg-3′; PAI-1: FW 5′-aagactcccttccccgactc-3′, RV 5′-cacagagacagtgctgccgt-3′; p21: FW 5′cctgcccaagctctaccttcc-3′, RV 5′-ggtccacatggtcttcctctgc-3′; HPRT: FW 5′-agactttgctttccttggtcagg-3′, RV 5′-gtctggcttatatccaacacttcg-3′) were designed using Beacon Design software (Premier Biosoft International, Palo Alto, CA). Real-time quantitative PCR was performed using GoTaq qPCR Master Mix (Promega, Madison, WI) on a MyIQ thermocycler (BioRad) and quantified using MyIQ software (BioRad) that analyzes the Ct value of real-time PCR data with the ΔΔCt method. HPRT, a housekeeping gene with constant expression, was used as an internal control to normalize input complementary DNA.
Results
Frequency and Association with Poor-Prognosis Predictors of p53-72 Genotypes
Genotype frequencies at codon 72 of p53 (Arg/Arg, Arg/Pro, and Pro/Pro) and other genetic and clinical characteristics of patients with NB included in this study are summarized in Table W1. The genotype frequencies in all groups were within the Hardy-Weinberg equilibrium. The frequency of the Pro/Pro genotype in tumor DNA was not significantly different from that of peripheral blood samples of control subjects (3.8% vs 6.6%, P = .190); no statistical significance was also noted if the comparison was limited to the Italian control and NB cohorts (not shown). In 40 cases tested, peripheral blood and tumor DNA genotypes were identical indicating that polymorphism frequency did not reflect somatic mutation or loss of heterozygosity/copy number gains which are uncommon in NB for region 17p13 [26], the chromosomal location of the p53 gene. In an attempt to validate independently the frequency of the p53-72 Pro/Pro genotype in a larger cohort of patients with NB, we searched the dbGaP database (http://www.ncbi.nlm.nih.gov/gap), which contains the data set of Wang et al. [27] with 1627 patients with NB and 3254 genetically matched disease-free controls. However, data for the p53-72 SNP (rs1042522) were not available because the array platforms used in the Wang et al. data set (Illumina HumanHap550v3.0, HumanHap550v1.1, and Human610_Quadv1_B) do not assay the rs1042522 SNP.
Of interest, the frequency of the Pro/Pro genotype was low (1/20, 5%) also in human NB cell lines (Table W2). We then assessed whether the p53-72 Pro/Pro genotype was associated with predictors of poor outcome such as advanced clinical stage (3 and 4), age at diagnosis 18 months or older, adrenal primary site, and MYCN amplification (Table 1). A statistically significant association of the Pro/Pro genotype with age 18 months or older (P = .031) was noted; although not statistically significant, the Pro/Pro genotype was more represented in patients with unfavorable stage (3 and 4), and seemed to be inversely associated with MYCN amplification (Table 1). In particular, of the seven patients with stage 4 disease with the Pro/Pro genotype, only one had MYCN amplification.
Table 1.
Association of p53-72 Polymorphism with Clinical and Genetic Characteristics of Patients with NB.
| Feature | p53 Arg72Pro | ||
| Arg/Arg or Arg/Pro (%) | Pro/Pro (%) | P* | |
| Stage | .338 | ||
| 1–2–4 S | 98 (98.0) | 2 (2.0) | |
| 3–4 | 172 (95.0) | 9 (5.0) | |
| MYCN | .304 | ||
| Non amp | 205 (95.3) | 10 (4.7) | |
| Amp | 69 (98.6) | 1 (1.4) | |
| Age | .031 | ||
| <18 mo | 115 (99.1) | 1 (0.9) | |
| ≥18 mo | 158 (94.0) | 10 (6.0) | |
| Site | .523† | ||
| Extra adrenal | 139 (97.2) | 4 (2.8) | |
| Adrenal | 121 (95.3) | 6 (4.7) | |
| Multiple sites | 1 (100.0) | ||
Amp indicates amplified; non amp, non amplified.
P: two-tailed Fisher exact test.
P value was calculated by comparing tumors with adrenal and extra-adrenal localization only.
