Abstract
Peanut/maize intercropping is a sustainable and effective agroecosystem to alleviate iron-deficiency chlorosis. Using suppression subtractive hybridization from the roots of intercropped and monocropped peanut which show different iron nutrition levels, a peanut gene, AhNRAMP1, which belongs to divalent metal transporters of the natural resistance-associated macrophage protein (NRAMP) gene family was isolated. Yeast complementation assays suggested that AhNRAMP1 encodes a functional iron transporter. Moreover, the mRNA level of AhNRAMP1 was obviously induced by iron deficiency in both roots and leaves. Transient expression, laser microdissection, and in situ hybridization analyses revealed that AhNRAMP1 was mainly localized on the plasma membrane of the epidermis of peanut roots. Induced expression of AhNRAMP1 in tobacco conferred enhanced tolerance to iron deprivation. These results suggest that the AhNRAMP1 is possibly involved in iron acquisition in peanut plants.
Keywords: AhNRAMP1, intercropping, iron acquisition, peanut, tobacco, transporter
Introduction
Iron (Fe) is an essential nutrient for plant growth and development, participating in a series of biochemical processes such as DNA biosynthesis, respiration, and photosynthesis. Although Fe is abundant in soil, the availability of Fe is often very limited because of the insoluble oxidized form (Guerinot and Yi, 1994). Higher plants have developed two unique mechanisms to acquire Fe in response to Fe deprivation (Römheld and Marschner, 1986). Graminaceous species secrete phytosiderophores to mobilize Fe in the rhizosphere (Takagi, 1976). Subsequently, the Fe(III)–phytosiderophore complexes are absorbed by specific membrane transporters yellow stripe1/yellow stripe like (YS1/YSL) (Curie et al., 2001; Murata et al., 2006; Inoue et al., 2009; Lee et al., 2009). Dicots and non-graminaceous monocots reduce Fe(III) to the more soluble Fe(II) by a membrane-bound ferric chelate reductase (Robinson et al., 1999) and the Fe(II) is then taken up via a high affinity Fe(II) uptake transporter, iron-regulated transporter 1 (IRT1) (Eide et al., 1996; Connolly et al., 2002; Vert et al., 2002; Varotto et al., 2002). IRT1 transports other divalent metals as well (Korshunova et al., 1999; Rogers et al., 2000). Disruption of AtIRT1 leads to a severe growth defect, which is rescued by application of Fe, indicating that IRT1 is the major transporter for Fe uptake from the soil (Vert et al., 2002). Interestingly, graminaceous plants such as rice can take up Fe2+ by OsIRT1 and OsIRT2 even without inducible Fe3+ chelate reductase activity, in addition to the Fe(III)–phytosiderophore system (Ishimaru et al., 2006).
In addition to IRT1, the NRAMP (natural resistance-associated macrophage protein) gene family encodes integral membrane proteins that transport a broad range of metal ions including Fe. The evolutionarily conserved NRAMP genes have been identified in various species from bacteria to human (Williams et al., 2000; Nevo and Nelson, 2006). In mammals, DCT1 (divalent cation transporter)/NRAMP2 is essential for intestinal Fe absorption and endosomal recycling of Fe (Fleming et al., 1997, 1998; Gunshin et al., 1997). SMF and MntH, NRAMP homologues in yeast and bacteria, respectively, are involved in manganese (Mn) accumulation (Makui et al., 2000; Portnoy et al., 2000). In plants, the NRAMP family transporters have been identified in various species and show diverse functions (Belouchi et al., 1997; Curie et al., 2000; Thomine et al., 2000; Bereczky et al., 2003; Kaiser et al., 2003; Mizuno et al., 2005; Xiao et al., 2008; Oomen et al., 2009; Wei et al., 2009; Xia et al., 2010; Takahashi et al., 2011; Ishimaru et al., 2012). In Arabidopsis, heterologous expression of AtNRAMP1, AtNRAMP3, and AtNRAMP4 in yeast mutants indicated that these proteins could transport Fe, Mn, and cadmium (Cd) (Curie et al., 2000; Thomine et al., 2000). AtNRAMP3 and AtNRAMP4 are responsible for mobilization of vacuolar Fe stores (Thomine et al., 2003; Lanquar et al., 2005). Meanwhile, both genes also function in export of vacuolar Mn into photosynthetic tissues of adult plants (Lanquar et al., 2010). AtNRAMP6 contributes to Cd toxicity (Cailliatte et al., 2009). Recently, it has been proposed that AtNRAMP1 is a high-affinity Mn transporter and essential for uptake of Mn from the soil in low Mn conditions (Cailliatte et al., 2010). The NRAMP1 transporters have also been identified in other plant species. In tomato, LeNRAMP1 localizes in the vascular parenchyma of the root hair zone as well as in the root epidermis and the cortex behind the root tip, and is thought to play a role in distribution of Fe in the vascular parenchyma upon Fe deficiency (Bereczky et al., 2003). The identification of MbNRAMP1 in a fruit tree (Malus baccata) suggested that MbNRAMP1 was involved in Fe, Mn, and Cd trafficking (Xiao et al., 2008). Rice OsNRAMP1 rescues the growth of an Fe-defective yeast mutant and is related to Cd accumulation in rice (Takahashi et al., 2011). Recent characterization of rice OsNRAMP5 revealed its involvement in Mn, Fe, and Cd uptake and transport (Ishimaru et al., 2012). Therefore, the biological functions of NRAMP are diverse in different plant species and need to be further clarified.
It has been reported that peanut/maize intercropping alleviates Fe-deficiency chlorosis of peanut compared with monocropped peanut (Zuo et al., 2000; Inal et al., 2007; Zuo and Zhang, 2008, 2009). In the present report, a peanut NRAMP gene, designated as AhNRAMP1, was isolated by suppression subtractive hybridization (SSH) from the roots of intercropped and monocropped peanut, which represented different Fe nutrition levels. Further functional identification of AhNRAMP1 showed that AhNRAMP1 is a functional Fe transporter, and might be responsible for Fe acquisition and distribution in peanut plants.
Materials and methods
Plant materials and growth conditions
The seedlings of peanut (Arachis hypogaea L.cv. Luhua 14) were grown in 5 litre boxes containing continuously aerated nutrient solution. The composition of the nutrient solution was as follows: 0.70 mM K2SO4, 0.10 mM KCl, 0.10 mM KH2PO4, 2.0 mM Ca(NO3)2, 0.50 mM MgSO4, 10 μM H3BO3, 0.50 μM MnSO4, 0.50 μM ZnSO4, 0.20 μM CuSO4, 0.01 μM (NH4)6Mo7O24, and 100 μM Fe(III)-EDTA. The plants were grown in a greenhouse with 30 °C light/25 °C dark cycles under natural light conditions.
