Abstract
The role of IκB kinase (IKK)-induced proteolysis of NF-κB1 p105 in innate immune signaling was investigated using macrophages from Nfkb1SSAA/SSAA mice, in which the IKK target serines on p105 are mutated to alanines. We found that the IKK/p105 signaling pathway was essential for TPL-2 kinase activation of extracellular signal-regulated kinase (ERK) mitogen-activate protein (MAP) kinase and modulated the activation of NF-κB. The Nfkb1SSAA mutation prevented the agonist-induced release of TPL-2 from its inhibitor p105, which blocked activation of ERK by lipopolysaccharide (LPS), tumor necrosis factor (TNF), CpG, tripalmitoyl-Cys-Ser-Lys (Pam3CSK), poly(I · C), flagellin, and R848. The Nfkb1SSAA mutation also prevented LPS-induced processing of p105 to p50 and reduced p50 levels, in addition to decreasing the nuclear translocation of RelA and cRel. Reduced p50 in Nfkb1SSAA/SSAA macrophages significantly decreased LPS induction of the IκBζ-regulated Il6 and Csf2 genes. LPS upregulation of Il12a and Il12b mRNAs was also impaired although specific blockade of TPL-2 signaling increased expression of these genes at late time points. Activation of TPL-2/ERK signaling by IKK-induced p105 proteolysis, therefore, induced a negative feedback loop to downregulate NF-κB-dependent expression of the proinflammatory cytokine interleukin-12 (IL-12). Unexpectedly, TPL-2 promoted soluble TNF production independently of IKK-induced p105 phosphorylation and its ability to activate ERK, which has important implications for the development of anti-inflammatory drugs targeting TPL-2.
INTRODUCTION
Following infection, macrophages are rapidly activated by pathogen-associated molecules, such as lipopolysaccharide (LPS), a cell wall component of Gram-negative bacteria that binds to Toll-like receptor 4 (TLR4) (23). The transcription of several hundred genes is induced following LPS stimulation of macrophages, the gene products of which regulate multiple aspects of the inflammatory and innate immune responses, including cell migration, phagocytosis, antimicrobial defense, tissue remodeling, and the adaptive immune response (28, 39). LPS induction of these genes involves activation of nuclear factor κB (NF-κB) transcription factors, interferon-regulatory factors, and each of the major mitogen-activated protein (MAP) kinase subtypes (extracellular signal-regulated kinases 1 and 2 [ERK1/2], Jun amino-terminal kinases [JNKs], and p38α/β) (23).
NF-κB transcription factors are composed of homo- and heterodimeric complexes of Rel proteins (16). This family of proteins, which is characterized by the presence of an N-terminal Rel homology domain (RHD), comprises NF-κB1 p50, NF-κB2 p52, RelA (also called p65), cRel, and RelB. RelA, cRel, and RelB are translated in their mature forms and contain C-terminal transcription activation domains. In contrast, NF-κB1 and NF-κB2 are synthesized as large precursors of 105-kDa (p105) and 100-kDa (p100) proteins, respectively (5). These are partially proteolysed (processed) by the proteasome to produce smaller, mature forms which lack transactivation domains and promote transcription when complexed with transactivating Rel subunits or other nuclear proteins (42). For example, p50 homodimers associated with IκBζ induce the transcription of a subset of NF-κB target genes in LPS-stimulated macrophages (47).
NF-κB dimers are held in the cytoplasm of unstimulated cells through their interaction with a family of inhibitory proteins (termed IκBs), which includes IκBα, IκBβ, and IκBε (16). In response to stimulation with agonists such as LPS, IκBα is phosphorylated by the IκB kinase (IKK) complex, which is composed of IKK1 (IKKα) and IKK2 (IKKβ) kinase subunits and the ubiquitin-binding adaptor NEMO (IKKγ). This phosphorylation creates a binding site for the ubiquitin E3 ligase SCFβ-TrCP, which catalyzes IκBα K48-linked polyubiquitination and targets IκBα for degradation by the proteasome, releasing associated p50-RelA and p50-cRel heterodimers to translocate into the nucleus. Proteolysis of IκBβ and IκBε is controlled in a similar fashion (22).
NF-κB1 p105 also functions as a cytoplasmic IκB, binding to preformed NF-κB dimers via its C-terminal ankyrin repeat region and to Rel monomers via its N-terminal RHD (37). Following LPS stimulation, the IKK complex phosphorylates p105 on serine residues 927 and 932 (17, 26, 30). This creates a binding site for SCFβ-TrCP, which catalyzes the subsequent K48-linked polyubiquitination of p105, triggering its proteolysis by the proteasome. NF-κB1 p105 is also a precursor for p50, which is produced constitutively from p105 by partial proteolysis (processing) by the proteasome. Constitutive processing of NF-κB1 p105 to p50 requires the monoubiquitination of p105 on multiple lysine residues (25). While in vitro experiments with recombinant p105 have suggested that IKK can induce p105 processing to p50 (30), cellular analyses have indicated that IKK promotes the complete degradation of p105 (17, 26). Both outcomes potentially allow p105 to regulate expression of NF-κB target genes.
In addition to its role in regulating the transcriptional activity of NF-κB, p105 has been shown to be a major regulator of ERK MAP kinase activity in innate immune responses. NF-κB1 p105 forms a high-affinity, stoichiometric complex with tumor progression locus 2 (TPL-2) (4, 7), a member of the MAP-3 kinase family of proteins. TPL-2 functions as a MEK kinase, which mediates TLR activation of ERK MAP kinase in macrophages (13, 14). TPL-2 plays an important regulatory role in innate immune responses and is essential for LPS induction of tumor necrosis factor (TNF), interleukin-1β (IL-1β), and cyclo-oxygenase 2 (COX-2) in macrophages (10, 11, 29). Binding to p105 maintains steady-state levels of TPL-2 protein, and LPS stimulation fails to activate ERK in Nfkb1−/− macrophages due to the virtual absence of TPL-2 (46).
Association with NF-κB1 p105 also negatively regulates TPL-2 activation of ERK by preventing TPL-2 phosphorylation of MEK (4, 46). LPS activation of TPL-2 MEK kinase activity in primary macrophages requires TPL-2 release from p105 (6, 45). Pharmacological experiments have indicated that proteasome and IKK2 activity are essential for LPS activation of TPL-2/ERK signaling. Analysis of RAW264.7 macrophages stably expressing wild-type (WT) FLAG-tagged p105 (FL-p105) or FL-p105S927A,S932A (FL-p105SSAA) (lacking IKK phosphorylation sites in human p105) has suggested that maximal LPS activation of TPL-2 MEK kinase activity and release from p105 require IKK phosphorylation of p105. Consistent with this, retroviral overexpression of FL-p105S927A,S932A in Nfkb1−/− macrophages cannot rescue LPS activation of ERK, in contrast to WT FL-p105 (6). Together, these results suggested a role for IKK phosphorylation of p105 in regulating TPL-2 activation (6, 45). However, the physiological significance of these data has remained unclear since the experiments involved overexpression of epitope-tagged p105 proteins.
The potential role for NF-κB1 p105 to regulate both NF-κB activity and TPL-2/ERK signaling suggests that p105 is a major effector protein through which the IKK complex regulates immune responses. However, the consequences of this regulation are largely unknown. To investigate this, we previously generated Nfkb1SSAA/SSAA knock-in mice (40), which express mutant p105SSAA (S930A and S935A mutations in mouse p105) that cannot be phosphorylated by IKK. In the present study, we used these mice to investigate the function of IKK-induced NF-κB1 p105 proteolysis in NF-κB and TPL-2/ERK activation in macrophages during an innate immune response. The Nfkb1SSAA mutation was found to reduce LPS activation of NF-κB, both by inhibiting processing of p105 to p50 and by p105SSAA-mediated retention of Rel subunits in the cytoplasm. Consequently, LPS induction of a panel of NF-κB target genes was reduced in Nfkb1SSAA/SSAA macrophages compared to induction in WT cells, with IκBζ-p50-regulated genes being particularly affected. The Nfkb1SSAA mutation also completely blocked LPS-induced TPL-2 release from p105 and TPL-2 activation of MEK and ERK MAP kinase, providing definitive genetic evidence that TLR4 activation of TPL-2/ERK signaling is dependent on IKK-induced p105 proteolysis. Surprisingly, the Nfkb1SSAA mutation did not affect TPL-2-dependent induction of soluble TNF by LPS (33). Therefore, TPL-2 complexed with p105 stimulated the production of soluble TNF independently of ERK activation, which has important implications for the development of anti-inflammatory drugs targeting TPL-2.
MATERIALS AND METHODS
Mouse strains.
Mouse strains were bred in a specific-pathogen-free environment at the National Institute for Medical Research (NIMR; London, United Kingdom), and all experiments were done in accordance with regulations of the Home Office of the United Kingdom. Nfkb1SSAA/SSAA, Nfkb1−/−, and Map3k8−/− mouse strains have been described previously (10, 31, 38, 40) and were all fully backcrossed on to a C57BL/6 background. Nfkb1SSAA/− and Nfkb1SSAA/SSAA Map3k8−/− mice were generated by intercrossing these strains. The generation of Map3k8D270A/D270A mice, which express catalytically inactive TPL-2, will be described in a separate study. These were intercrossed with Nfkb1SSAA/SSAA mice to generate the Nfkb1SSAA/SSAA Map3k8D270A/D270A mouse strain. Mice homozygous for deletion of the C terminus coding sequence in Nfkb1 (Nfkb1ΔCT/ΔCT) (19) were kindly provided by Bristol Meyers Squibb. After being backcrossed eight times onto a C57BL/6 background, Nfkb1ΔCT/ΔCT mice were crossed with Nfkb1SSAA/SSAA mice to generate Nfkb1SSAA/ΔCT experimental mice.
Antibodies.
The p105C antibody was used for immunoprecipitation of p105 (35), while anti-p50 (Delta Biolabs) was used for immunoblotting NF-κB1 p50/p105. The ABIN-2 antibody used for detection of ABIN-2 on immunoblots and the 70-mer TPL-2 antibody used for immunoprecipitation of TPL-2 have been described previously (6, 27). Antibodies against MEK-1 and MEK-2 (MEK-1/2), MEK-1/2 phosphorylated at S217 and S221 [phospho(S217/S221)-MEK-1/2] activated MEK-1/2; phospho-MEK), p38, phospho(T180/Y182)-p38 (activated p38; phospho-p38), JNK, and phospho(T183/Y185)-JNK (phospho-JNK) were purchased from Cell Signaling Technology. Antibodies to TPL-2 (M20; H-7), ERK-1 and ERK-2, Egr-1, and IκBα were obtained from Santa Cruz, and phospho(T185/Y187)-ERK1/2 (activated ERK; phospho-ERK) antibody was from Biosource. Antibodies to Rel family proteins were purchased from Cell Signaling Technology while anti-TNF was purchased from R&D Systems. Tubulin was detected with TAT-1 α-tubulin monoclonal antibody (MAb; kindly provided by Keith Gull, University of Oxford, United Kingdom) and used as a loading control protein for immunoblotting of total cell lysates. Glutathione S-transferase (GST)–MEKK207A was generously donated by Richard Marais (Cancer Research UK, London, United Kingdom). IL-10R antibody and control IgG were kindly provided by Anne O'Garra (NIMR, London, United Kingdom).