Effect of p53-72 Genotypes on Survival
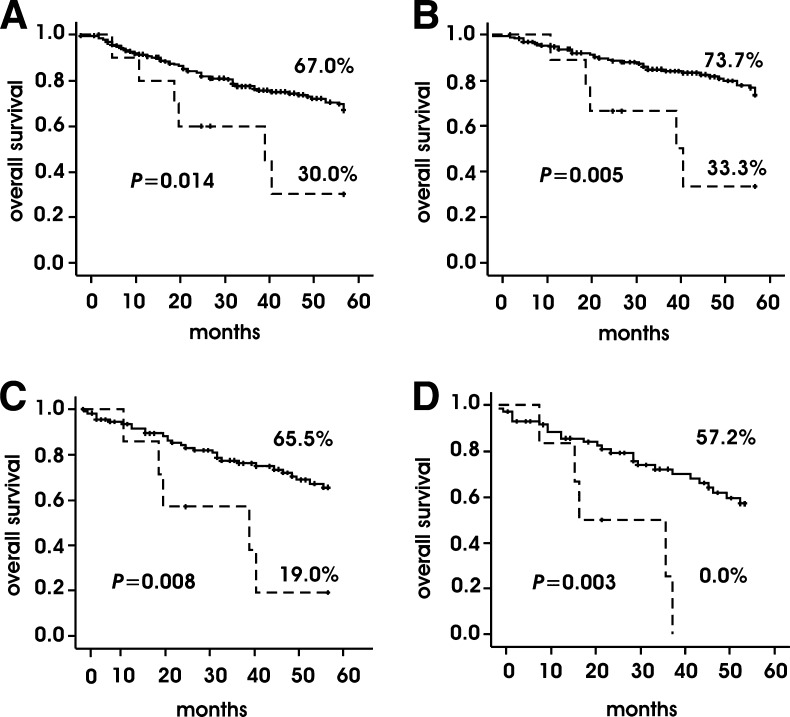
Cumulative Kaplan-Meier 5-year overall survival of patients with NB with the Pro/Pro genotype was shorter than that of patients with the Arg/Arg (P = .042, log-rank test) or the Arg/Pro genotype (P =.003, log-rank test). Because survival of patients with the Arg/Arg and the Arg/Pro genotype was undistinguishable, we decided to combine these two groups and compare to the Pro/Pro group in all the analyses. As shown in Figure 1A, patients with the Pro/Pro genotype had a shorter survival that those with the Arg/Arg and Arg/Pro genotypes (P =.014, log-rank test). Moreover, in the NB subgroup with normal MYCN status, patients with the Pro/Pro genotype had a shorter overall survival than those with the Arg/Arg and Arg/Pro genotypes (P = .005, log-rank test; Figure 1B). In this subgroup, overall survival of patients with the Pro/Pro genotype was also shorter when limiting the analysis to those either at stage 3 and 4 or at stage 4 only (P =.008 and P = .003, respectively, log-rank test; Figure 1, C and D). We also observed a shorter overall survival in patients with the Pro/Pro genotype in the subgroups at stage 4, with age 18 months or older, and with normal MYCN status (P = .013; Figure W1).
Figure 1.
Kaplan-Meier 5-year cumulative overall survival on the basis of p53-72 genotypes in all NBs (A), NBs with normal MYCN status (B), stage 3 and stage 4 NBs with normal MYCN status (C), stage 4 NBs with normal MYCN status (D). Solid line indicates Arg/Arg or Arg/Pro genotype; dashed line, Pro/Pro genotype.
The p53 Codon 72 Polymorphism Is an Independent Prognostic Factor
Because the p53 codon 72 Pro/Pro genotype was associated with a shorter overall survival, we asked whether it might represent an independent factor distinct from other common NB prognostic indicators such as clinical stage, MYCN status, and age at diagnosis. Thus, we carried out a Cox multiple regression analysis for overall survival that included stage, MYCN status, age at diagnosis, and p53 codon 72 genotypes as covariates. Interestingly, the hazard ratio between Pro/Pro and Arg/Arg or Arg/Pro genotypes was significant (2.74; 95% confidence interval [CI] = 1.14–6.55, P = .024; Table 2), suggesting that the p53-72 Pro/Pro genotype may represent an additional independent prognostic marker in NB.
Table 2.
Five-Year Overall Survival Analysis.
| Feature | Univariate (Kaplan-Meier) | Cox Multiple Regression | |||
| % Survival | P | P | Hazard Ratio | 95% CI | |
| Stage | |||||
| 1–2–4 S | 85.0 | ||||
| 3–4 | 54.8 | <.001 | .03 | 3.09 | 1.47–6.48 |
| MYCN status | |||||
| Non amp | 71.9 | <.001 | <.001 | 3.15 | 1.86–5.36 |
| Amp | 41.0 | ||||
| Age | |||||
| <18 mo | 86.5 | <.001 | .013 | 2.31 | 1.19–4.48 |
| ≥.18 mo | 51.1 | ||||
| p53 codon 72 | |||||
| Arg/Arg or Arg/Pro | 67.0 | .014 | .024 | 2.74 | 1.14–6.55 |
| Pro/Pro | 30.0 | ||||
CI indicates confidence interval.
Biologic Effects of p53-72R and p53-72P in NB Cells
To address potential mechanisms associated with the more aggressive disease in patients with NB with the p53-72 Pro/Pro genotype, we established p53-null LAN-1 derivative cell lines expressing the tamoxifen (4-HT)-regulated p53 72R-ER or p53 72P-ER isoform (Figure W2) and assessed whether their activation modulates differentially the effects of cytotoxic drugs or IR on apoptosis, survival, and senescence.
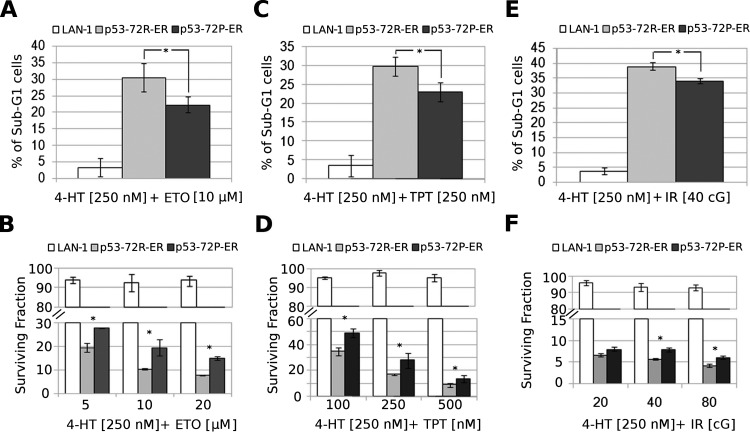
For the experiments assessing the effects of cytotoxic drugs on apoptosis and survival, we selected etoposide and topotecan because they are used to treat patients with NB and function, in part, through DNA damage-dependent p53 activation and also taxol because its mechanism of action is p53 independent [28,29].