Cloning of the AhNRAMP1 gene
Peanut monocropping and intercropping with maize in a pot experiment were performed as previously described (Zuo and Zhang, 2008). The roots of intercropped and monocropped peanut were harvested after intercropping with maize for ∼1 month. Total RNA was extracted with Trizol reagent (Invitrogen, USA) and then mRNA was isolated using an Oligotex mRNA Purification Kit (Qiagen, http://www.qiagen.com). SSH between intercropped and monocropped peanut roots was performed using a Clontech PCR-Select™ cDNA Subtraction Kit (Clontech, http://www.clontech.com) following the manufacturer’s instructions. The clones were sequenced and one clone containing the fragment of the NRAMP gene was found by BLAST alignment (http://www.ncbi.nlm.nih.gov/). Based on the sequence of the cloned fragment, the full-length cDNA, which was designated as AhNRAMP1, was generated by the rapid amplification of cDNA ends (RACE) technique (SMART™ RACE cDNA amplification kit; Clontech) using the following primers: 5′-GCCAATCCACGAAGGCAGTGATGAGG-3′ (5′ RACE) and 5′-AACACAGCAATGCAAACCCATGTGGA-3′ (3′ RACE). The open reading frame (ORF) of AhNRAMP1 cDNA was amplified by PCR using primers AhNRAMP1-F, 5′-GACTCATCACTTGGATTGACTGT-3′ and AhNRAMP1-R, 5′-CTCATACATACATAGCTCAAGTCACT-3′. After cloning, the plasmid content of AhNRAMP1 was confirmed by sequencing. The phylogenic tree was constructed after multiple alignment using BioEdit v7.0.5 (Hall, 1999). The accession number of AhNRAMP1 in GenBank is JQ581595.
Quantitative real-time PCR
Total RNA was extracted from the roots and leaves of Fe-deficient or Fe-sufficient peanut in hydroponics by the SDS/phenol method and then treated with RNase-free DNase I (Takara, Tokyo, Japan) to remove genomic DNA contamination. First-strand cDNA was synthesized by ReverTra Ace reverse transcriptase (Toyobo, Tokyo, Japan) by priming with the d(T)17-adaptor primer. Quantitative real-time PCR was performed using the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA) and SYBR Premix Ex Taq (Perfect Real Time) reagent (Takara) using gene-specific primers QAhNRAMP1-F, 5′-CCTCATCACTGCCTTCGT-3′ and QAhNRAMP1-R, 5′-ATTGCTGTGTTATCCTTGGTC-3′. The PCR products were confirmed by DNA sequencing (3130 Genetic Analyzer, Applied Biosystems, Tokyo, Japan). The transcript abundance was normalized against peanut Ubiquitin (Luo et al., 2005) transcript levels.
Yeast functional complementation
The full-length coding sequence of AhNRAMP1 with restriction sites was amplified by PCR using primers XbaI-NRAMP-F, 5′-TCTAGAATGGCAAGCGTTCTTAGACA-3′ and SacI-NRAMP-R, 5′-GAGCTCTTATTCCGGTAGTGGGATAT-3′. The PCR product of AhNRAMP1 cDNA was then subcloned into pCR®-Blunt II-TOPO® vector (Invitrogen) and the sequence was confirmed. After digestion, the AhNRAMP1 cDNA was introduced into yeast expression vector pDR195 (Schaaf et al., 2005; kindly provided by Dr Nicolaus von Wirén, University of Hohenheim, Germany). Saccharomyces cerevisiae strain DEY1453 (fet3fet4 mutant, MAT_/MAT_ ade2/_ can1/can1 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 fet3-2::His3/fet3-2::HIS3 fet4-1::LEU2/fet4-1::LEU2) (Dix et al., 1994; kindly provided by Dr David Eide, University of Minnesota School of Medicine, USA and Dr Toshihiro Yoshihara, CRIEPI, Japan) was transformed with pDR195-AhNRAMP1, empty vector pDR195, and pDR195-OsNRAMP1 (Takahashi et al., 2011) as a positive control, using the LiAc/SS-DNA/PEG method (Gietz and Schiestl, 1995). The yeast cells were selected on solid synthetic defined (SD) media plates without uracil. The transformed cells were diluted to OD 1 to 0.001 at 600 nm and spotted onto the plate containing SD medium at pH 5, 6, and 7. The spotted yeast cells were then incubated at 30 °C for 2–3 d.
Subcellular localization of AhNRAMP1
The AhNRMAP1 ORF without a stop codon and containing XhoI and BglII restriction sites was amplified by PCR using primers 5′-CTCGAGATGGCAAGCGTTCTTAGACA-3′ and 5′-AGATCTGTTCCGGTAGTGGGATATCAG-3′. The PCR product was then subcloned into the Cauliflower mosaic virus (CaMV) 35S-sGFP(S65T)-NOS3′ vector (kindly provided by Dr Yasuo Niwa, University of Shizuoka, Japan). The AhNRAMP1–sGFP (synthetic green fluorescent protein) fusion construct or the CaMV35S–sGFP construct was transiently expressed in onion epidermal cells transformed by DNA particle bombardment as described by Mizuno et al. (2003). After 4–6 h, FM4-64 (Invitrogen, Molecular Probes, www.invitrogen.com) was added to the onion epidermal cells at a final concentration of 2 μM. The fluorescent cells were imaged by confocal microscopy (LSM5Pascal; Carl Zeiss, Göttingen, Germany).
Laser microdissection (LMD) and expression analysis
LMD was performed as previously described (Takahashi et al., 2010). Briefly, peanut roots from Fe sufficiency and deficiency conditions were dissected into 5 mm strips in the fixative solution (ethanol:acetic acid 3:1) on ice. The fixative was infiltrated into the tissues under vacuum three times for 5 min on ice and then overnight at 4 °C. The samples in fixative solution were further fixed by microwave at 37 °C for 15 min, and this was repeated three times with fresh and pre-chilled fixative solution. The samples were then dehydrated by 70, 80, 90, and 100% ethanol at 58 °C (1.5 min each time) in the microwave. The paraffin-embedded blocks were prepared by gradually exchanging butanol with melted paraffin wax at 58 °C. The sections were laser-microdissected using the Veritas Laser Microdissection System LCC1704 (Molecular Devices) and the dissected content was confirmed. Total RNA was extracted from the laser-microdissected samples using a Pico-Pure™ RNA isolation kit (Molecular Devices) and quantified by a Quant-iT™ RiboGreen RNA reagent and kit (Invitrogen). About 10 ng of RNA was used for quantitative real-time PCR.