In vitro generation and stimulation of macrophages.
Bone marrow-derived macrophages (BMDM) were prepared as described previously (44). Briefly, bone marrow cells were plated in 10 ml of complete BMDM medium (RPMI 1640 medium [Sigma] supplemented with 10% fetal bovine serum [FBS], antibiotics, 20% L-cell conditioned medium, and 50 μM β-mercaptoethanol) at 5 × 106 cells per 90-mm bacterial petri dish (Sterilin). After 4 days of culture, 10 ml of complete BMDM medium was added, and cells were cultured a further 3 days. Nonadherent cells were aspirated, and remaining adherent cells were harvested by incubation with 5 ml of phosphate-buffered saline (PBS) supplemented with 5% FBS and 2.5 mM EDTA. Flow cytometric analysis indicated that >95% of these cells were positive for the macrophage markers F4/80 and CD11b. The levels of expression of F4/80, CD11b, major histocompatibility complex (MHC) class II, CD86, and TLR4/MD2 were similar between macrophages generated from WT, Nfkb1SSAA/SSAA, Nfkb1SSAA/SSAA Map3k8−/−, and Nfkb1SSAA/SSAA Map3k8D270A/D270A mice (data not shown). Resident peritoneal macrophages were obtained by intraperitoneal injection of mice with 4 ml of RPMI medium supplemented with 10% FBS and aspiration. Macrophages were purified from the resulting cell suspension by adherence after 3 h of culture at 37°C and removal of nonadherent cells by washing with PBS. The cell populations produced were >95% F4/80+, as determined by flow cytometry (results not shown).
For experiments, harvested BMDM were replated in Nunc tissue culture dishes (six-well plates, 1 × 106 cells/well; 60-mm dishes, 3 × 106 cells; 90-mm dishes, 8 × 106 cells) in BMDM medium plus 1% FBS minus L-cell supplement. Peritoneal macrophages were replated in six-well plates (1 × 106 cells/well) and cultured in RPMI medium supplemented with 1% FBS. BMDM and peritoneal macrophages were cultured overnight prior to stimulation with LPS (Salmonella enterica serovar Minnesota R595; Alexis Biochemicals) at a final concentration of 10 ng/ml, unless otherwise indicated. Cells were stimulated with TNF (R&D Systems) at 20 ng/ml, with CpG (InvivoGen) at 500 nM, with tripalmitoyl-Cys-Ser-Lys (Pam3CSK; InvivoGen) at 1 μg/ml, with poly(I · C) (InvivoGen) at 10 μg/ml, with flagellin (InvivoGen) at 1 μg/ml, with R848 (InvivoGen) at 1 μg/ml, and with phorbol myristate acetate (PMA; Sigma) at 10 ng/ml. Control cells were left untreated (time zero). For experiments in which ERK activation was blocked pharmacologically, cells were preincubated with 2 μM PD184352 for 1 h prior to LPS stimulation, unless specified otherwise in figure legends.
Protein analyses.
Cells were washed once in PBS before lysis. For immunoblotting of total lysates, cells (3 × 106 cells per point) were lysed in buffer A (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 1 mM Na3VO4, 100 nM okadaic acid [Calbiochem], and 2 mM Na4P2O7 plus protease inhibitors [Roche Molecular Biochemicals]) containing 1% Nonidet-P40 or in radioimmunoprecipitation assay (RIPA) buffer, which additionally contained 0.5% deoxycholate and 0.1% SDS. After centrifugation to remove particulate matter, lysates were mixed with an equal volume of 2× SDS-PAGE sample buffer and then subjected to immunoblotting. For immunoprecipitations, lysis (2 × 107 to 4 × 107 cells per point) was carried out using buffer B (10 mM HEPES, pH 7.6, 1 mM EGTA, 10 mM KCl, 1 mM dithiothreitol [DTT], 20 mM NaF, and 1 mM Na4P2O7 plus a mixture of protease inhibitors [Roche Molecular Biochemicals]) containing 0.5% Nonidet-P40. Covalent coupling of antibodies to protein A-Sepharose (Amersham Biosciences) was performed as described previously (20). To detect release of TPL-2 from p105, BMDM lysates were precleared of p105 by immunodepletion with p105C antibody coupled to protein A-Sepharose (6). Complete removal of p105 from lysates was confirmed by immunoblotting.
TPL-2 MEK kinase activity was assayed using a modification of a previously published method (32). A total of 16 × 106 cells per point were either stimulated for 15 min with LPS (100 ng/ml) or left unstimulated and then lysed using kinase assay lysis buffer (buffer A containing 0.5% NP-40, 5 mM β-glycerophosphate, 1 mM DTT). Immunoprecipitation was carried out using a 1:1 mixture of 70-mer and M20 (Santa Cruz) TPL-2 antisera. Immunoprecipitates were washed four times in kinase assay lysis buffer, followed by two washes in kinase buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM β-glycerophosphate, 100 nM okadaic acid, 1 mM dithiothreitol, 0.1 mM sodium vanadate, 10 mM MgCl2, 1 mM EGTA, 0.03% Brij 35). Beads were then resuspended in 50 μl of kinase buffer plus 1 mM ATP and 1 μg of GST-MEK1K207A. Reaction mixtures were incubated at 30°C for 30 min and terminated by the addition of 50 μl of 2× SDS sample buffer. Phosphorylation of GST-MEK1K207A was assessed by immunoblotting using phospho-MEK antibody (166F8; Cell Signaling). To measure p105-associated TPL-2 kinase activity (2, 32), p105 was immunoprecipitated from cell lysates with p105C antibody coupled to protein A-Sepharose for 3 h. After an extensive washing step, beads were resuspended in 50 μl of kinase buffer plus 0.1 mM ATP and 2.5μCi of [γ-32P]ATP (Amersham Biosciences) and incubated for 30 min at 30°C. Labeled protein was visualized by autoradiography after 10% SDS-PAGE.
To analyze p105 processing, BMDM (1 × 106 cells per well; Nunc 12-well plate) were cultured in complete medium for 18 h. Cells were then washed with methionine- and cysteine-free minimal essential Eagle's medium (Sigma) supplemented with 1% dialyzed FBS and cultured in the same medium for 60 min to starve. Cells were pulse-labeled by addition of 100 μCi/ml of [35S]methionine-cysteine (EasyTag Express Protein Labeling Mix; PerkinElmer) and culture for 60 min. Labeling was stopped by the addition of complete medium (RPMI medium plus 10% FCS), supplemented with LPS (100 ng/ml) to activate IKK. Triplicate cultures of cells were lysed in RIPA buffer, pooled, and precleared three times with protein A-Sepharose beads. p105 was immunoprecipitated with p50 antibody (1263-TB4) obtained from the Biological Resources Branch of the National Cancer Institute (Maryland). After an extensive washing step in RIPA buffer, isolated proteins were eluted in sample buffer and resolved by SDS-PAGE. Labeled bands were visualized by autoradiography.
To analyze nuclear translocation of Rel subunits, cells were washed in PBS and then lysed on ice in a buffer containing 0.2% NP-40, 10 mM HEPES, pH 7.6, 0.1 mM EGTA, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 20 mM NaF, 1 mM Na-pyrophosphate, and protease inhibitors. After 2 min, nuclei were pelleted by centrifugation, samples were washed twice in lysis buffer, and nuclear proteins were extracted with RIPA buffer. Rel proteins in extracts were quantified by immunoblotting and densitometry. Data were normalized against SAM68 content of nuclear extracts, determined by immunoblotting with anti-SAM68 (Santa Cruz).
NF-κB activation assays.
For enzyme-linked immunosorbent assay (ELISA) of NF-κB activity, nuclear extracts were prepared using a commercial kit (5 × 106 cells/point; Active Motif). For greater sensitivity, cRel binding was assayed with anti-c-Rel (sc-70X; Santa Cruz). Five micrograms of extract per point was then assayed in duplicate, using a TransAm NF-κB family kit (Active Motif). Data were normalized against the SAM68 content of nuclear extracts.
qRT-PCR.
Macrophages were plated in six-well plates (Nunc) at 5 × 105/ml in 2 ml of RPMI 1640 medium supplemented with 1% FBS, 50 μM 2-mercaptoethanol (2-ME), and antibiotics. After overnight culture, cells were stimulated for the times indicated in the figures with 10 ng/ml LPS. RNA from stimulated and unstimulated cells was isolated using an RNeasy kit, and contaminating DNA was removed using an RNase-free DNase set (Qiagen, Hilden, Germany), according to the manufacturer's instructions, and DNase treated (Roche, East Sussex, United Kingdom). cDNA was produced using a SuperScript III First-Strand Synthesis SuperMix for quantitative real-time PCR (qRT-PCR) (Life Technologies) and standard protocols. Expression of target genes was determined by real-time PCR using a Perkin Elmer ABI Prism 7000 Sequence Detection System. Commercial 6-carboxyfluorescein (FAM)-labeled probes (Applied Biosystems) were used with a TaqMan Gene Expression Master Mix (Applied Biosystems). Target gene mRNA levels were normalized against Hprt mRNA levels.
ELISA of cytokines.
Macrophages were plated either at 5 × 105/ml in 250 μl of RPMI 1640 medium supplemented with 1% FBS and antibiotics in a 96-well plate (Nunc) or at 1 × 106 cells in 1 to 2 ml of RPMI 1640 medium in six-well plates (Nunc). After overnight culture, cells were stimulated with 10 ng/ml LPS for the times indicated in the figures. Levels of TNF and IL-12 p40 in culture supernatants were determined using commercial enzyme-linked immunosorbent assay (ELISA) kits (eBioscience), according to the manufacturer's instructions.
To measure cytokine production in vivo, 6- to 8-week-old mice were injected intraperitoneally with 100 μg of LPS/mouse. Mice were sacrificed after 1 h for TNF and after 6 h for IL-12 p40 determinations. Blood was collected by cardiac puncture and allowed to clot at 4°C for 2 h. Resulting sera were stored at −20°C until cytokine assay by ELISA.