As expected, the frequency of apoptotic cells (measured by hypo-diploid DNA content) on treatment with etoposide or topotecan and 4-HT was higher in LAN-1-p53-72R-ER or in LAN-1-p53-72P-ER than in parental cells (Figure 2, A and C). However, LAN-1-p53-72P-ER cells showed less apoptosis than LAN-1-p53-72R-ER cells did (Figure 2, A and C); such lower apoptosis correlated with an increase in the number of surviving cells measured by clonogenic assays (Figure 2, B and D). As expected, cytotoxic drug plus 4-HT-treated parental cells were more clonogenic than the LAN-1-p53-72R/P-ER counterpart (Figure 2, B and D). The frequency of apoptosis and the number of clonogenic/surviving cells were comparable in LAN-1 cells (parental, p53 72R-ER, p53 72P-ER) treated with etoposide or topotecan only (not shown).
Figure 2.
Frequency of apoptotic (A, C, and E) and clonogenic cells (B, D, and F) in cytotoxic- or IR-treated LAN-1 (parental, p53-72R, or p53-72P) cells on 4-HT-dependent activation of p53. In B, D, and F, histograms show residual colony formation of cytotoxic- or IR- and 4-HT-treated LAN-1 (parental, p53-72R, or p53-72P) cells compared with that of cells treated with cytotoxic drug or IR only. Results are presented as the mean ± SD of three experiments performed in triplicate. *P < .05.
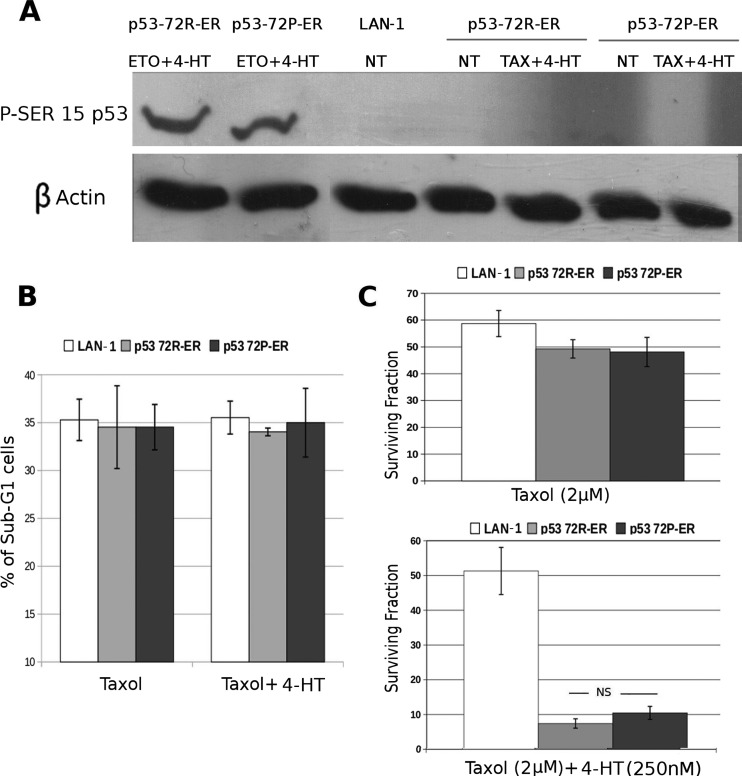
Compared with parental cells, treatment of LAN-1-p53-72R-ER or p53-72P-ER cells with 4-HT did not enhance taxol-induced apoptosis (measured by DNA content in cells treated with 4-HT for 24 hours), probably because of the lack of p53-Ser 15 phosphorylation (Figure 3, A and B). Likewise, the surviving/clonogenic fraction of taxol-treated parental or LAN-1-p53-ER cells was essentially identical (Figure 3C, upper panel). Compared with parental cells, cotreatment with 4-HT and taxol markedly suppressed colony formation/survival of LAN-1-p53-ER cells, but there was no statistically significant difference in the effects of p53-72R and p53-72P (Figure 3C, lower panel).
Figure 3.
Frequency of apoptotic and clonogenic cells in taxol- or taxol and 4-HT-treated LAN-1 (parental, p53-72R, or p53-72P) cells. (A) Western blot shows levels of p53 Ser-15 phosphorylation in LAN-1 (parental, p53-72R, or p53-72P) cells treated with taxol or etoposide (positive control) and 4-HT. (B) Histograms show percent apoptosis (hypodiploid DNA content). (C) Residual colony formation after treatment of LAN-1 (parental, p53-72R, p53-72) cells with taxol or taxol and 4-HT. Results are presented as the mean ± SD of three (B) or two (C) experiments performed with three different seeding (C). NS indicates not significant, P > .05.
As with etoposide and topotecan, treatment with IR (40 cGy) led to higher apoptosis in 4-HT-treated LAN-1-p53-72R-ER and LAN1-p53-72P-ER than parental cells (Figure 2E). Likewise, apoptosis was more frequent in 4-HT-treated LAN-1-p53-72R-ER than in LAN-1-p53 72P-ER cells (Figure 2E). Conversely, the number of surviving cells measured by clonogenic assays was significantly higher in 4-HT- and IR-(40 or 80 cGy) treated LAN-1-p53-72P-ER than LAN-1-p53-72R-ER cells (Figure 2F). No statistically significant differences were noted in the frequency of apoptosis and clonogenic cells after treatment of LAN-1 cells (parental, p53 72R-ER,-p53 72P-ER) with IR only (not shown).