In situ hybridization
A gene-specific fragment of AhNRAMP1 at the 3′-untranslated region was amplified by primers IAhNRAMP-F, 5′-ACCACTTCACACTGCTTTTTAGG-3′ and IAhNRAMP-R, 5′-GATCTTCAAAGGAGAAATTGTCAC-3′. The PCR product was then subcloned into pCR®-Blunt II-TOPO® vector (Invitrogen) and the content was confirmed by sequencing. The correct plasmids with two directions of insert fragments for sense or antisense probes were individually linearized with BamHI. The sense or antisense probe was generated by T7 RNA polymerase and labelled with digoxigenin-11-UTP (Roche, Mannheim, Germany). Peanut plants were treated without Fe for 1 week. The roots were fixed in FAA (50% ethanol:acetic acid:37% formaldehyde solution 18:1:1) and embedded in paraffin after dehydration with an ethanol and t-butanol series. The root tissues were sectioned to 10 μm and mounted on slides. In situ hybridization was performed according to Ishimaru et al. (2005) with some modifications. In brief, after pre-treatment of sections with proteinase K and acetylation, the slides were hybridized for 16 h with sense or antisense probes at 50 °C and washed. The tissues were then incubated with anti-digoxigenin–alkaline phosphatase conjugate (Roche) and stained with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche).
Generation of transgenic plants
The coding region of the AhNRAMP1 gene was amplified with the following primers: 5′-TCTAGAATGGCAAGCGTTCTTAGACAG-3′ and 5′-GAGCTCTTATTCCGGTAGTGGGATATC-3′. XbaI and SacI restriction sites were used to replace AhNRAMP1 cDNA with the GUS (β-glucuronidase) gene of E-90Ω plasmid (Kobayashi et al., 2004). The resultant plasmid has the backbone of the pIG121Hm binary vector (Hiei et al., 1994) and drives AhNRAMP1 cDNA under the control of the –272/–131 region of the barley IDS2 gene containing Fe-deficiency-responsive element 1 (IDE1) and IDE2, flanked by the –90/+8 region of the CaMV35S promoter and the Tobacco mosaic virus 5′ leader (Ω) sequence. The constructed plasmid was introduced into Agrobacterium tumefaciens strain C58 by electroporation and then transformed to tobacco (Nicotiana tabacum L. cv. Petit-Havana SR1) according to the method of Helmer et al. (1984).
Analysis of transgenic tobacco plants
Nine T2 transgenic tobacco lines were generated, and germinated on Murashige and Skoog (MS) medium containing hygromycin B (50 mg l−1). Non-transgenic seeds were germinated on MS medium lacking hygromycin B. After 1 week the seedlings were transferred to MS medium without Fe for 6 d. The expression level of the AhNRAMP1 gene in all lines treated with Fe deficiency was analysed by quantitative real-time PCR. The transcript abundance was normalized against tobacco Actin (Yoshihara et al., 2006) transcript levels. Two lines of transgenic tobacco plants with a high expression level of AhNRAMP1 were selected for further analysis. For hydroponics, after 2–3 weeks of growth in MS medium followed by an acclimation period of 3 d, the plantlets were transferred to nutrient solution (the same composition as described above for peanut) in a greenhouse under natural light conditions at 25 °C. The transgenic lines and wild-type plants with at least three replicates were grown in hydroponics for ∼1 week and then subjected to Fe deficiency for 9 d. Roots and young leaves were harvested for metal content measurement. Samples were dried for 2–3 d at 80 °C, and 100–200 mg portions were then wet-ashed with 4 ml of 4.4 M HNO3 and 6.5 M H2O2 for 30 min at 220 °C using a MarsXpress oven (CEM, http://www.cem.com/). Metal concentrations were measured by using inductively coupled plasma optical emission spectrometry SPS3000 (Seiko, Tokyo, Japan).
Results
Identification of the NRAMP1 gene from peanut
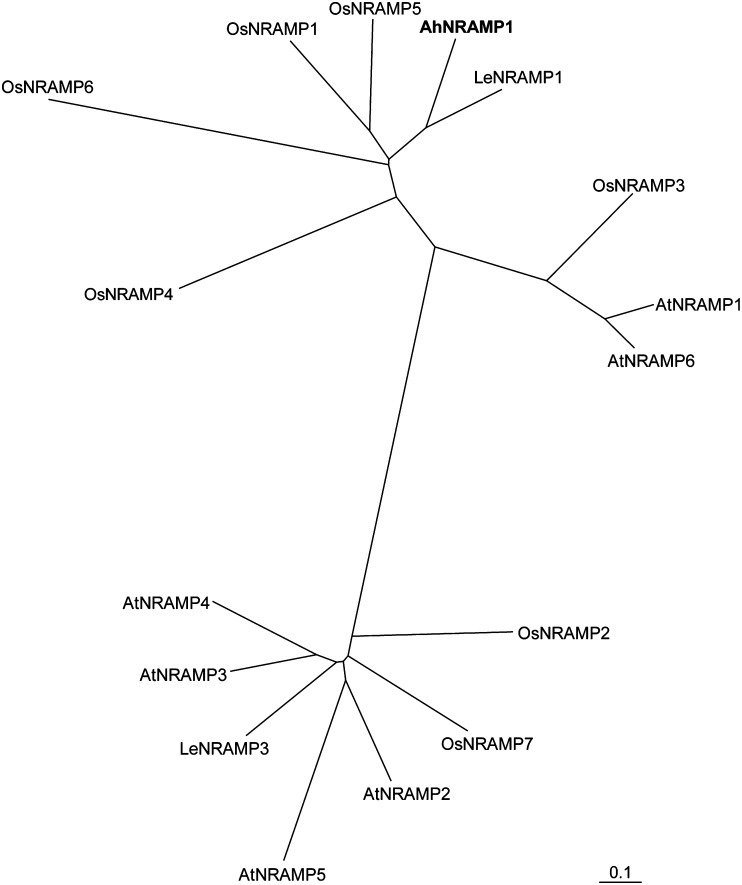
Using SSH between intercropped (with maize) and monocropped peanut, a partial sequence of a 490 bp cDNA fragment showing 73% identity with LeNRAMP1 was obtained. By additional 5' and 3' RACE analysis of cDNA from Fe-deficient peanut roots, a 2033 bp cDNA sequence containing the full-length ORF was identified, and designated as AhNRAMP1. The AhNRAMP1 gene was predicted to encode 545 amino acids, consisting of 12 putative transmembrane domains (TMs) (Supplementary Fig. S1 available at JXB online). Phylogenetic tree analysis showed that among several NRAMP genes from Arabidopsis, rice, and tomato, AhNRAMP1 was most similar to LeNRAMP1, with 78% identity (Fig. 1).
Fig. 1.
Phylogenetic tree of the NRAMP proteins from Arabidopsis, rice, tomato, and peanut. Accession numbers are as follows: AhNRAMP1, JQ581595; AtNRAMP1, AEE36455.1; AtNRAMP2, AEE32142.1; AtNRAMP3, AAF13278.1; AtNRAMP4, AAF13279.1; AtNRAMP5, NP_193614.1; AtNRAMP6, AEE29390.1; OsNRAMP1, AAB36424.1; OsNRAMP2, Q10Q65; OsNRAMP3, Q653V6; OsNRAMP4, Os02g0131800; OsNRAMP5, Os07g0257200; OsNRAMP6, Os01g0503400; OsNRAMP7, Os12g0581600; LeNRAMP1, NP_001234318; LeNRAMP3, NP_001233770.