Flow cytometric analyses.
For analysis of ERK phosphorylation, peritoneal macrophages were aspirated 10 min after LPS injection (200 μg per mouse) and fixed in an equal volume of Intracellular (IC) Fixation Buffer (eBioscience) for 10 min on ice. Cells were then stained with F4/80-allophycocyanin (APC) (BD Pharmingen), permeabilized with 90% methanol, and sequentially stained with phospho-ERK antibody (4377; Signal Transduction Laboratories) and anti-rabbit Ig–fluorescein isothiocyanate (FITC). Labeled cells were detected by flow cytometry on a FACSCalibur (BD) by standard methods, using FlowJo software (Treestar). Surface pre-TNF was analyzed as previously described (33).
Statistical analysis.
All data analyses were performed using GraphPad software (GraphPad Software, Inc., San Diego, CA). Data were compared using Student's t test (two-tailed and unpaired test). P values of less than 0.05 or 0.005 were considered significant. Error bars represent standard errors of the means (SEM).
RESULTS
The Nfkb1SSAA mutation reduces TLR4 activation of NF-κB.
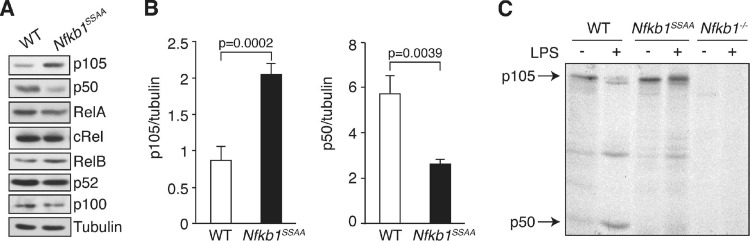
NF-κB p105 is the precursor of p50 and also functions as an IκB (5), binding to NF-κB dimers and Rel monomers via its ankyrin repeat region and RHD, respectively (37). Nfkb1SSAA/SSAA bone marrow-derived macrophages (BMDM) were used to investigate the extent to which IKK-induced p105 proteolysis contributes to total NF-κB activity in macrophages. Immunoblotting demonstrated that steady-state expression levels of RelA, cRel, RelB, and NF-κB2 p100/p52 were similar between wild-type (WT) and Nfkb1SSAA/SSAA BMDM (Fig. 1A). Surprisingly, p50 levels were significantly reduced in Nfkb1SSAA/SSAA BMDM even though steady-state levels of the p105 precursor were elevated (Fig. 1A and B). A similar phenotype was detected in Nfkb1SSAA/SSAA peritoneal macrophages (data not shown).
Fig 1.
Nfkb1SSAA mutation reduces LPS-induced p105 processing to p50. (A) Lysates from BMDM were immunoblotted. The Nfkb1SSAA/SSAA genotype is shown in this figure and in subsequent figures as Nfkb1SSAA. (B) p105 and p50 levels in BMDM were assayed by immunoblotting. Bands were quantified by laser densitometry and are expressed relative to tubulin loading control (error bars, SEM). (C) BMDM were metabolically pulse-labeled for 1 h with [35S]methionine-cysteine (−) and then chased for 1 h with nonradioactive medium containing 100 ng/ml LPS (+). p105/p50 were immunoprecipitated from cell lysates with p105N antibody and resolved by SDS-PAGE, and labeled bands were detected by autoradiography. In panels A and C, results are representative of three independent experiments. In panel B, data were pooled from five independent experiments.
Pulse-chase metabolic labeling of BMDM with [35S]methionine-cysteine was used to investigate the effect of the Nfkb1SSAA mutation on p105 processing to p50. WT p105 and p105SSAA were synthesized at similar rates during the 1-h pulse. In WT cells, addition of LPS in the chase promoted a reduction in p105 levels and the appearance of p50. The Nfkb1SSAA mutation blocked both LPS-induced p105 proteolysis and p50 production (Fig. 1C). IKK phosphorylation of p105, therefore, induced the processing of p105 to p50 in LPS-stimulated macrophages. These data implied that low levels of IKK activity induced p105 processing to p50 in unstimulated WT BMDM. However, this was impaired in unstimulated Nfkb1SSAA/SSAA BMDM, resulting in reduced steady-state levels of p50 and elevated p105.
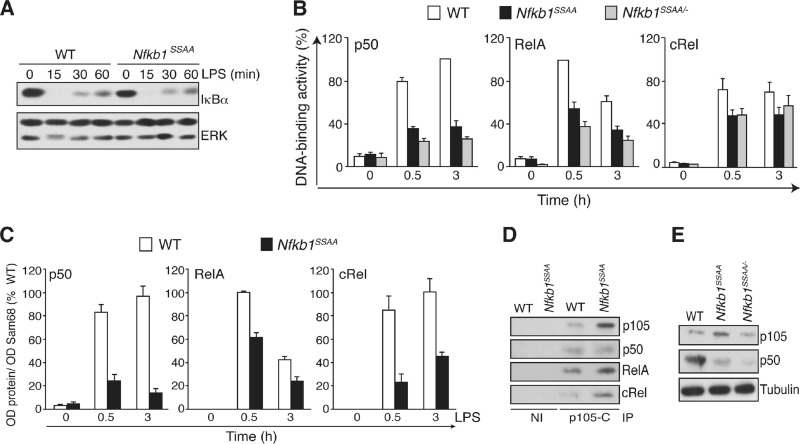
LPS-induced IκBα degradation in Nfkb1SSAA/SSAA BMDM was similar in extent to that in WT cells (Fig. 2A). Gel filtration of BMDM lysates also showed that the gross distribution of Rel subunits between low-molecular-weight (MW) fractions containing IκBα and high-MW fractions containing p105 (37) was not affected by the Nfkb1SSAA mutation (data not shown). However, an NF-κB ELISA demonstrated that LPS activation of p50 was substantially reduced in Nfkb1SSAA/SSAA BMDM while activation of RelA and cRel was decreased to a lesser extent (Fig. 2B). Subcellular fractionation revealed that LPS-induced nuclear translocation of p50, RelA, and cRel was significantly reduced in Nfkb1SSAA/SSAA BMDM compared to levels in WT cells (Fig. 2C). Anti-p105 immunoprecipitates from WT BMDM lysates copurified with p50, RelA, and cRel, and the amounts of RelA and cRel in complex with p105 in Nfkb1SSAA/SSAA BMDM were higher than in WT cells due to the increased steady-state levels of p105 (Fig. 2D). The Nfkb1SSAA mutation, therefore, impaired LPS activation of NF-κB by retention of Rel subunits in the cytoplasm, in addition to reduction of p50 production.
Fig 2.
Nfkb1SSAA mutation impairs LPS activation of NF-κB. (A) BMDM were stimulated for the indicated times with LPS. Cell lysates were immunoblotted. (B) BMDM, cultured in triplicate, were stimulated with LPS (1 μg/ml) for 0.5 and 3 h. Binding of p50, RelA, and cRel to an NF-κB oligonucleotide was determined by ELISA (error bars, SEM). (C) BMDM were cultured in triplicate and stimulated with LPS (1 μg/ml). The amounts of the indicated Rel subunits in nuclear fractions were determined by immunoblotting and densitometry (error bars, SEM). (D) BMDM lysates were immunoprecipitated (IP) with p105C antiserum or nonimmune (NI) serum. Isolated proteins were immunoblotted. (E) Lysates from BMDM were immunoblotted. In panels A, D, and E, results are representative of at least three independent experiments. In panels B and C, results were pooled from five independent experiments.
It was possible that the effect of the Nfkb1SSAA mutation on nuclear translocation of RelA and cRel was primarily due to the increased steady-state levels of p105 compared to levels in WT control cells rather than to acute blockade of IKK-induced p105 proteolysis (Fig. 1A and B). To investigate this, Nfkb1SSAA/SSAA mice were crossed with Nfkb1−/− mice. BMDM generated from the resulting Nfkb1SSAA/− mice expressed levels of p105 comparable to those of WT cells (Fig. 2E). LPS activation of p50 in Nfkb1SSAA/− BMDM was reduced compared to WT BMDM levels (Fig. 2B), at least partly due to the lower steady-state amounts of p50 in Nfkb1SSAA/− BMDM (Fig. 2E). LPS-induced RelA and cRel binding activities were also decreased in Nfkb1SSAA/− BMDM compared to WT BMDM levels (Fig. 2B). Therefore, the inhibitory effect of the Nfkb1SSAA mutation on the activation of RelA and cRel did not result from p105SSAA overexpression.
The Nfkb1SSAA mutation reduces LPS-induced expression of p50-dependent NF-κB target genes.
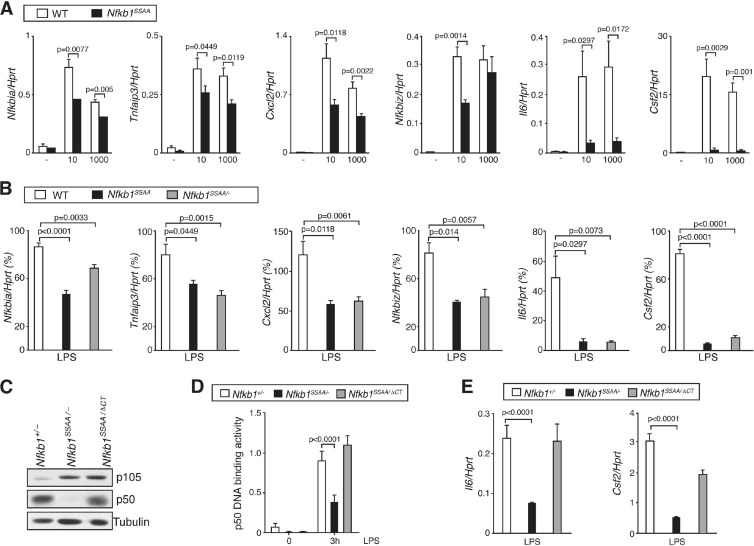
Since the Nfkb1SSAA mutation reduced NF-κB activation following LPS stimulation, it was expected that expression of NF-κB target genes would also be affected. To investigate this, Nfkb1SSAA/SSAA and WT BMDM were stimulated with LPS at two different concentrations, and mRNA levels of selected NF-κB target genes were measured by quantitative real-time PCR (qRT-PCR). Upregulation of Nfkbia (encoding IκBα), Tnfaip3 (encoding A20), and Cxcl2 mRNAs was partially reduced by the Nfkb1SSAA mutation, while Nfkbiz (encoding IκBζ) mRNA was only decreased at the lower LPS dose (Fig. 3A). In contrast, LPS upregulation of Il6 and Csf2 mRNAs, which are dependent on IκBζ and p50 for their induction (47), was essentially blocked by the Nfkb1SSAA mutation at both LPS doses. Experiments with Map3k8−/− BMDM, which do not express TPL-2 (10), revealed that LPS-induced mRNA expression of all of the genes analyzed was unaffected by TPL-2 deficiency (data not shown). Analysis of Nfkb1SSAA/− BMDM revealed that the inhibitory effects of the Nfkb1SSAA mutation on NF-κB target gene expression were not due to overexpression of p105SSAA (Fig. 3B). Together, these results suggested that IKK-induced p105 proteolysis was required for maximal expression of NF-κB-dependent genes in macrophages and was particularly important for expression of genes regulated by IκBζ-p50.