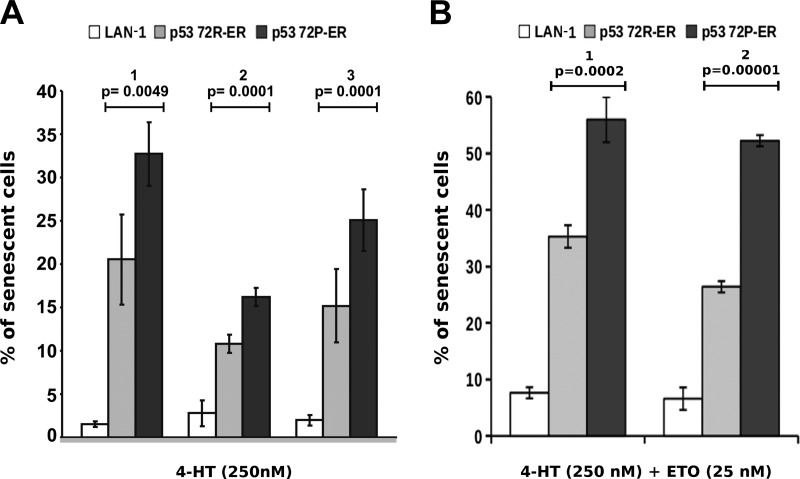
The effect of p53-72R or p53-72P activation on accelerated senescence was investigated upon treatment of parental and LAN-1-p53-ER cells with 4-HT alone or with a low concentration of etoposide (25 nM). Treatment with 4-HT alone induced a higher frequency of SA-β-gal-positive cells in LAN-1-p53-72P than in LAN-1-p53-72R cultures (Figure 4A); such a difference was also observed in LAN-1-p53-72 cells co-treated with 4-HT and etoposide (Figure 4B). By contrast, SA-β-gal positivity was barely detectable or clearly lower in parental cells treated with 4-HT only or with 4-HT and etoposide (Figure 4, A and B).
Figure 4.
Frequency of senescent (SA-β-gal-positive) cells in parental or LAN-1-p53-ER cultures treated with 4-HT alone (A) or co-treated with etoposide (B). In A, parental, LAN-1-p53-72R-, or p53-72P-ER cells were treated with 4-HT (250 nM; 48 hours) and percent of SA-β-gal-positive cells was determined by counting 600 cells; histograms represent the results (mean ± SD) of three independent experiments performed in triplicate. In B, parental, LAN-1-p53-72R-, or p53-72P-ER cells were treated with a suboptimal concentration of etoposide (25 nM; 3 hours), washed and treated with 4-HT (250 nM; 48 hours). In each experiment, 1000 cells were scored for SA-β-gal positivity. Histograms represent the results (mean ± SD) of two experiments performed in duplicate.
Differential Expression of p53-Regulated Genes in LAN-1-p53-72R and LAN-1-p53P Cells
To assess whether the differences in survival and senescence of cytotoxic drug- or IR-treated LAN-1-p53-72 isoform-specific cells may be explained by differences in the levels of p53-regulated genes, the expression of some of these genes was assessed by real-time PCR in 4-HT-treated LAN-1-p53-72R and LAN-1-p53-72P-ER cells. Among the genes tested, levels of BAX and MDM2 were unchanged; by contrast, the expression of NOXA and PERP was higher in 4-HT-treated LAN-1-p53-72R cells, whereas the levels of the CDK inhibitor p21 and PA-I (at least at 12 hours) were higher in 4-HT-treated LAN-1-p53-72P cells (Figure W3).
The difference in p21 expression between 4-HT-treated LAN-1p53-72P and LAN-1-p53-72R cells was confirmed by Western blot analysis (Figure W4).