AhNRAMP1 is induced by Fe deficiency in both roots and leaves
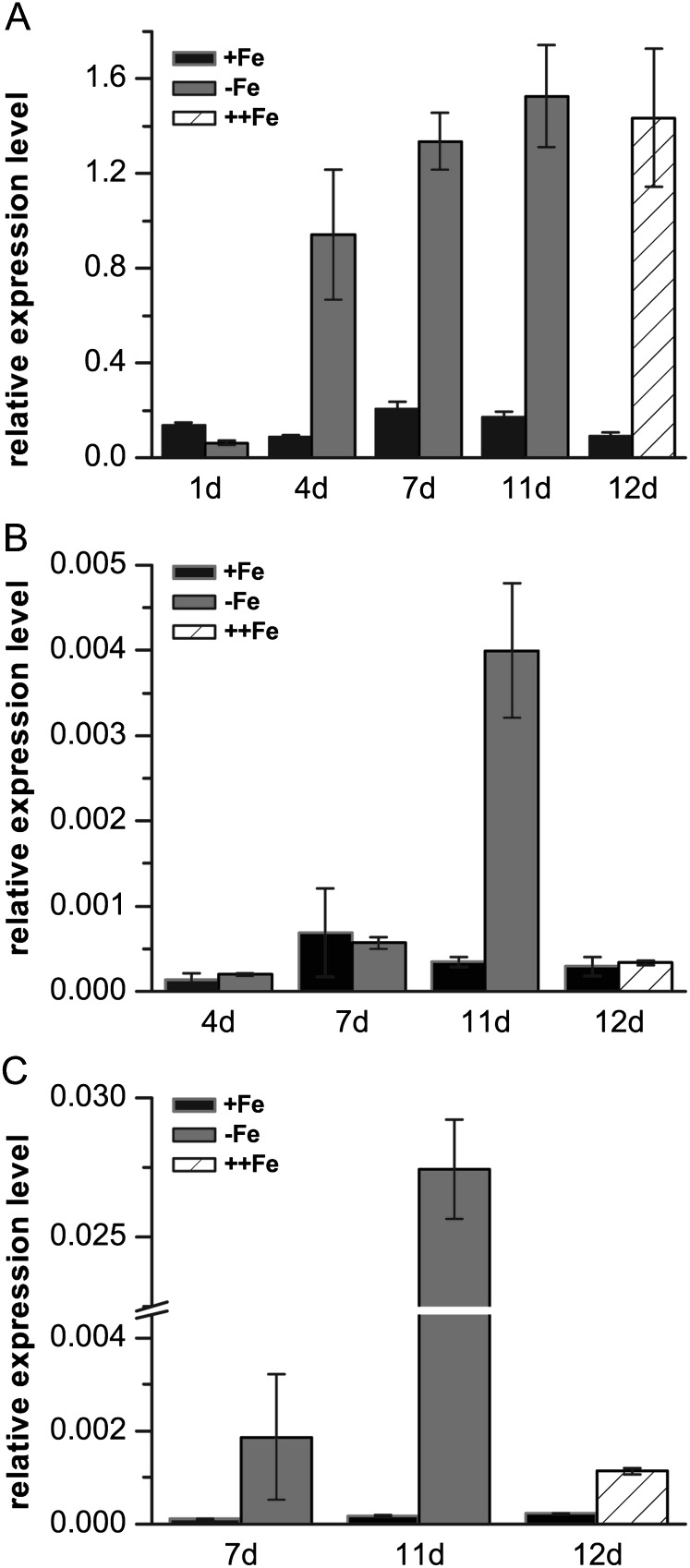
The transcriptional abundance of AhNRAMP1 was assessed in peanut supplemented or not with Fe. The expression of AhNRAMP1 was higher in roots than in leaves, and markedly induced by Fe deficiency in both roots and leaves (Fig. 2). The transcript level of AhNRAMP1 increased at least 5-fold after treatment without Fe for 4, 7, and 11 d in roots (Fig. 2A). In comparison with roots, the induction time of expression was delayed in young and old leaves. In leaves, only at 11 d did AhNRAMP1 show remarkably higher expression under Fe limitation (Fig. 2B, C). Interestingly, after transferring the Fe-starved plants to a solution containing an excess amount of Fe (500 μM) for 1 d, the expression of the AhNRAMP1 gene was not down-regulated in roots (Fig. 2A), but was significantly suppressed in young and old leaves (Fig. 2B, C).
Fig. 2.
Expression pattern of AhNRAMP1 in response to Fe-deficient and Fe-sufficient conditions. Peanut plants were treated without Fe or with normal amounts of Fe (100 μM) for 1, 4, 7, and 11 d. 1d, 4d, 7d, and 11d represent the day of treatments. On day 12 (12d), the Fe-deficient peanut plants were transferred to 500 μM Fe for the treatment with excess Fe (++Fe) for 1 d. The samples were harvested from roots (A), young leaves (B), and old leaves (C) of the treated peanut plants. The vertical bars indicate the expression level of the genes relative to that of the control AhUbiquitin gene. Values are the means of three replications. Error bars indicate the SD.
AhNRAMP1 functionally complements the growth defect of Fe-deficient yeast mutant
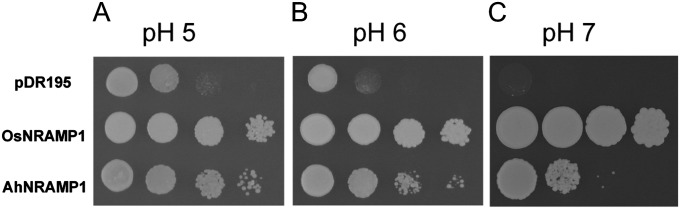
In order to investigate whether AhNRAMP1 transports Fe, a yeast functional complementation assay was performed by using the fet3fet4 yeast double mutant (strain DEY1453; Dix et al., 1994), which is defective in high- and low-affinity Fe-uptake systems. The full-length cDNA of the AhNRAMP1 gene was subcloned into the yeast expression vector pDR195. Meanwhile, the rice OsNRAMP1 gene (Curie et al., 2000) was used as a positive control. Expression of AhNRAMP1, as well as OsNRAMP1, significantly improved yeast growth compared with the control strain transformed with empty vector under various pH values tested (pH 5, 6, and 7) (Fig. 3), suggesting that the peanut AhNRAMP1 transports Fe.
Fig. 3.
Functional complementation of the fet3fet4 yeast mutant with AhNRAMP1. Empty pDR195 vector was used as a negative control and OsNRAMP1 cDNA as a positive control. Serial dilutions of the transformed yeast cells with OD600 nm 1 to 0.001 were plated onto SD medium at pH 5 (A), pH 6 (B), and pH 7 (C).