Fig 3.
Nfkb1SSAA mutation reduces LPS-induced expression of NF-κB-dependent genes. (A) Triplicate cultures of BMDM were stimulated for 1 h with LPS at 10 or 1000 ng/ml. Expression of the mRNAs shown was determined by quantitative RT-PCR (error bars, SEM). (B) BMDM were stimulated for 1 h with LPS at 10 ng/ml and analyzed as described in panel A. (C) Lysates of BMDM were immunoblotted. (D) BMDM were stimulated with LPS (1 μg/ml) for 3 h. Binding of p50 to an NF-κB oligonucleotide was determined by ELISA (error bars, SEM). (E) Expression of the mRNAs shown was determined as described in panel A, following stimulation of BMDM with 1 μg/ml LPS for 1 h. All results are representative of at least three independent experiments.
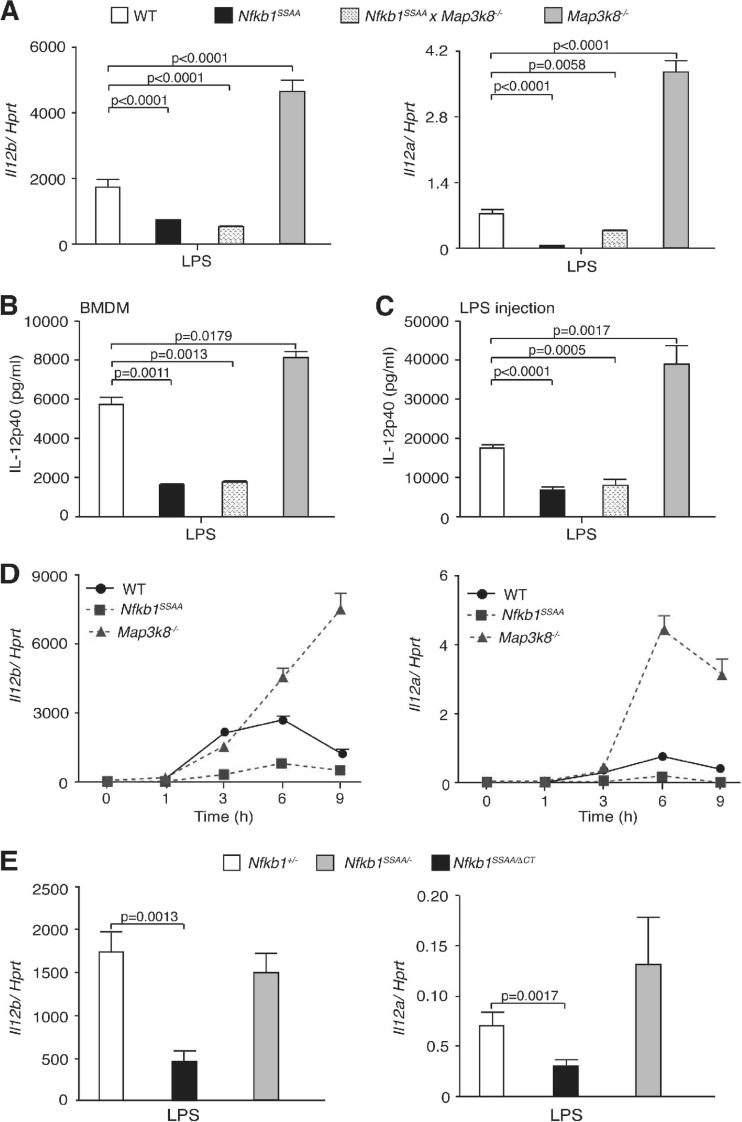
To investigate the role of reduced p50 in the induction of p50-dependent gene expression in Nfkb1SSAA/SSAA BMDM, Nfkb1SSAA/SSAA mice were crossed with Nfkb1ΔCT/ΔCT mice, which express elevated amounts of p50 compared to WT cells and no p105 (19). For comparison with these Nfkb1SSAA/ΔCT mice, Nfkb1+/− and Nfkb1SSAA/− strains were also generated. Nfkb1SSAA/ΔCT BMDM expressed steady-state levels of p50 equivalent to Nfkb1+/− cells, while p50 levels were very low in Nfkb1SSAA/− BMDM (Fig. 3C). LPS activation levels of p50 DNA binding were similar between Nfkb1SSAA/ΔCT BMDM and control Nfkb1+/− cells, while p50 activation was significantly decreased in Nfkb1SSAA/− BMDM (Fig. 3D). LPS-induced expression of Il6 and Csf2 mRNAs was significantly reduced in Nfkb1SSAA/− BMDM compared to Nfkb1+/− BMDM levels (Fig. 3E). However, expression of these genes was increased in Nfkb1SSAA/ΔCT compared to Nfkb1SSAA/− BMDM, although not to the same levels as that detected in Nfkb1+/− BMDM. The defects in LPS-induced expression of Il6 and Csf2 in Nfkb1SSAA/SSAA macrophages were therefore principally due to defective p50 generation.
Nfkb1SSAA mutation blocks activation of ERK MAP kinase by TNF receptor 1 (TNFR1) and multiple TLRs.
Our earlier studies overexpressing FL-p105 or FL-p105SSAA in RAW264.7 cells and Nfkb1−/− macrophages suggested an important role for IKK-induced p105 proteolysis in LPS activation of the MEK kinase activity of TPL-2 and subsequent activation of ERK (6). However, their physiological relevance remained uncertain, and it was unclear whether IKK-induced p105 proteolysis was essential for TPL-2 activation of ERK or simply modulated the amplitude of TPL-2 signaling.
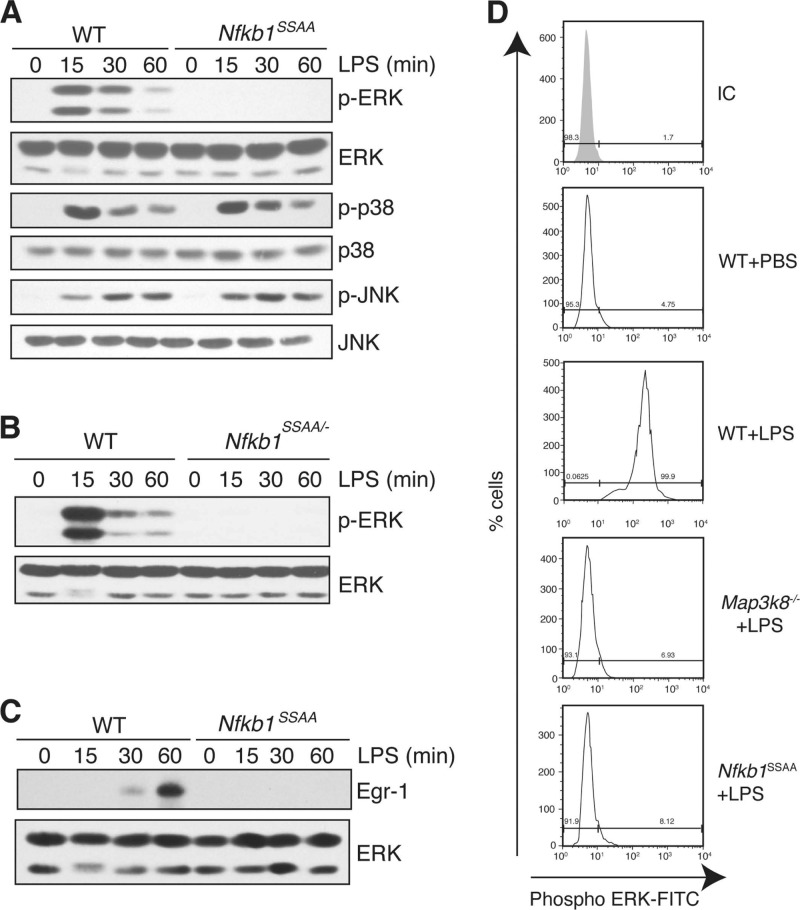
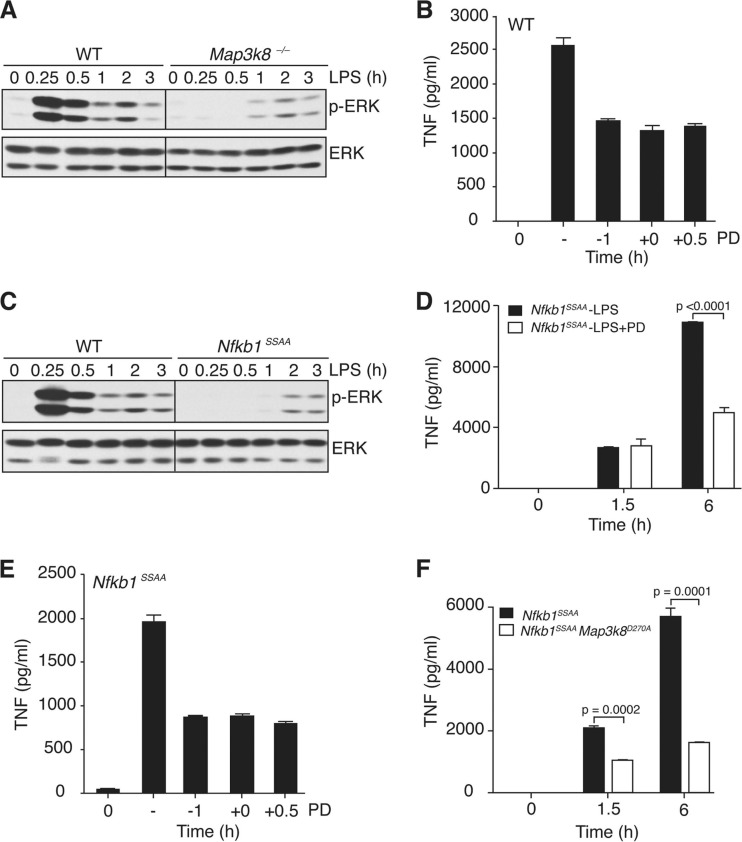
The availability of Nfkb1SSAA/SSAA BMDM allowed us to investigate the effect of endogenous p105SSAA on TPL-2 activation of ERK. LPS did not induce ERK phosphorylation in Nfkb1SSAA/SSAA BMDM, while phosphorylation of p38 and JNK was similar to that of WT cells (Fig. 4A). LPS activation of ERK was also ablated in Nfkb1SSAA/− BMDM (Fig. 4B), demonstrating that the inhibitory effects of Nfkb1SSAA mutation were not due to increased levels of p105SSAA. Furthermore, the Nfkb1SSAA mutation blocked LPS-stimulated upregulation of the transcription factor Egr-1 (Fig. 4C), which is positively regulated by TPL-2/ERK signaling (46). To investigate whether the Nfkb1SSAA mutation was also required for activation of ERK in macrophages in vivo, mice were injected intraperitoneally with LPS, and peritoneal macrophages were aspirated after 10 min. Intracellular staining clearly detected phospho-ERK in WT peritoneal macrophages after LPS injection (Fig. 4D). In contrast, no phospho-ERK signal was detected in Map3k8−/− or Nfkb1SSAA/SSAA peritoneal macrophages, demonstrating that IKK-induced p105 proteolysis was essential for TPL-2-dependent activation of ERK in vivo.