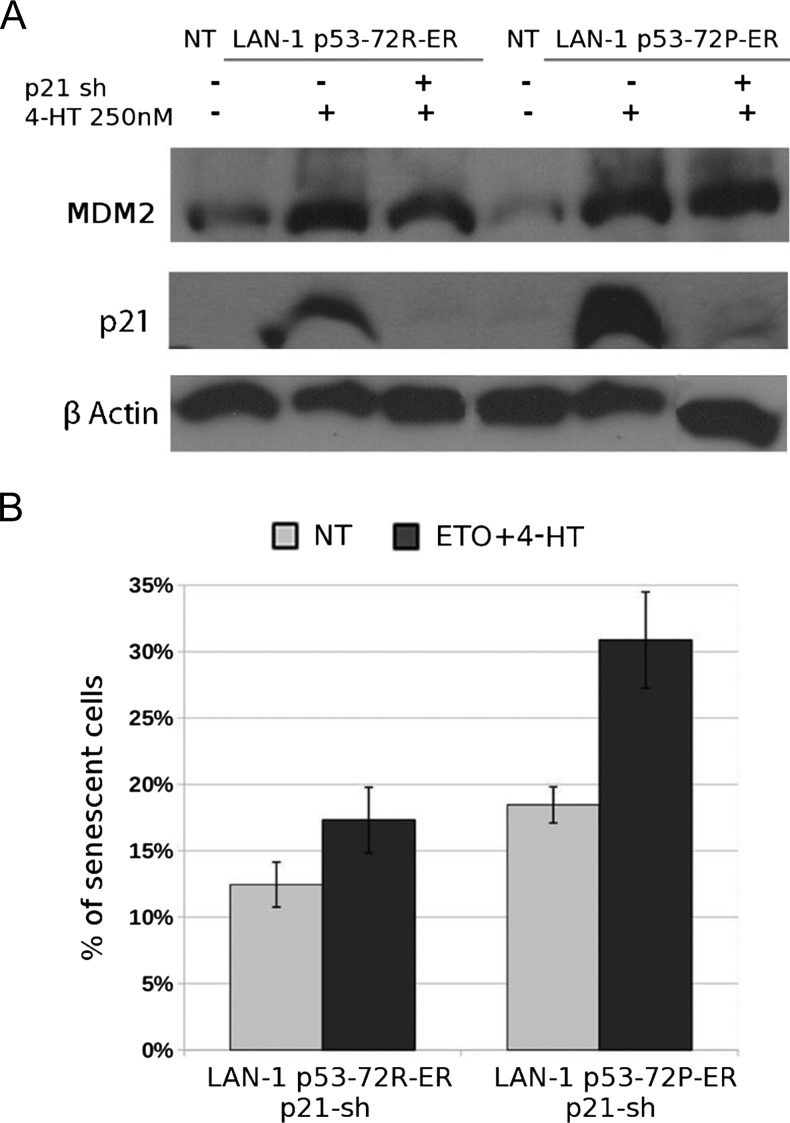
To assess whether the differential expression of p21 in 4-HT-treated LAN-1-p53-72P- and LAN-1-p53-72R-ER cells had any effect on the biologic response of etoposide-treated cells, we measured apoptosis, colony formation, and senescence (SA-β-gal positivity) in p21-silenced LAN-1-p53-72R- and LAN-1-p53-72P-ER cells (Figure 5A). Down-regulation of p21 expression had no effect on the apoptosis and survival of etoposide/4-HT-treated LAN-1-p53-72R and LAN-1-p53-72P cells (Figure W5); by contrast, SA-β-gal-positive cells were markedly reduced on down-regulation of p21 expression (compare Figure 4B with Figure 5B; % SA-β-gal positivity in p53-72-ER vs p53-72-ER p21-sh cells: 30.85% ± 6% vs 17.32% ± 2% in cells expressing p53-72R and 54.24% ± 3% vs 30.87% ± 4% in cells expressing p53-72P, respectively). However, senescent cells remained more numerous in etoposide/4-HT-treated p53-72P-than p53-72R-ER-LAN-1 cultures (Figure 5B), consistent with p53 isoform-specific regulatory mechanisms independent of p21.
Figure 5.
Frequency of SA-β-gal-positive cells in p21-silenced LAN-1-p53-72R- and LAN-1-p53-72P-ER cell cultures. (A) Western blot shows expression of p21, MDM2, and β-actin in 4-HT-treated parental or p21 shRNA-lentivirally transduced LAN-1-p53-72R- and LAN-1-p53-72P-ER cells. Levels of MDM2 and β-actin were measured as control for specificity and loading, respectively. (B) Histograms (mean ± SD of two experiments) show percent SA-β-gal positivity in 4-HT/etoposide-treated p21 shRNA-lentivirally transduced LAN-1-p53-72R- and LAN-1-p53-72P-ER cells.
Discussion
Genetic characteristics associated with tumor initiation and/or progression and predictive of clinical outcome are a subject of intense investigation. Several recent studies have analyzed the relation between the polymorphism of p53 codon 72 and cancer development, disease progression, response to chemotherapy, and overall survival. In several studies, the p53-72Arg/Arg genotype was associated with increased risk for breast [30,31], gastric [32], ovary [33], oral [34], skin cancer [35], and sporadic colorectal adenocarcinoma [36]. On the contrary, other studies have demonstrated an association between the Pro/Pro genotype and increased risk for non-small cell lung [37], nasopharyngeal [38,39], thyroid [40], and skin cancer [41]. The reasons for these discrepancies might depend on methodological differences in the studies (laboratory and statistical analyses, selection of patients, and control subjects) and/or true geographic and ethnic variations. Instead, more consistent results were obtained when assessing the correlation between p53 codon 72 allelic variants and tumor progression, patients' response to chemotherapy, and overall survival [42–45]. In these studies, patients with the Pro/Pro genotype had a worse outcome than those with the Arg/Arg genotype, probably reflecting different biologic properties of the two p53 variants. The p53-72R isoform has been shown to be more effective of the 72P isoform in inducing apoptosis, a difference possibly explained by its more efficient mitochondrial localization and/or reduced affinity for iASPP, a member of the ASPP family which inhibits the transactivating and apoptotic functions of p53 [17,18].
Thus, we analyzed the frequency of p53-72 genotypes in DNA samples of patients with NB and their association with disease extent and clinical outcome. Because the p53 gene is rarely mutated in NB, it is conceivable that the expression of a p53 isoform with reduced proapoptotic activity may have a negative impact on cancer risk and clinical outcome. Although there was no association between any of the p53 codon 72 genotypes and risk to develop NB, patients with the p53-72 Pro/Pro genotype had a shorter overall survival compared to those with the Arg/Arg and Arg/Pro genotypes. Of interest, overall survival of patients with the Arg/Arg and the Arg/Pro genotypes was undistinguishable; this observation is consistent with a number of possibilities (which can be tested experimentally) such as functional dominance of the p53-72R over the p53-72P isoform or unequal levels of isoform expression when both p53-72 alleles are present.