AhNRAMP1 is a plasma membrane protein
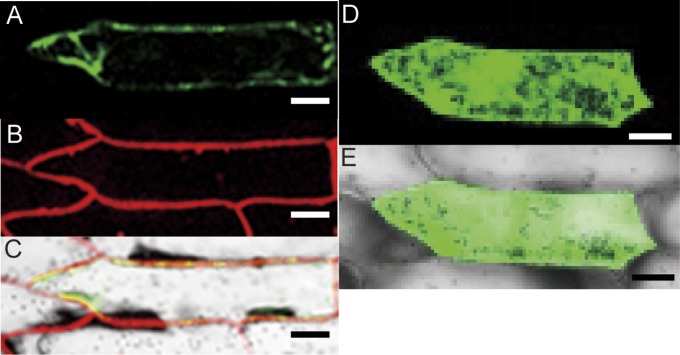
The subcellular localization of AhNRAMP1 was investigated by expressing an AhNRAMP1–GFP fusion protein in onion epidermal cells. The dye FM4-64 was used as a plasma membrane marker (Uraguchi et al., 2011). AhNRAMP1–GFP fluorescence was observed in the plasma membrane, overlapping the FM4-64 fluorescence immediately after staining (Fig. 4A–C). The green fluorescence of GFP alone, as control, was observed mainly in the cytosol and nucleus (Fig. 4D, E).
Fig. 4.
Subcellular localization of AhNRAMP1 in onion epidermal cells. (A) AhNRAMP1 fused to GFP was transiently expressed in onion epidermal cells. (B) The red fluorescence detected from the FM4-64 dye (2 μM), as a plasma membrane marker. (C) Confocal microscopy images illustrating AhNRAMP1–GFP co-localization with the FM4-64 dye at the plasma membrane. (D) GFP alone was transiently expressed in onion epidermal cells. (E) Overlay with the transmission image shown in (D). Scale bars represent 50 μm.
AhNRAMP1 is mainly localized in the epidermis of peanut roots
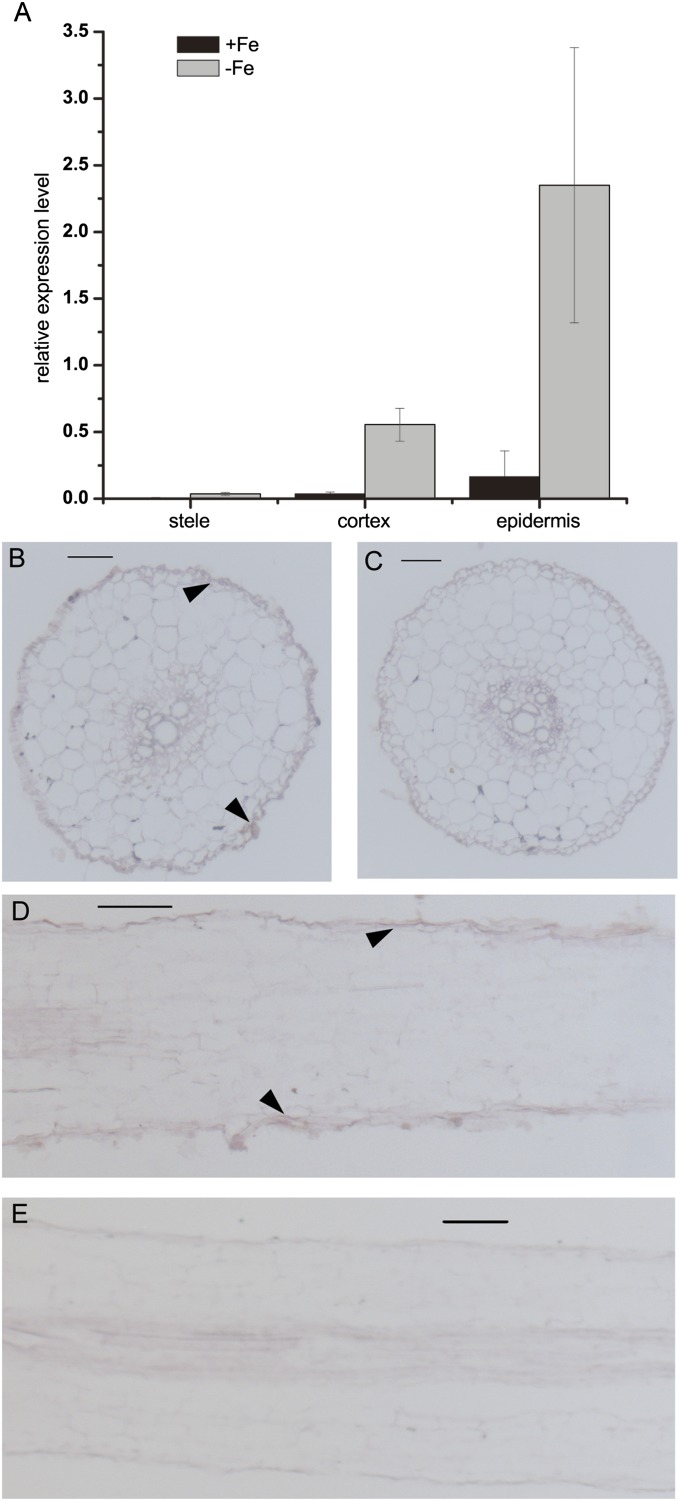
The peanut root tissues from Fe-deficient and Fe-sufficient conditions were separated into epidermis/exodermis, cortex, and stele by LMD. Quantitative real-time PCR was performed to identify the expression abundance of the AhNRAMP1 gene in the different parts of root tissues. The expression abundance of AhNRAMP1 was the highest in the epidermis/exodermis, and the lowest in the stele, irrespective of the Fe status (Fig. 5A). Consistent with the expression pattern in whole roots (Fig. 2A), the expression of AhNRAMP1 was strongly induced by Fe deprivation in all parts of root tissues (Fig. 5A).
Fig. 5.
Tissue localization of AhNRAMP1 in peanut roots by LMD analysis and in situ hybridization. (A) LMD and expression analysis of AhNRAMP1. Peanut plants were cultured with or without Fe for 7 d. Stele, cortex, and epidermis/endodermis of peanut roots were separated by LMD. The vertical bars indicate the relative expression level of AhNRAMP1 as compared with the control AhUbiquitin gene. Three biological replications were performed for each treatment. Error bars represent the SD. (B–E) In situ hybridization analysis of AhNRAMP1 in the roots of Fe-deficient peanut. AhNRAMP1 antisense probes were hybridized in a cross-section (B) or longitudinal section (D) of peanut root. Sense probes of AhNRAMP1 were used as a control (C, E). Bars = 100 μm. Arrowheads indicate representative hybridization signals.