Fig 4.
Nfkb1SSAA mutation blocks LPS activation of ERK. (A to C) BMDM were stimulated for different times with LPS. Lysates were immunoblotted. (D) WT, Map3k8−/−, and Nfkb1SSAA/SSAA mice were injected intraperitoneally with LPS (200 μg) or PBS. Peritoneal cells were aspirated after 10 min, and intracellular phospho-ERK (p-ERK) in F4/80+ cells was monitored by flow cytometry. All results are representative of at least three independent experiments.
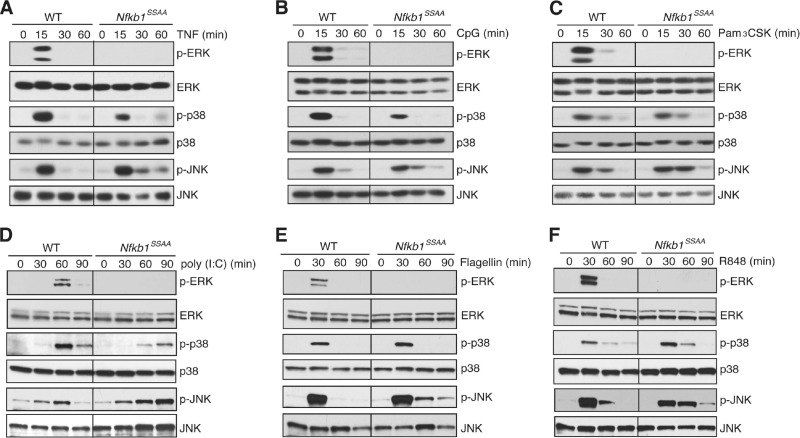
The Nfkb1SSAA mutation additionally blocked induction of ERK phosphorylation in BMDM by TNF, CpG (TLR9), Pam-3-Cys (TLR2), poly(I · C) (TLR3), flagellin (TLR5), and R848 (TLR7/TLR8) (Fig. 5A to F), which all signal to ERK via TPL-2 (3, 12, 21, 29). However, activation of ERK by phorbol ester and zymosan, which are both independent of TPL-2 (10, 29), was unaffected by the Nfkb1SSAA mutation (data not shown). Together, these data provided clear genetic evidence that TPL-2-dependent activation of ERK in macrophages required IKK-induced p105 proteolysis.
Fig 5.
Nfkb1SSAA mutation blocks activation of ERK by TNFR1 and multiple TLRs. Lysates from BMDM, stimulated for the indicated times with TNF (A), CpG (B), Pam3CSK (C), poly(I · C) (D), flagellin (E), or R848 (F) or left unstimulated (0 min), were immunoblotted. All results are representative of at least two independent experiments.
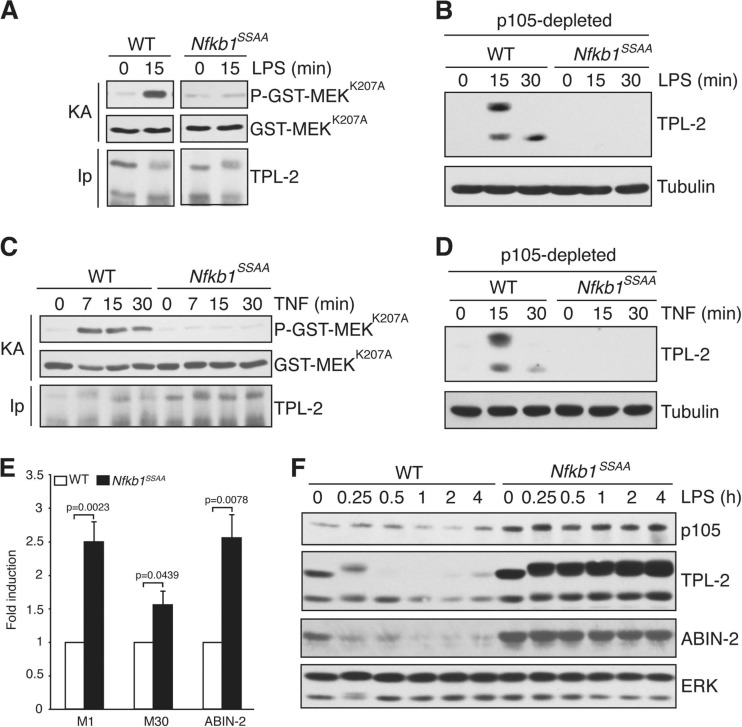
To determine whether the Nfkb1SSAA mutation prevented ERK activation by affecting TPL-2 activation, TPL-2 was immunoprecipitated from BMDM lysates, using limiting amounts of antibody to ensure that similar amounts of TPL-2 were isolated. MEK kinase activity was measured in vitro using GST-MEK as the substrate. LPS did not induce TPL-2 MEK kinase activity in Nfkb1SSAA/SSAA BMDM, in contrast to WT cells (Fig. 6A). To determine whether the Nfkb1SSAA mutation impaired the release of TPL-2 from p105 as shown previously in RAW264.7 cells (6), p105 was removed from BMDM lysates by anti-p105 immunoprecipitation. LPS stimulated release of p105-free TPL-2 in WT BMDM, but this was completely blocked by the Nfkb1SSAA mutation (Fig. 6B). The Nfkb1SSAA mutation blocked TNF activation of TPL-2 MEK kinase activity and the generation of p105-free TPL-2 in a similar fashion (Fig. 6C and D). Together, these data demonstrated that IKK-induced p105 proteolysis was essential for LPS and TNF to induce release of TPL-2 from p105 and activate TPL-2 MEK kinase activity in primary macrophages.
Fig 6.
Nfkb1SSAA mutation prevents activation of TPL-2 MEK kinase activity. (A and C) TPL-2 was immunoprecipitated (IP) from lysates of BMDM with or without LPS (A) or TNF (C) stimulation and assayed for its ability to phosphorylate GST-MEKK207A (KA) in vitro. To ensure that equivalent amounts of TPL-2 were assayed for each condition, immunoprecipitations were carried out with cell lysates in excess. (B and D) Lysates of BMDM, stimulated with LPS (B) or TNF (D), were depleted of p105 by immunoprecipitation and then immunoblotted. (E and F) Lysates of BMDM, unstimulated (E) or stimulated for the indicated times with LPS (F), were immunoblotted. Due to alternative translational initiation on a second methionine (at residue 30), TPL-2 is expressed as two isoforms, M1-TPL-2 and M30-TPL-2 (1). In panels A, B, C, D, and F, results are representative of three independent experiments. In panel E, data were pooled from six independent experiments.
Steady-state levels of p105 were increased in Nfkb1SSAA/SSAA BMDM compared to levels in WT cells (Fig. 1B and 6F). Resting concentrations of TPL-2 and ABIN-2, which are both associated with p105 (7, 27), were also elevated by the Nfkb1SSAA mutation (Fig. 6E and F). These increases were not attributed to alterations in the steady-state amounts of Nfkb1, Map3k8 (encoding TPL-2), or Tnip2 (encoding ABIN-2) mRNAs (data not shown). Rather, the Nfkb1SSAA mutation blocked “tonic” p105 turnover, as observed previously in CD4+ T cells (40). This could also explain the increases in TPL-2 and ABIN-2 proteins in macrophages. The Nfkb1SSAA mutation had comparable effects on steady-state levels of p105, TPL-2, and ABIN-2 in ex vivo peritoneal macrophages (data not shown), suggesting that tonic IKK signaling also occurred in vivo.
LPS stimulated the proteolysis of p105 in WT BMDM, and this was blocked in Nfkb1SSAA/SSAA cells, as expected (Fig. 6F). LPS-induced proteolysis of TPL-2 and ABIN-2 by the proteasome (6, 27) was also reduced by the Nfkb1SSAA mutation (Fig. 6F). IKK-induced p105 proteolysis therefore regulated steady-state levels of TPL-2 and ABIN-2 in unstimulated BMDM and the proteolysis of both of these proteins by the proteasome after TLR4 stimulation.
Nfkb1SSAA mutation inhibits TLR4 induction of Il12a and Il12b.
Pathogen infection induces macrophage production of the proinflammatory cytokine IL-12 (interleukin-12), a heterodimer of IL-12α (p35) and IL-12β (p40) proteins (41), which promotes differentiation of T helper 1 (Th1) effector cells. LPS induction of Il12a and Il12b mRNAs in macrophages is positively regulated by NF-κB and negatively regulated by TPL-2/ERK signaling (21, 24, 36). Since IKK-induced p105 proteolysis promoted nuclear translocation of Rel subunits and facilitated TPL-2 activation of ERK, it was interesting to study the expression of Il12a and Il12b mRNAs. Comparison of WT and Nfkb1SSAA/SSAA BMDM revealed that the effect of the Nfkb1SSAA mutation on LPS upregulation of both Il12a and Il12b mRNAs was inhibitory (Fig. 7A). In contrast, LPS-induced mRNA expression of both genes was substantially increased in Map3k8−/− BMDM relative to levels in WT controls, as reported previously (21). Analysis of Nfkb1SSAA/SSAA Map3k8−/− BMDM indicated that the inhibitory effect of the Nfkb1SSAA mutation on LPS upregulation of Il12a and Il12b mRNAs was TPL-2 independent, ruling out a contribution of residual TPL-2 signaling. Nfkb1SSAA mutation and TPL-2 deficiency had comparable inhibitory and stimulatory effects, respectively, on the production of IL-12 p40 protein by LPS-stimulated BMDM (Fig. 7B). Similarly, the Nfkb1SSAA mutation reduced the production of IL-12 p40 in serum after intraperitoneal injection of LPS compared to WT controls, while TPL-2 deficiency increased IL-12 p40 production (Fig. 7C).