Although caution is necessary because of the groups' small size, a decrease in overall survival in patients with the Pro/Pro genotype was also observed in the subgroup with no MYCN amplification, including patients at stages 3 and 4 or at stage 4 only. These findings are intriguing and potentially important because amplification of MYCN is the most common and reliable marker of poor outcome, especially in infants (children <1 year at diagnosis) with stage 4 disease [46]. Nevertheless, a substantial part of older children with stage 4 NB has a dismal outcome despite the absence of MYCN amplification [47]. In these patients, the genetic trait(s) responsible for tumor aggressiveness are still under investigation [47]. The association of the p53-72 Pro/Pro genotype with shorter overall survival in the subgroup of patients with stage 3 and 4 disease without MYCN amplification raises the possibility that it may be one of the factors promoting tumor aggressiveness in stage 4 disease with normal MYCN status. That the p53-72 Pro/Pro genotype might be a marker of poor outcome in NB is further supported by the results of the Cox multiple regression analysis, which suggest the independence of this polymorphism from other commonly used prognostic indicators such as clinical stage, age at diagnosis, and MYCN status. Consistent with these data, of 20 NB cell lines examined, the only one with the p53-72 Pro/Pro genotype is the NBL-S line, which has an aggressive behavior in mice and normal MYCN status [48], although the half-life of N-myc protein in NBL-S cells is longer than normal [48].
Experiments carried out using etoposide, topotecan, or IR-treated p53-null LAN-1 derivative cell lines expressing the 4-HT-regulated p53-72R-ER or p53-72P-ER isoform support the hypothesis that NB cells from patients with the p53-72 Pro/Pro genotype exhibit an increased survival and a reduced propensity to undergo apoptosis. However, in the p53-72-ER-LAN-1 cell lines treated with taxol, a drug with p53-independent mechanism of action [27,28], the effects of the two p53 isoforms were essentially undistinguishable. Based on these data, it is reasonable to speculate that patients with NB with the p53-72 Pro/Pro genotype may benefit more from co-treatment with drugs that induce DNA damage and increase p53 expression (e.g., topoisomerase I or II inhibitors or IR) and drugs with p53-independent mechanisms of action.
Of interest, p53-72P was more effective than p53-72R in promoting the appearance of senescent (β-gal-positive) cells in vitro. Although it is unknown whether enhanced senescence would also be associated with activation of p53 in NB with the p53-72 Pro/Pro genotype, it is conceivable that undergoing p53-dependent accelerated senescence rather than apoptosis may allow some NB cells to escape cytotoxic drug-induced killing through cell-autonomous and/or paracrine mechanisms [49,50].
In summary, although relatively rare, the p53 codon 72 Pro/Pro genotype might be relevant for improving risk stratification and, perhaps, selecting more personalized therapeutic protocols. In light of the results of this pilot study, further investigations in a larger number of patients and in mouse models of NB are warranted to validate the impact of this p53 polymorphism as a prognostic marker and modulator of chemotherapy sensitivity.
Supplementary Material
Acknowledgments
The authors thank Adam Ertel (Department of Cancer Biology, Thomas Jefferson University) for analysis of SNP array data set. The authors thank members of the Labgen Laboratory (University of Modena e Reggio Emilia) for sequence analysis. The authors also thank Alberto Garaventa, responsible for the Italian Neuroblastoma study, and clinicians and pathologists of the Associazione Italiana Ematologia Oncologia Pediatrica.
Abbreviations
- 4-HT
4-hydroxy-tamoxifen
- ER
estrogen receptor
- IR
ionizing radiation
- MDM2
murine double minute
- MYCN
v-myc myelocytomatosis viral-related oncogene neuroblastoma derived
- NB
neuroblastoma
- SNP
single nucleotide polymorphism
Footnotes
This work was supported by grants of the Fondazione Cassa di Risparmio di Modena, Fondazione Italiana per la Lotta al Neuroblastoma, Fondazione Guido Berlucchi, Associazione “Amici di Lino,” the Italian Ministry of Education, University and Research, and “Io….domani” Associazione per la Lotta contro i Tumori Infantili. S.C. was supported by fellowships of the Associazione Italiana Ricerca sul Cancro (AIRC) and the Fondazione Cassa di Risparmio di Vignola and is currently supported by a fellowship of the Fondazione Cassa di Risparmio di Modena. G.F.-A. was supported by Fellowship of AIRC and is currently supported by the Fondazione Angela Serra. R.D. was supported by a fellowship of the Fondazione per la lotta al Neuroblastoma. A.R.S. and G.M. are supported by a fellowship of the AIRC. S.C. was supported by a fellowship of “Io…. domani” Associazione per la Lotta contro i Tumori Infantili.