Further, in situ hybridization was employed to confirm the tissue localization of the AhNRAMP1 gene in peanut root using a specific probe. Young roots of peanut under Fe deficiency were analysed. In good agreement with the expression in LMD sections, the staining of AhNRAMP1 antisense probe was more visible on the cells of the root epidermis in both transverse (Fig. 5B) and longitudinal (Fig. 5D) sections compared with that of the sense probe as a negative control (Fig. 5C, E).
Induced expression of AhNRAMP1 in tobacco results in tolerance to Fe deprivation
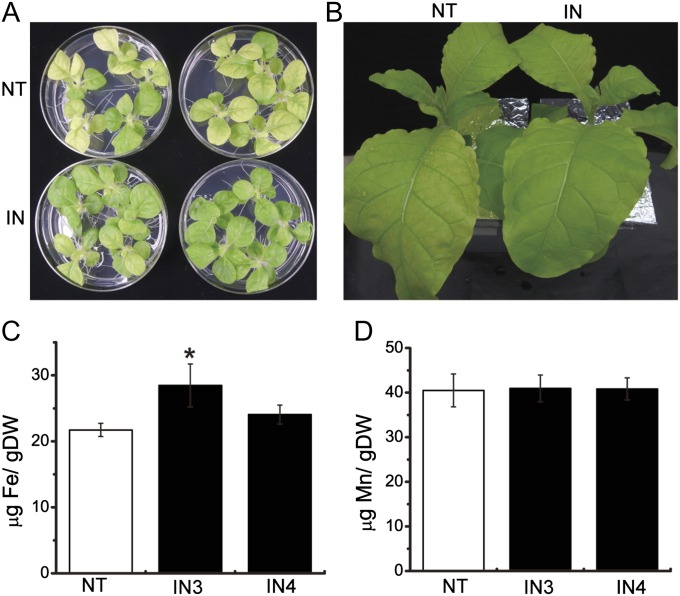
To evaluate further the function of AhNRAMP1 in planta, this gene was introduced into tobacco, a model dicot plant. Since constitutive overexpression of NRAMP genes can cause a growth defect (Takahashi et al., 2011; Ishimaru et al., 2012), an Fe deficiency-inducible artificial promoter (Kobayashi et al., 2004) was utilized to drive high expression of AhNRAMP1 under Fe deficiency in tobacco. When grown in Fe-deprivation conditions on both MS medium and hydroponics, non-transgenic (NT) plants exhibited more chlorosis than AhNRAMP1-induced transformants (Fig. 6A, B). The Fe concentration in young leaves of AhNRAMP1-induced plants was higher than in NT plants (Fig. 6C). The Mn and Zn levels of young leaves were similar between the induced lines and NT plants (Fig. 6D, Supplementary Fig. S3 at JXB online). In the roots, no significant changes in the level of any metals were observed (Supplementary Fig. S4 at JXB online). Taken together, these results indicate that the induced expression of AhNRAMP1 in tobacco facilitates the availability and accumulation of Fe in young leaves and thus leads to tolerance under Fe deficiency.
Fig. 6.
The response to Fe deficiency in AhNRAMP1-induced tobacco lines. The phenotype of AhNRAMP1-induced lines grown on MS medium (A) or hydroponics (B) without Fe. NT represents non-transformed tobacco. IN is AhNRAMP1-induced tobacco. (C) Fe and (D) Mn concentration in the new leaves of NT and IN tobacco in hydroponics treated under Fe deficiency for 9 d. The results are presented as the means ±SD of triplicate samples. Asterisks indicate a significant difference at P < 0.05 by t-test.
Discussion
SSH is a powerful approach for isolation of differentially expressed genes between two samples (Diatchenko et al., 1996). In the present study the AhNRAMP1 cDNA fragment was obtained by SSH between intercropped and monocropped peanut roots in calcareous soils. It has been clearly elucidated that peanut intercropped with maize enhances the Fe nutrition of peanut in calcareous soils (Zuo et al., 2000; Zuo and Zhang, 2008, 2009). The phytosiderophores released by maize may play a role in improving the Fe nutrition of peanut in the intercropping system (Zuo and Zhang, 2009), and the molecular mechanisms involved in this phytosiderophore-based Fe improvement are being clarified (H. Xiong et al., unpublished results). Since the monocropped peanut suffers from less Fe availability, peanut under monocropping changes the expression of Fe-responsive genes, such as AhIRT1, encoding an Fe-uptake transporter (Ding et al., 2010), and AhFRO1, encoding ferric-chelate reductase (Ding et al., 2009). Therefore, the distinctly different expression levels of AhNRAMP1 between intercropped and monocropped peanut roots lead us to hypothesize that AhNRAMP1 plays a role in Fe nutrition. Moreover, the transcript level of AhNRAMP1 was specifically higher in the roots and increased at least 5-fold after treatment without Fe for 4, 7, and 11 d in the roots (Fig. 2A). In tomato, Arabidopsis, M. baccata, and rice, LeNRAMP1, AtNRAMP1, MbNRAMP1, and OsNRAMP1 genes also show higher expression in the roots under Fe deficiency compared with the Fe-sufficient condition (Curie et al., 2000; Bereczky et al., 2003; Xiao et al., 2008; Takahashi et al., 2011). Therefore, these NRAMP genes are thought to belong to an Fe deficiency-induced subclass, suggesting a possible conserved function in Fe homeostasis.
By clustal analysis, AhNRAMP1 was closer to LeNRAMP1, with 78% identity (Fig. 1). Similar to LeNRAMP1, expression of AhNRAMP1 restored growth of the high- and low- affinity Fe uptake-defective yeast mutant (Fig. 3), suggesting that AhNRAMP1 is also a functional Fe transporter. AhNRAMP1 expression was strongly induced under Fe deficiency, and rapidly decreased to nearly basal levels after 1 d of Fe resupply only in leaves but not in roots (Fig. 2). It has been reported that the expression level of AhIRT1 increased ∼70-fold after exposure to Fe deficiency for 6 d in the roots and was suppressed rapidly by transfer to Fe-sufficient conditions (Ding et al., 2010). Comparing the expression pattern of AhNRAMP1 with that of AhIRT1, AhIRT1 is thought to be more sensitive in response to Fe deficiency and resupply. AhNRAMP1 is a plasma membrane protein (Fig. 4) and is mainly localized in the epidermis of peanut roots, as indicated by LMD and in situ hybridization assay (Fig. 5). LeNRAMP1, on the other hand, is expressed in the vascular parenchyma of the root hair zone and root epidermis and the cortex behind the root tip, and is thought to distribute Fe in the vascular parenchyma upon Fe deficiency (Bereczky et al., 2003). Therefore, in contrast to LeNRAMP1, the localization of AhNRAMP1 in the epidermis suggested that the Fe-regulated transporter AhNRAMP1 might function in Fe acquisition from the soil, in addition to the major Fe uptake system AhIRT1. This speculation is supported by a previous study showing that Arabidopsis AtNRAMP1 partially rescued the growth and Fe content of the irt1 mutant, which is defective in essential Fe uptake (Cailliatte et al., 2010). AhNRAMP1 expression was also induced by Fe starvation in the leaves (Fig. 2B, C), suggesting that AhNRAMP1 might also be involved in Fe distribution in the leaves.