Fig 7.
Nfkb1SSAA mutation reduces LPS induction of IL-12. (A and B) BMDM were stimulated for 6 h with LPS (10 ng/ml). In panel A, expression of Il12a and Il12b mRNAs was determined by quantitative RT-PCR (error bars, SEM). In panel B, IL-12 p40 in culture supernatants was measured by ELISA (error bars, SEM). (C) WT, Nfkb1SSAA/SSAA, Map3k8−/−, and Nfkb1SSAA/SSAA Map3k8−/− mice were injected intraperitoneally with LPS. After 6 h, IL-12 p40 in serum was measured by ELISA (error bars, SEM of 6 mice). (D) Triplicate cultures of BMDM were stimulated with LPS for the times shown. Expression of Il12a and Il12b mRNAs was determined by quantitative RT-PCR (error bars, SEM). (E) Levels of Il12a and Il12b mRNAs in LPS-stimulated BMDM were measured as described in panel A (error bars, SEM). All results are representative of at least three independent experiments.
A time course experiment showed that the Nfkb1SSAA mutation reduced LPS-induced Il12a and Il12b mRNA expression in BMDM at all time points between 1 and 9 h (Fig. 7D). In contrast, TPL-2 deficiency did not affect mRNA expression of these genes at 1 and 3 h poststimulation, while expression of both genes was enhanced with more prolonged stimulation. Comparison of Nfkb1+/−, Nfkb1SSAA/−, and Nfkb1SSAA/ΔCT BMDM indicated that the Nfkb1SSAA mutation decreased the early expression of Il12a and Il12b mRNAs largely by reducing p50 levels (Fig. 7E). The addition of blocking IL-10R antibodies to cultures demonstrated that the inhibitory effect of the Nfkb1SSAA mutation on Il12a and Il12b mRNA expression was IL-10 independent (data not shown). IKK-induced p105 proteolysis, therefore, promoted the initial transcription of Il12a and Il12b genes via activation of p50-containing NF-κB dimers, while stimulation of TPL-2/ERK signaling downregulated their expression at later time points.
Nfkb1SSAA mutation does not affect TLR4 induction of sTNF.
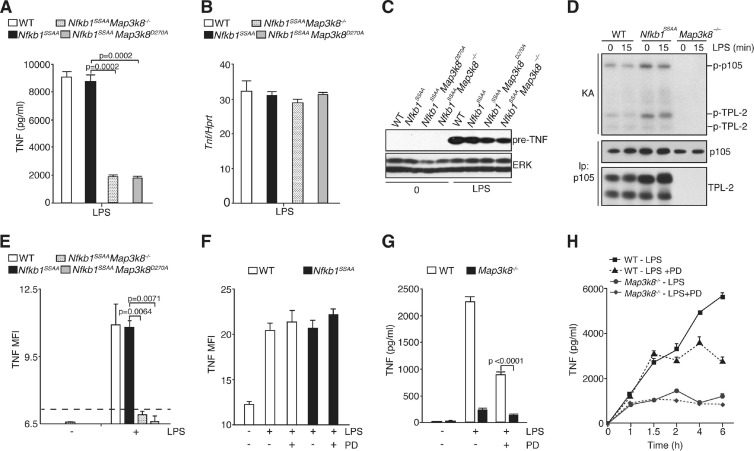
Activation of the TPL-2/ERK signaling pathway is essential for the posttranslational regulation of soluble TNF (sTNF) production in LPS-stimulated macrophages (10, 33). In WT and Nfkb1SSAA/SSAA BMDM, LPS induced comparable levels of Tnf mRNA and of pre-TNF protein, the 26-kDa transmembrane precursor that is proteolytically cleaved to produce the mature secreted 17-kDa form of TNF (Fig. 8B and C). Surprisingly, however, LPS stimulation of sTNF production was not blocked by the Nfkb1SSAA mutation (Fig. 8A) although TPL-2 was not able to activate ERK (Fig. 4A). In contrast, LPS stimulation induced very little detectable sTNF by Nfkb1SSAA/SSAA Map3k8−/− BMDM (Fig. 8A) although LPS induction of Tnf mRNA and pre-TNF protein in these cells was comparable to levels in WT cells (Fig. 8B and C). LPS similarly failed to induce detectable sTNF production by primary peritoneal macrophages isolated from Nfkb1SSAA/SSAA Map3k8−/− mice, whereas equivalent amounts of sTNF were produced by WT and Nfkb1SSAA/SSAA peritoneal macrophages (data not shown). Furthermore, intraperitoneal LPS injection induced serum sTNF in WT and Nfkb1SSAA/SSAA mice but no detectable TNF in Nfkb1SSAA/SSAA Map3k8−/− mice (data not shown). The Nfkb1SSAA mutation, therefore, did not block the posttranslational stimulation of sTNF production by TPL-2.
Fig 8.
Nfkb1SSAA mutation does not inhibit LPS induction of sTNF production. (A and B) BMDM were cultured in triplicates and stimulated with LPS for 6 h (A) or 1 h (B). The Tpl2D270A/D270A genotype is shown here and subsequently as Tpl2D270A. In panel A, TNF in culture supernatants was assayed by ELISA (error bars, SEM). In panel B, Tnf mRNA was quantified as described in the legend of Fig. 3A (error bars, SEM). (C) Lysates were prepared from unstimulated or LPS-stimulated BMDM of the indicated genotypes and immunoblotted. (D) p105 was quantitatively immunoprecipitated from BMDM lysates of the indicated genotypes with or without LPS stimulation (100 ng/ml) and subjected to an in vitro kinase assay (KA). 32P-labeled bands were revealed by autoradiography after SDS-PAGE. The amounts of TPL-2 and p105 isolated were determined by immunoblotting. (E) BMDM were stimulated for 1 h with LPS (1 μg/ml) and fixed, and surface pre-TNF expression was assayed by flow cytometry. The dotted line shows the mean fluorescence intensity (MFI) for negative-control antibody staining. (F) WT or Nfkb1SSAA/SSAA BMDM were stimulated with LPS (1 μg/ml) with or without PD184352 (PD) for 1 h, and surface pre-TNF expression was determined by flow cytometry. (G) BMDM were cultured for 6 h with LPS with or without PD184352 or with control medium (0). Culture supernatants were assayed for TNF (triplicate cultures; error bars, SEM). (H) BMDM were stimulated for the indicated times with LPS with or without PD or with control medium (0). PD was added 1 h before LPS stimulation. TNF in culture supernatants was assayed (error bars, SEM). All results are representative of at least three independent experiments.
The role of TPL-2 kinase activity in regulating sTNF production in Nfkb1SSAA/SSAA BMDM was investigated by intercrossing Nfkb1SSAA/SSAA mice with Map3k8D270A/D270A mice, which expressed the catalytically inactive mutant TPL-2D270A (34). Stimulation of Nfkb1SSAA/SSAA Map3k8D270A/D270A BMDM induced very low levels of sTNF compared to those in WT and Nfkb1SSAA/SSAA cells (Fig. 8A) and levels equivalent to those detected with Nfkb1SSAA/SSAA Map3k8−/− BMDM. Tnf mRNA and pre-TNF protein levels were similar between WT and Nfkb1SSAA/SSAA Map3k8D270A/D270A BMDM (Fig. 8B and C). Posttranslational regulation of sTNF production in Nfkb1SSAA/SSAA BMDM, therefore, required TPL-2 kinase activity.
The above data implied that TPL-2 catalytic activity was not blocked by the Nfkb1SSAA mutation. To investigate this directly, endogenous p105 was immunoprecipitated from Nfkb1SSAA/SSAA BMDM, and the activity of associated TPL-2 was monitored by in vitro kinase assay. TPL-2 phosphorylated associated p105 and also autophosphorylated (Fig. 8D), consistent with previous experiments with recombinant TPL-2 and p105 (2, 32). However, as shown previously (2), LPS stimulation did not alter p105-associated TPL-2 catalytic activity. No 32P-labeled bands were detected in anti-p105 immunoprecipitates from Map3k8−/− cells, demonstrating that the kinase activity associated with p105 was due to TPL-2 (Fig. 8D).
TPL-2 is required for LPS to induce the transport of pre-TNF to the plasma membrane (33). Consistent with the normal production of sTNF by Nfkb1SSAA/SSAA and WT BMDM, surface expression levels of pre-TNF were similar between these BMDM after 1 h of LPS stimulation (Fig. 8E). However, pre-TNF was not detected at the cell surfaces of Nfkb1SSAA/SSAA Map3k8−/− and Nfkb1SSAA/SSAA Map3k8D270A/D270A BMDM although the total levels of cellular pre-TNF were comparable to those in Nfkb1SSAA/SSAA BMDM (Fig. 8C). Furthermore, PD184352 did not affect detection of cell surface pre-TNF in WT or Nfkb1SSAA/SSAA BMDM following LPS stimulation (Fig. 8F). TPL-2 therefore controlled the production of sTNF by regulating the intracellular transport of pre-TNF to the cell surface, and this was independent of IKK-induced p105 proteolysis and TPL-2 regulation of ERK activation but required the kinase activity of TPL-2.
Distinct temporal regulation of sTNF production by TPL-2 and ERK.
Pharmacological inhibition of MEK-1/2 and TPL-2 deficiency correlates with inhibition of LPS-induced ERK phosphorylation and sTNF production, supporting the hypothesis that TPL-2 regulates sTNF production exclusively via activation of ERK (10). However, our previous analyses demonstrated that TPL-2 catalytic activity could stimulate sTNF production independently of ERK activation. To further investigate the relevance of ERK activation in TPL-2-dependent regulation of sTNF production in primary macrophages, we initially compared the effects of MEK inhibition and TPL-2 deficiency on LPS induction of sTNF.
Pretreatment of BMDM with the MEK-1/2 inhibitor PD184352 blocked LPS induction of ERK phosphorylation (data not shown) but only partially reduced the abundance of sTNF (Fig. 8G). However, TPL-2 deficiency had a significantly more pronounced inhibitory effect on the production of sTNF protein (>90% inhibition), than PD184352 treatment of WT cells (40 to 50% inhibition) (Fig. 8G). Neither PD184352 nor TPL-2 deficiency altered the level of Tnf mRNA or pre-TNF protein induced by LPS (data not shown). These data suggested that posttranslational regulation of sTNF production by TPL-2 was not solely mediated via activation of ERK.