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.London WB, Castleberry RP, Matthay KK, Look AT, Seeger RC, Shimada H, Thorner P, Brodeur G, Maris JM, Reynolds CP, et al. Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children's Oncology Group. J Clin Oncol. 2005;23:6459–6465. doi: 10.1200/JCO.2005.05.571. [DOI] [PubMed] [Google Scholar]
- 3.Brodeur GM, Pritchard J, Berthold F, Carisen NL, Castel V, Castelberry RP, De Bernardi B, Evans AE, Favrot M, Hedborg F, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 4.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK, et al. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 5.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 7.Maris JM, Matthay KK. Molecular biology of neuroblastoma. J Clin Oncol. 1999;17:2264–2279. doi: 10.1200/JCO.1999.17.7.2264. [DOI] [PubMed] [Google Scholar]
- 8.Plantaz D, Mohapatra G, Matthay KK, Pellarin M, Seeger RC, Feuerstein BG. Gain of chromosome 17 is the most frequent abnormality detected in neuroblastoma by comparative genomic hybridization. Am J Pathol. 1997;150:81–89. [PMC free article] [PubMed] [Google Scholar]
- 9.Komuro H, Hayashi Y, Kawamura M, Hayashi K, Kaneko Y, Kamoshita S, Hanada R, Yamamoto K, Hongo T, Yamada M, et al. Mutations of the p53 gene are involved in Ewing's sarcomas but not in neuroblastomas. Cancer Res. 1993;53:5284–5288. [PubMed] [Google Scholar]
- 10.Imamura J, Bartram CR, Berthold F, Harms D, Nakamura H, Koeffler HP. Mutation of the p53 gene in neuroblastoma and its relationship with N-myc amplification. Cancer Res. 1993;53:4053–4058. [PubMed] [Google Scholar]
- 11.Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell. 2002;110:9–12. doi: 10.1016/s0092-8674(02)00818-8. [DOI] [PubMed] [Google Scholar]
- 12.Vazquez A, Bond EE, Levine AJ, Bond LG. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 13.Piette J, Neel A, Marechal V. Mdm2: keeping p53 under control. Oncogene. 1997;15:1001–1010. doi: 10.1038/sj.onc.1201432. [DOI] [PubMed] [Google Scholar]
- 14.Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 16.Matlashewski GJ, Tuck S, Pim D, Lamb P, Schneider J, Crawford LV. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7:961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumont P, Leu JI, Della Pietra AC, III, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 18.Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet. 2006;38:1133–1141. doi: 10.1038/ng1879. [DOI] [PubMed] [Google Scholar]
- 19.Cattelani S, Defferrari R, Marsilio S, Bussolari R, Candini O, Corradini F, Ferrari-Amorotti G, Guerzoni C, Pecorari L, Menin C, et al. Impact of a single nucleotide polymorphism in the MDM2 gene on neuroblastoma development and aggressiveness: results of a pilot study on 239 patients. Clin Cancer Res. 2008;14:3248–3253. doi: 10.1158/1078-0432.CCR-07-4725. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 21.Cox DR. Regression models and life tables. J R Stat Soc. 1972;34:187–220. [Google Scholar]
- 22.Ferrari-Amorotti G, Keeshan K, Zattoni M, Guerzoni C, Iotti G, Donato NJ, Calabretta B. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBPα. Blood. 2006;108:1353–1362. doi: 10.1182/blood-2006-01-011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidoff AM, Pence JC, Shorter NA, Iglehart JD, Marks JR. Expression of p53 in human neuroblastoma- and neuroepithelioma-derived cell lines. Oncogene. 1992;7:127–133. [PubMed] [Google Scholar]
- 24.Carr J, Bell E, Pearson ADJ, Kees UR, Beris H, Lunec J, Tweddle DA. Increased frequency of aberrations in the p53/MDM2/p14ARF pathway in neuroblastoma cell lines established at relapse. Cancer Res. 2006;66:2138–2145. doi: 10.1158/0008-5472.CAN-05-2623. [DOI] [PubMed] [Google Scholar]
- 25.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubeli I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, Fox EA, Meyerson M, Diller L, Fortina P, et al. Genome-wide analysis of neuroblastoma using high-density single nucleotide polymorphism arrays. PLoS One. 2007;2(2):e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debernardis D, Sire EG, De FP, Vikhanskaya F, Valenti M, Russo P, Parodi S, D'Incalci M, Broggini M. p53 status does not affect sensitivity of human ovarian cancer cell lines to paclitaxel. Cancer Res. 1997;57:870–874. [PubMed] [Google Scholar]
- 29.Vikhanskaya F, Vignati S, Beccaglia P, Ottoboni C, Russo P, D'Incalci M, Broggini M. Inactivation of p53 in a human ovarian cancer cell line increases the sensitivity to paclitaxel by inducing G2/M arrest and apoptosis. Exp Cell Res. 1998;241:96–101. doi: 10.1006/excr.1998.4018. [DOI] [PubMed] [Google Scholar]
- 30.Langerød A, Bukholm IR, Bregård A, Lønning PE, Andersen TI, Rognum TO, Meling GE, Lothe RA, Borresen-Dale AL. The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas. Cancer Epidemiol Biomarkers Prev. 2002;11:1684–1688. [PubMed] [Google Scholar]
- 31.Buyru N, Tigli H, Dalay N. P53 codon 72 polymorphism in breast cancer. Oncol Rep. 2003;10:711–714. [PubMed] [Google Scholar]