To clarify its physiological function in planta, AhNRAMP1 was introduced into tobacco plants by using an artificial promoter containing IDE1 and IDE2, which are cis-acting elements conferring Fe-deficiency-specific expression in tobacco roots (Kobayashi et al., 2003), fused to the –90/+8 region of the CaMV35S promoter and the 5′ leader (Ω) sequence of Tobacco mosaic virus to enhance the expression level (Kobayashi et al., 2004). The AhNRAMP1-induced tobacco accumulated a higher concentration of Fe in young leaves and was more tolerant to Fe deficiency compared with the NT plants (Fig. 6). It has been shown that the IDE1 and IDE2 elements fused to the –90/+8 region of the 35S promoter drive expression in the whole root tissues under Fe deficiency in tobacco, including the epidermis (Kobayashi et al., 2003). Therefore, it is reasonable to consider that the high expression of AhNRAMP1 in the whole roots of tobacco results in the acquisition of more Fe and thus these plants are resistant to Fe starvation.
Recent studies showed that AtNRAMP1 and OsNRAMP5 transporters play important roles in Mn uptake from the soil (Cailliatte et al., 2010; Ishimaru et al., 2012). In rice, OsNRAMP1 and OsNRAMP5 transporters are also involved in Cd accumulation (Takahashi et al., 2011; Ishimaru et al., 2012). It has been well established that the NRAMP gene family transports a broad range of metal ions. Yeast complementation tests suggested that AtNRAMP1, AtNRAMP3, and AtNRAMP4 transport Fe, Mn, and Cd (Curie et al., 2000; Thomine et al., 2000). AtNRAMP3 and AtNRAMP4 function in mobilization of vacuolar Fe pools and export of vacuolar Mn into photosynthetic tissues of adult plants (Thomine et al., 2003; Lanquar et al., 2005). Hence, it is possible that AhNRAMP1 also transports other metals, such as Mn and Cd.
In conclusion, in the present study novel functions for AhNRAMP1 in peanut are proposed. Several lines of evidence indicate a putative function for AhNRAMP1 in acquisition of Fe, namely: (i) AhNRAMP1 is an Fe transporter and is strongly induced by Fe deficiency in the roots; (ii) AhNRAMP1 is a plasma membrane protein and is mainly localized in the epidermis of peanut roots; and (iii) induced expression of AhNRAMP1 in tobacco results in Fe accumulation and tolerance of Fe deprivation.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Multiple alignments of the amino acid sequences of AhNRAMP1, LeNRAMP1, AtNRAMP1, and OsNRAMP1.
Figure S2 . The expression level of AhNRAMP1 in non-transformant (NT) and the AhNRAMP1-induced (IN) tobacco lines treated under Fe-deficient MS medium for 6 d.
Figure S3 . Zn concentration in new leaves of NT and AhNRAMP1-induced (IN) tobacco lines treated under Fe deficiency for 9 d in hydroponics.
Figure S4 . Fe (A), Mn (B), Zn (C), and Cu (D) concentration in roots of NT and AhNRAMP1-induced (IN) tobacco lines treated under Fe deficiency for 9 d in hydroponics.
Acknowledgments
We thank Dr Rui Proenca (Johns Hopkins University, USA) for critical reading of the manuscript and Dr Ryuichi Takahashi (Graduate School of Agricultural and Life Sciences, The University of Tokyo) for providing the OsNRAMP1 vector. We also thank Dr Satoshi Mori (NPO-WINEP) and Dr Tomoko Nozoye (Graduate School of Agricultural and Life Sciences, The University of Tokyo) for valuable discussion. This work was supported by the National Natural Science Foundation of China (Grant no. 31071840), the PhD Programs Foundation of the Ministry of Education of China (Grant no. 20100008110001), and the Innovative Group Grant of the National Science Foundation of China (Grant no. 31121062).
References
- Belouchi A, Kwan T, Gros P. Cloning and characterization of the OsNramp family from Oryza sativa, a new family of membrane proteins possibly implicated in the transport of metal ions. Plant Molecular Biology. 1997;33:1085–1092. doi: 10.1023/a:1005723304911. [DOI] [PubMed] [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P. Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. Journal of Biological Chemistry. 2003;278:24697–24704. doi: 10.1074/jbc.M301365200. [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Lapeyre B, Briat JF, Mari S, Curie C. The NRAMP6 metal transporter contributes to cadmium toxicity. Biochemical Journal. 2009;422:217–228. doi: 10.1042/BJ20090655. [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. The Plant Cell. 2010;22:904–917. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. The Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochemical Journal. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences, USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Duan L, Li J, Yan H, Zhao M, Zhang F, Li WX. Cloning and functional analysis of the peanut iron transporter AhIRT1 during iron deficiency stress and intercropping with maize. Journal of Plant Physiology. 2010;167:996–1002. doi: 10.1016/j.jplph.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Ding H, Duan L, Wu H, Yang R, Ling H, Li WX, Zhang F. Regulation of AhFRO1, an Fe(III)-chelate reductase of peanut, during iron deficiency stress and intercropping with maize. Physiologia Plantarum. 2009;136:274–283. doi: 10.1111/j.1399-3054.2009.01219.x. [DOI] [PubMed] [Google Scholar]
- Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. Journal of Biological Chemistry. 1994;269:26092–26099. [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences, USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proceedings of the National Academy of Sciences, USA. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MD, Trenor CC, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nature Genetics. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Transforming yeast with DNA. Methods in Molecular and Cellular Biology. 1995;5:255–269. [Google Scholar]
- Guerinot ML, Yi Y. Iron: nutritious, noxious, and not readily available. Plant Physiology. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Helmer G, Casadaban M, Bevan M, Kayes L, Chilton MD. A new chimeric gene as a marker for plant transformation: the expression of Escherichia coli [beta]-galactosidase in sunflower and tobacco cells. Nature Biotechnology. 1984;2:520–527. [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Inal A, Gunes A, Zhang F, Cakmak I. Peanut/maize intercropping induced changes in rhizosphere and nutrient concentrations in shoots. Plant Physiology and Biochemistry. 2007;45:350–356. doi: 10.1016/j.plaphy.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. Journal of Biological Chemistry. 2009;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. OsZIP4, a novel zinc-regulated zinc transporter in rice. Journal of Experimental Botany. 2005;56:3207–3214. doi: 10.1093/jxb/eri317. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, et al. Rice plants take up iron as an Fe3+–phytosiderophore and as Fe2+ . The Plant Journal. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Takahashi R, Bashir K, et al. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Scientific Reports. 2012;2:286. doi: 10.1038/srep00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA. The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. The Plant Journal. 