A time course experiment showed that TPL-2 deficiency reduced TNF production at all the time points tested between 1 and 6 h (Fig. 8H). In contrast, MEK inhibition had no effect at earlier time points and only started to reduce TNF production compared to the vehicle control level at 2 to 4 h after LPS stimulation. These results demonstrated that the major regulatory effect of TPL-2 on TNF production was initiated early after LPS stimulation, whereas ERK control of TNF production was a delayed, and possibly secondary, process.
TPL-2-independent, delayed activation of ERK stimulates sTNF production.
Prolonged time course experiments revealed that LPS induced two waves of ERK phosphorylation in WT BMDM (Fig. 9A). The first major wave was maximal at 15 min and was absent in TPL-2-deficient cells. In contrast, the second minor wave, which peaked at 2 h, was still evident in TPL-2-deficient cells. Pretreatment of cells with cycloheximide revealed that the second wave of ERK phosphorylation required protein synthesis while early ERK phosphorylation did not (data not shown).
Fig 9.
Delayed regulation of sTNF production by ERK following LPS stimulation. (A and C) BMDM were stimulated with LPS. Lysates were subjected to immunoblotting. (B and E) WT or Nfkb1SSAA/SSAA BMDM were treated with PD184352 (PD) 1 h prior to LPS addition (−1), coincident with LPS addition (0), or 0.5 h after LPS addition. Vehicle control (−) was added 1 h prior to LPS stimulation. After 6 h of culture, TNF in supernatants was assayed by ELISA (error bars, SEM). (D) Nfkb1SSAA/SSAA BMDM were stimulated with LPS with or without PD184352. Supernatants were assayed for TNF (error bars, SEM). (F) Nfkb1SSAA and Nfkb1SSAA/SSAA Tpl2D270A/D270A (labeled as Nfkb1SSAA Tpl2D270A) BMDM were stimulated with LPS, and supernatants were assayed for TNF (error bars, SEM). All results are representative of at least three independent experiments.
To investigate the contribution of the late phase of ERK activation to sTNF production, PD184352 was added to WT BMDM 30 min after LPS stimulation, thereby leaving the early TPL-2-dependent phase of ERK activation unaffected. This treatment reduced sTNF production by approximately 50% (Fig. 9B). Addition of PD184352 at 1 h prior to LPS stimulation (our normal methodology) or at the same time as LPS had an equivalent inhibitory effect on sTNF production. These results indicated that PD184352 predominantly mediated its inhibitory effect on sTNF production by blocking the late-phase activation of ERK, which was largely TPL-2 independent.
Analysis of lysates of Nfkb1SSAA/SSAA BMDM stimulated with LPS over an extended time course revealed that the Nfkb1SSAA mutation blocked the early TPL-2-dependent phase of ERK activation while the late TPL-2-independent phase was relatively unaffected (Fig. 9C). Pretreatment of Nfkb1SSAA/SSAA BMDM with PD184352 had no effect on sTNF levels detected at 1.5 h (Fig. 9D). However, the production of sTNF at 6 h was reduced by approximately 60% compared to vehicle control. Addition of PD184352 30 min after LPS stimulation inhibited sTNF production by Nfkb1SSAA/SSAA BMDM to a similar degree to addition 1 h prior to, or coincident with, LPS stimulation (Fig. 9E). This ruled out the potential contribution of very low (undetectable) levels of TPL-2-dependent ERK activation to sTNF production by Nfkb1SSAA/SSAA BMDM. Similar to the mechanism in WT cells, therefore, PD184352 mediated its inhibitory effect on sTNF production by Nfkb1SSAA/SSAA BMDM by blocking the late, TPL-2-independent, activation of ERK. In contrast, comparison of Nfkb1SSAA/SSAA and Nfkb1SSAA/SSAA Map3k8D270A/D270A BMDM revealed that TPL-2 catalytic activity was required for optimal production of TNF at both 1.5 and 6 h (Fig. 9F). These data support the hypothesis that TPL-2 mainly induced sTNF production independently of its stimulatory effects on ERK activation.
DISCUSSION
Previous experiments comparing the effects of overexpression of FL-p105 and FL-p105SSAA in RAW264.7 cells and Nfkb1−/− macrophages have suggested that IKK-induced p105 proteolysis is an important regulatory step in the activation of ERK by TPL-2 following LPS stimulation (6, 45). In the present study, we have provided definitive genetic support for this model, showing that the Nfkb1SSAA knock-in mutation completely prevented LPS activation of TPL-2 MEK kinase activity and consequently ERK. LPS activation of ERK was also blocked in Nfkb1SSAA/− macrophages, showing that the inhibitory effects of the mutation were not simply due to p105SSAA overexpression. Furthermore, the Nfkb1SSAA mutation prevented activation of ERK in peritoneal macrophages in vivo, following intraperitoneal LPS injection. IKK therefore directly regulates both NF-κB and ERK activation in inflammatory responses of macrophages.
LPS-induced rapid degradation of the high-molecular-weight M1 form of TPL-2 (M1-TPL-2) by the proteasome (5) was completely blocked by the Nfkb1SSAA mutation. Proteolysis of M1-TPL-2 is therefore dependent on IKK-induced p105 degradation. Since IKK-induced p105 proteolysis was required for release of TPL-2 from p105, this may indicate that only p105-free TPL-2 is susceptible to proteasome-mediated proteolysis, perhaps for steric reasons. The Nfkb1SSAA mutation significantly increased the steady-state levels of M1-TPL-2 and, to a lesser extent, M30-TPL-2, expressed in macrophages, without affecting Map3k8 mRNA levels. Constitutive IKK phosphorylation of p105, therefore, controls steady-state levels of TPL-2 in macrophages and consequently the amplitude of TPL-2 signaling following agonist stimulation. Steady-state levels of ABIN-2 were also increased in Nfkb1SSAA/SSAA macrophages compared to WT levels, suggesting that constitutive IKK-induced p105 proteolysis may also control the degree of signaling by ABIN-2, a ubiquitin-binding protein (43).
Analyses of Nfkb1SSAA/SSAA mice have previously demonstrated an essential role for IKK-induced p105 proteolysis for optimal activation of NF-κB in CD4+ T cells and mature CD4+ T cell helper function (40). The Nfkb1SSAA mutation does not affect steady-state expression of p50 or other Rel subunits in peripheral CD4+ T cells. However, the amplitude of NF-κB activation after T-cell receptor (TCR)/CD28 costimulation is reduced due to cytoplasmic retention of the major Rel dimers (p50/p50, p50/RelA, and p50/cRel) by mutant p105SSAA, which is expressed in larger steady-state amounts than WT p105 and is insensitive to IKK-induced proteolysis. Reduced nuclear translocation of Rel proteins due to cytoplasmic retention by p105SSAA could also explain the impairment in TLR4 activation of RelA and cRel in Nfkb1SSAA/SSAA macrophages. Moreover, analysis of Nfkb1SSAA/− macrophages, which expressed p105 at levels similar to WT cells, indicated that it was the blockade in IKK-induced p105 proteolysis that reduced RelA and cRel activation rather than simply p105SSAA overexpression. Since the Nfkb1SSAA mutation did not affect LPS-induced IκBα degradation, this implies that IKK-induced p105 proteolysis contributes significantly to the degree of NF-κB activation in LPS-stimulated macrophages by releasing associated Rel subunits.
Earlier experiments using cell lines have indicated that IKK phosphorylation of p105 does not affect processing to p50 but rather triggers the complete degradation of p105 (18, 26). However, in vitro experiments have suggested that IKK can induce p105 degradation via SCFβ-TrCP-mediated ubiquitination and processing to p50 via an unknown E3 ligase (9). Analyses of cells from Nfkb1SSAA/SSAA mice suggest that the effects of IKK phosphorylation on p105 proteolysis may be cell type specific. Thus, the Nfkb1SSAA mutation does not affect p50 levels in CD4+ T cells (40). In contrast, in macrophages, the Nfkb1SSAA mutation substantially decreased steady-state p50 levels and blocked the processing of p105 to p50 induced by LPS stimulation. IKK phosphorylation of p105, therefore, can stimulate the production of p50 and facilitate the nuclear translocation of associated Rel subunits in macrophages during innate immune responses. Why the Nfkb1SSAA mutation has different effects on p50 generation in T cells and macrophages is not known. One possibility is that CD4+ T cells and macrophages express different amounts of the E3 ligase that triggers IKK-dependent p105 processing. Identification of this E3 ligase will be necessary to determine whether this explanation is correct.
The Nfkb1SSAA mutation fractionally reduced the induction of Nfkbia, Tnfaip3, Cxcl2, and Nfkbiz mRNAs after stimulation at a low LPS dosage. After stimulation with a higher dose of LPS, the inhibitory effect of the Nfkb1SSAA mutation on the expression of the majority of these genes was less pronounced. The effect of partial reduction in NF-κB activation in Nfkb1SSAA/SSAA macrophages on expression of these NF-κB target genes, therefore, was largely overcome if the strength of stimulus was increased. In contrast, LPS upregulation of Il6 and Csf2 genes, which are positively regulated by IκBζ-p50 (47), was essentially blocked by the Nfkb1SSAA mutation even at high-LPS dosing, which induced normal IκBζ expression. Analysis of Nfkb1SSAA/ΔCT BMDM revealed that the inhibitory effect of the Nfkb1SSAA mutation on the expression of Il6 and Csf2 genes following high-dose LPS stimulation was largely caused by the reduction in p50 levels. The IKK/p105 pathway is therefore critical for LPS induction of IκBζ-p50-regulated target genes.
Analysis of the effect of the Nfkb1SSAA mutation on LPS-induced IL-12 expression in BMDM suggested that IKK-induced p105 proteolysis facilitated the upregulation of Il12a and Il12b mRNAs by controlling p50 production, consistent with the known transcriptional regulation of these genes by NF-κB (24, 36). IKK-induced p105 proteolysis was also essential for activation of the TPL-2/ERK signaling pathway, which inhibits the expression of these genes (21). An explanation for these apparently paradoxical effects was revealed in time course experiments. The negative regulatory effect of TPL-2 was only evident several hours after LPS stimulation, and at early time points TPL-2 deficiency had no effect on the abundance of Il12a and Il12b mRNAs. In contrast, the stimulatory effect of NF-κB was observed early after LPS addition. IKK activation of TPL-2/ERK signaling, therefore, induced a negative feedback loop to switch off NF-κB-dependent transcription of Il12a and Il12b genes. Consequently, linking the activation of TPL-2 to NF-κB activation via IKK-induced p105 proteolysis limits the proinflammatory effects of IL-12.