- 32.Shen H, Solari A, Wang X, Zhang Z, Xu Y, Wang L, Hu X, Guo J, Wei Q. p53 codon 72 polymorphism and risk of gastric cancer in a Chinese population. Oncol Rep. 2004;11:1115–1120. [PubMed] [Google Scholar]
- 33.Pegoraro RJ, Rom L, Lanning PA, Moodley M, Naiker S, Moodley J. p53 codon 72 polymorphism and human papillomavirus type in relation to cervical cancer in South African women. Int J Gynecol Cancer. 2002;12:383–388. doi: 10.1046/j.1525-1438.2002.01109.x. [DOI] [PubMed] [Google Scholar]
- 34.Bau DT, Tsai MH, Lo YL, Hsu CM, Tsai Y, Lee CC, Tsai FJ. Association of p53 and p21(CDKN1A/WAF1/CIP1) polymorphisms with oral cancer in Taiwan patients. Anticancer Res. 2007;27:1559–1564. [PubMed] [Google Scholar]
- 35.de Oliveira WR, Rady PL, Grady J, Hughes TK, Neto CF, Rivitti EA, Tyring SK. Association of p53 arginine polymorphism with skin cancer. Int J Dermatol. 2004;43:489–493. doi: 10.1111/j.1365-4632.2004.02289.x. [DOI] [PubMed] [Google Scholar]
- 36.Dakouras A, Nikiteas N, Papadakis E, Perakis M, Valis D, Rallis G, Tzanakis N, Peros G, Tsigkris C, Kittas C, et al. p53Arg72 homozygosity and its increased incidence in left-sided sporadic colorectal adenocarcinomas, in a Greek-Caucasian population. Anticancer Res. 2008;28:1039–1043. [PubMed] [Google Scholar]
- 37.Szymanowska E, Jassem E, Dziadziuszko A, Borg J, Limon G, Kobierska-Gulida W, Rzyman W, Jassem J. Increased risk of non-small cell lung cancer and frequency of somatic TP53 gene mutations in Pro72 carriers of TP53 Arg72Pro polymorphism. Lung Cancer. 2006;1:9–14. doi: 10.1016/j.lungcan.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Tiwawech D, Srivatanakul P, Karaluk A, Ishida T. The p53 codon 72 polymorphism in Thai nasopharyngeal carcinoma. Cancer Lett. 2003;198:69–75. doi: 10.1016/s0304-3835(03)00283-0. [DOI] [PubMed] [Google Scholar]
- 39.Hadhri-Guiga B, Toumi N, Khabir A, Sellami-Boudawara T, Ghorbel A, Daoud J, Frikha M, Gargouri A, Mokdad-Gargouri R. Proline homozygosity in codon 72 of TP53 is a factor of susceptibility to nasopharyngeal carcinoma in Tunisia. Cancer Genet Cytogenet. 2007;178:89–93. doi: 10.1016/j.cancergencyto.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 40.Granja F, Morari J, Morari EC, Correa LA, Assumpção LV, Ward LS. Proline homozygosity in codon 72 of p53 is a factor of susceptibility for thyroid cancer. Cancer Lett. 2004;210(2):151–157. doi: 10.1016/j.canlet.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Chen YC, Xu L, Guo YL, Su HJ, Hsueh YM, Smith TJ, Ryan LM, Lee MS, Chaor SC, Lee JM, et al. Genetic polymorphism in p53 codon 72 and skin cancer in southwestern Taiwan. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2003;38:201–211. doi: 10.1081/ese-120016889. [DOI] [PubMed] [Google Scholar]
- 42.Csejtei A, Tibold A, Varga Z, Koltai K, Ember A, Orsos Z, Feher G, Horvath OP, Ember I, Kiss I. GSTM, GSTT and p53 polymorphisms as modifiers of clinical outcome in colorectal cancer. Anticancer Res. 2008;28:1917–1922. [PubMed] [Google Scholar]
- 43.Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, Hiller L, Farrel PJ, Smith P, Lu X, et al. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–3337. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Yao L, Ouyang T, Li J, Wang T, Fan Z, Lin B, Lu Y, Xie Y. p53 codon 72 polymorphism predicts the pathologic response to neoadjuvant chemotherapy in patients with breast cancer. Clin Cancer Res. 2005;11:7328–7333. doi: 10.1158/1078-0432.CCR-05-0507. [DOI] [PubMed] [Google Scholar]
- 45.Xu Y, Yao L, Zhao A, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Lu Y, et al. Effect of the p53 codon 72 genotype on breast cancer survival depends on p53 gene status. Int J Cancer. 2008;122:2761–2766. doi: 10.1002/ijc.23454. [DOI] [PubMed] [Google Scholar]
- 46.Friedman GK, Castleberry RP. Changing trends of research and treatment in infant neuroblastoma. Pediatr Blood Cancer. 2007;49:1060–1065. doi: 10.1002/pbc.21354. [DOI] [PubMed] [Google Scholar]
- 47.Brodeur GM, Ambros PF. Genetic and biological markers of prognosis in neuroblastoma. In: Brodeur GM, Sawada T, Tsuchida Y, Voute PA, editors. Neuroblastoma. 2000. pp. 355–365. [Google Scholar]
- 48.Cohn SL, Salwen H, Quasney MW, Ikegaki N, Cowan JM, Herst CV, Kenneth RH, Rosen ST, DiGiuseppe JA, Brodeur GM. Prolonged N-myc protein half-life in a neuroblastoma cell line lacking N-myc amplification. Oncogene. 1990;5:1821–1827. [PubMed] [Google Scholar]
- 49.Gewirtz DA, Holt SE, Elmore LW. Accelerated senescence: an emerging role in tumor cell response to chemotherapy and radiation. Biochem Pharmacol. 2008;76:947–957. doi: 10.1016/j.bcp.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 50.Canino C, Mori F, Cambria A, Diamantini A, Germoni S, Alessandrini G, Borsellino G, Galati R, Battistini L, Blandino R, et al. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene. 2011 Oct 24; doi: 10.1038/onc.2011.485. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.