2003;35:295–304. doi: 10.1046/j.1365-313x.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nakayama Y, Itai RN, Nakanishi H, Yoshihara T, Mori S, Nishizawa NK. Identification of novel cis-acting elements, IDE1 and IDE2, of the barley IDS2 gene promoter conferring iron-deficiency-inducible, root-specific expression in heterogeneous tobacco plants. The Plant Journal. 2003;36:780–793. doi: 10.1046/j.1365-313x.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nakayama Y, Takahashi M, Inoue H, Nakanishi H, Yoshihara T, Mori S, Nishizawa NK. Construction of artificial promoters highly responsive to iron deficiency. Soil Science and Plant Nutrition. 2004;50:1167–1175. [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Molecular Biology. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelievre F, Bolte S, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. The EMBO Journal. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Ramos MS, Lelièvre F, Barbier-Brygoo H, Krieger-Liszkay A, Krämer U, Thomine S. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiology. 2010;152:1986–1999. doi: 10.1104/pp.109.150946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chiecko JC, Kim SA, Walker EL, Lee Y, Guerinot ML, An G. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiology. 2009;150:786–800. doi: 10.1104/pp.109.135418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dang P, Bausher MG, Holbrook CC, Lee RD, Lynch RE, Guo BZ. Identification of transcripts involved in resistance responses to leaf spot disease caused by Cercosporidium personatum in peanut (Arachis hypogaea) Phytopathology. 2005;95:381–387. doi: 10.1094/PHYTO-95-0381. [DOI] [PubMed] [Google Scholar]
- Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MF. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Molecular Microbiology. 2000;35:1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- Mizuno D, Higuchi K, Sakamoto T, Nakanishi H, Mori S, Nishizawa NK. Three nicotianamine synthase genes isolated from maize are differentially regulated by iron nutritional status. Plant Physiology. 2003;132:1989–1997. doi: 10.1104/pp.102.019869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Usui K, Horie K, Nosaka S, Mizuno N, Obata H. Cloning of three ZIP/Nramp transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni2+-transport abilities. Plant Physiology and Biochemistry. 2005;43:793–801. doi: 10.1016/j.plaphy.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T. A specific transporter for iron(III)–phytosiderophore in barley roots. The Plant Journal. 2006;46:563–572. doi: 10.1111/j.1365-313X.2006.02714.x. [DOI] [PubMed] [Google Scholar]
- Nevo Y, Nelson N. The NRAMP family of metal-ion transporters. Biochimica et Biophysica Acta. 2006;1763:609–620. doi: 10.1016/j.bbamcr.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Oomen RJFJ, Wu J, Lelièvre F, Blanchet S, Richaud P, Barbier-Brygoo H, Aarts MGM, Thomine S. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytologist. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- Portnoy ME, Liu XF, Culotta VC. Saccharomyces cerevisiae expresses three functionally distinct homologues of the Nramp family of metal transporters. Molecular and Cellular Biology. 2000;20:7893–7902. doi: 10.1128/mcb.20.21.7893-7902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Eide DJ, Guerinot ML. Altered selectivity in an Arabidopsis metal transporter. Proceedings of the National Academy of Sciences, USA. 2000;97:12356–12360. doi: 10.1073/pnas.210214197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiology. 1986;80:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G, Schikora A, Häberle J, Vert G, Ludewig U, Briat JF, Curie C, von Wirén N. A putative function for the Arabidopsis Fe–phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant and Cell Physiology. 2005;46:762–774. doi: 10.1093/pcp/pci081. [DOI] [PubMed] [Google Scholar]
- Takagi S. Naturally occurring iron-chelating compounds in oat- and rice-root washing. I. Activity measurement and preliminary characterization. Soil Science and Plant Nutrition. 1976;22:423–433. [Google Scholar]
- Takahashi H, Kamakura H, Sato Y, Shiono K, Abiko T, Tsutsumi N, Nagamura Y, Nishizawa N, Nakazono M. A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. Journal of Plant Research. 2010;123:807–813. doi: 10.1007/s10265-010-0319-4. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. Journal of Experimental Botany. 2011;62:4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Lelièvre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. The Plant Journal. 2003;34:685–695. doi: 10.1046/j.1365-313x.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proceedings of the National Academy of Sciences, USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S, Kamiya T, Sakamoto T, Kasai K, Sato Y, Nagamura Y, Yoshida A, Kyozuka J, Ishikawa S, Fujiwara T. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proceedings of the National Academy of Sciences, USA. 2011;108:20959–20964. doi: 10.1073/pnas.1116531109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. The Plant Journal. 2002;31:589–599. doi: 10.1046/j.1365-313x.2002.01381.x. [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. The Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Chai T, Zhang Y, Han L, Xu J, Guan Z. The Thlaspi caerulescens NRAMP homologue TcNRAMP3 is capable of divalent cation transport. Molecular Biotechnology. 2009;41:15–21. doi: 10.1007/s12033-008-9088-x. [DOI] [PubMed] [Google Scholar]
- Williams LE, Pittman JK, Hall JL. Emerging mechanisms for heavy metal transport in plants. Biochimica et Biophysica Acta. 2000;1465:104–126. doi: 10.1016/s0005-2736(00)00133-4. [DOI] [PubMed] [Google Scholar]
- Xia J, Yamaji N, Kasai T, Ma JF. Plasma membrane-localized transporter for aluminum in rice. Proceedings of the National Academy of Sciences, USA. 2010;107:18381–18385. doi: 10.1073/pnas.1004949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Yin L, Xu X, Li T, Han Z. The iron-regulated transporter, MbNRAMP1, isolated from Malus baccata is involved in Fe, Mn and Cd trafficking. Annals of Botany. 2008;102:881–889. doi: 10.1093/aob/mcn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T, Hodoshima H, Miyano Y, Shoji K, Shimada H, Goto F. Cadmium inducible Fe deficiency responses observed from macro and molecular views in tobacco plants. Plant Cell Reports. 2006;25:365–373. doi: 10.1007/s00299-005-0092-3. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Zhang F. Effect of peanut mixed cropping with gramineous species on micronutrient concentrations and iron chlorosis of peanut plants grown in a calcareous soil. Plant and Soil. 2008;306:23–36. [Google Scholar]
- Zuo Y, Zhang F. Iron and zinc biofortification strategies in dicot plants by intercropping with gramineous species. A review. Agronomy for Sustainable Development. 2009;29:63–71. [Google Scholar]
- Zuo Y, Zhang F, Li X, Cao Y. Studies on the improvement in iron nutrition of peanut by intercropping with maize on a calcareous soil. Plant and Soil. 2000;220:13–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.