TPL-2 expression in macrophages is essential for LPS activation of ERK phosphorylation and for induction of sTNF (10, 33). However, blockade of TPL-2-dependent activation of ERK by the Nfkb1SSAA mutation did not prevent normal production of sTNF. Only genetic ablation of TPL-2 or its kinase activity substantially reduced the production of sTNF by LPS-stimulated Nfkb1SSAA/SSAA macrophages. Therefore, TPL-2 catalytic activity regulates TNF production in macrophages independently of IKK-induced p105 proteolysis and activation of ERK. Consistent with this hypothesis, comparative analyses of the effects of TPL-2 deficiency with pharmacological inhibition of ERK activation on LPS-induced TNF induction revealed that the extent and timing of TPL-2 regulation could be clearly distinguished from that of ERK. Thus, the effect of genetic ablation of TPL-2 on sTNF production was significantly greater than the effect of pharmacological inhibition of MEK. Furthermore, ERK activation regulated production of sTNF by LPS-stimulated BMDM only at later time points and was not required for the early transport of pre-TNF to the cell surface. In contrast, TPL-2 was essential for cell surface expression of pre-TNF at 1 h post-LPS stimulation and for sTNF production at both early and late time points. Clearly, an important area of future research will be the identification of substrates of the TPL-2/p105/ABIN-2 complex which promote TNF production by controlling the transport of pre-TNF to the cell surface. These studies may also identify novel physiological processes that are regulated by TPL-2 independently of MEK/ERK activation.
Due to its critical role in regulating TNF production, TPL-2 is regarded as a good anti-inflammatory drug target (15). Over the last decade, several large pharmaceutical companies have carried out high-throughput assays with a truncated recombinant form of TPL-2 (M30-TPL-2ΔC) to identify and develop small-molecule inhibitors. However, although inhibitors have been developed that can block TPL-2 signaling in cultured cells, there are very few data on whether these work in vivo, and no TPL-2 inhibitors have yet been tested in clinical trials. The present study shows that TPL-2 stimulates TNF production when it is still associated with p105 and suggests that inhibitor screens formatted using TPL-2 in complex with NF-κB1 p105 would be more physiologically relevant. Assays using TPL-2/p105 complex may identify novel classes of TPL-2 inhibitor since the TPL-2 kinase domain forms interactions with both p105 and its own C terminus (4, 8), which is lacking in M30-TPL-2ΔC used in earlier inhibitor screens and therefore may be folded differently.
In conclusion, this study demonstrated that IKK-induced p105 proteolysis positively regulated NF-κB-dependent gene transcription during an innate immune response and was particularly important for the upregulation of Il6 and Csf2 genes, whose expression required p50. The release of TPL-2 from its inhibitor p105 and, consequently, activation of MEK and ERK were also completely dependent on IKK-induced p105 proteolysis. However, TPL-2 signaling was not completely blocked by its association with p105 and induced the production of soluble TNF independently of ERK activation.
ACKNOWLEDGMENTS
We thank P. Tsichlis (Tufts University, Boston, MA) and Thomas Jefferson University, Philadelphia, PA, for the Map3k8−/− mice; Philip Cohen for MEK inhibitor (University of Dundee, Scotland); Emilie Jacque (NIMR), George Kassiotis (NIMR), Ben Seddon (NIMR), Steve Smale (UCLA), and Victor Tybulewicz (NIMR) for advice and critical reading of the manuscript; Hamish Allen (Abbott Bioresearch Center, Worcester, MA) for encouragement at the start of the project; the NIMR Photographics department, NIMR Biological Services, NIMR flow cytometry service, and other members of the Ley laboratory for help during the course of this work.
This work was supported by the United Kingdom Medical Research Council.
Footnotes
Published ahead of print 25 June 2012
REFERENCES
- 1. Aoki M, et al. 1993. The human cot proto-oncogene encodes two protein serine/threonine kinases with different transforming activities by alternative initiation of translation. J. Biol. Chem. 268: 22723– 22732 [PubMed] [Google Scholar]
- 2. Babu GR, et al. 2006. Phosphorylation of NF-κB1/p105 by oncoprotein kinase Tpl2: implications for a novel mechanism of Tpl2 regulation. Biochim. Biophys. Acta 1763: 174– 181 [DOI] [PubMed] [Google Scholar]
- 3. Banerjee A, et al. 2008. NF-kB1 and c-Rel cooperate to promote the survival of TLR4-activated B cells by neutralizing Bim via distinct mechanisms. Blood 112: 5063– 5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beinke S, et al. 2003. NF-κB p105 negatively regulates TPL-2 MEK kinase activity. Mol. Cell. Biol. 23: 4739– 4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beinke S, Ley SC. 2004. Functions of NF-κB1 and NF-κB2 in immune cell biology. Biochem. J. 382: 393– 409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beinke S, et al. 2004. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IκB kinase-induced proteolysis of NF-κB1 p105. Mol. Cell. Biol. 24: 9658– 9667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belich MP, Salmeron A, Johnston LH, Ley SC. 1999. TPL-2 kinase regulates the proteolysis of the NF-κB inhibitory protein NF-κB1 p105. Nature 397: 363– 368 [DOI] [PubMed] [Google Scholar]
- 8. Ceci JD, et al. 1997. TPL-2 is an oncogenic kinase that is activated by carboxy-terminal truncation. Gene Dev. 11: 688– 700 [DOI] [PubMed] [Google Scholar]
- 9. Cohen S, Achbert-Weiner H, Ciechanover A. 2004. Dual effects of IκB kinase β-mediated phosphorylation on p105 fate: SCFβ-TrCP-dependent degradation and SCFβ-TrCP-independent processing. Mol. Cell. Biol. 24: 475– 486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dumitru CD, et al. 2000. TNFα induction by LPS is regulated post-transcriptionally via a TPL2/ERK-dependent pathway. Cell 103: 1071– 1083 [DOI] [PubMed] [Google Scholar]
- 11. Eliopoulos AG, Dumitru CD, Wang C-C, Cho J, Tsichlis PN. 2002. Induction of COX-2 by LPS in macrophages is regulated by TPL2-dependent CREB activation signals. EMBO J. 21: 4831– 4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eliopoulos AG, Wang C-C, Dumitru CD, Tsichlis PN. 2003. TPL-2 transduces CD40 and TNF signals that activate ERK and regulates IgE induction by CD40. EMBO J. 22: 3855– 3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gantke T, Sriskantharajah S, Ley SC. 2011. Regulation and function of TPL-2, an IκB kinase-regulated MAP kinase kinase kinase. Cell Res. 21: 131– 145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gantke T, Sriskantharajah S, Sadowski M, Ley SC. 2012. IκB kinase regulation of the TPL-2/ERK MAPK pathway. Immunol. Rev. 246: 168– 182 [DOI] [PubMed] [Google Scholar]
- 15. George D, Salmeron A. 2009. Cot/TPL-2 protein kinase as a target for the treatment of inflammatory disease. Curr. Top. Med. Chem. 9: 611– 622 [DOI] [PubMed] [Google Scholar]
- 16. Ghosh S, May MJ, Kopp EB. 1998. NF-κB and Rel proteins: evolutionary conserved mediators of immune responses. Annu. Rev. Immunol. 16: 225– 260 [DOI] [PubMed] [Google Scholar]
- 17. Heissmeyer V, Krappmann D, Hatada EN, Scheidereit C. 2001. Shared pathways of IκB kinase-induced SCFβTrCP-mediated ubiquitination and degradation for the NF-κB precursor p105 and IκBα. Mol. Cell. Biol. 21: 1024– 1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heissmeyer V, Krappmann D, Wulczyn FG, Scheidereit C. 1999. NF-κB p105 is a target on IκB kinases and controls signal induction of BCL-3-p50 complexes. EMBO J. 18: 4766– 4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishikawa H, et al. 1998. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p)105 precursor (NF-κB1) but expressing p50. J. Exp. Med. 187: 985– 996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kabouridis PS, Magee AI, Ley SC. 1997. S-acylation of LCK protein tyrosine kinase is essential for its signalling function in T lymphocytes. EMBO J. 16: 4983– 4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaiser F, et al. 2009. TPL-2 negatively regulates interferon-beta production in macrophages and myeloid dendritic cells. J. Exp. Med. 206: 1863– 1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karin M, Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18: 621– 663 [DOI] [PubMed] [Google Scholar]
- 23. Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11: 373– 384 [DOI] [PubMed] [Google Scholar]
- 24. Kollet JI, Petro TM. 2006. IRF-1 and NF-κB p50 / cRel bind to distinct regions of the proximal murine IL-12 p35 promoter during costimulation with IFN-γ and LPS. Mol. Immunol. 43: 623– 633 [DOI] [PubMed] [Google Scholar]
- 25. Kravtsova-Ivantsiv Y, Cohen S, Ciechanover A. 2009. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-κB precursor. Mol. Cell 33: 496– 504 [DOI] [PubMed] [Google Scholar]
- 26. Lang V, et al. 2003. βTrCP-mediated proteolysis of NF-κB1 p105 requires phosphorylation of p105 serines 927 and 932. Mol. Cell. Biol. 23: 402– 413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lang V, et al. 2004. ABIN-2 forms a ternary complex with TPL-2 and NF-κB1 p105 and is essential for TPL-2 protein stability. Mol. Cell. Biol. 24: 5235– 5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Medzhitov R, Horng T. 2009. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9: 692– 703 [DOI] [PubMed] [Google Scholar]
- 29. Mielke LA, et al. 2009. Tumor progression locus 2 (Map3k8) is critical for host defense against Listeria monocytogenes and IL-1 production. J. Immunol. 183: 7984– 7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Orian A, et al. 2000. SCFβTrCP ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 19: 2580– 2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Papoutsopoulou S, et al. 2006. ABIN-2 is required for optimal activation of the TPL-2 / ERK MAP kinase pathway in innate immune responses. Nat. Immunol. 7: 606– 615 [DOI] [PubMed] [Google Scholar]
- 32. Robinson MJ, Beinke S, Kouroumalis A, Tsichlis PN, Ley SC. 2007. Phosphorylation of TPL-2 on serine 400 is essential for lipopolysaccharide activation of extracellular signal-regulated kinase in macrophages. Mol. Cell. Biol. 27: 7355– 7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rousseau S, et al. 2008. TPL2-mediated activation of ERK1 and ERK2 regulates the processing of pre-TNF alpha in LPS-stimulated macrophages. J. Cell Sci. 121: 149– 154 [DOI] [PubMed] [Google Scholar]
- 34. Salmeron A, et al. 1996. Activation of MEK-1 and SEK-1 by Tpl-2 proto-oncoprotein, a novel MAP kinase kinase kinase. EMBO J. 15: 817– 826 [PMC free article] [PubMed] [Google Scholar]
- 35. Salmeron A, et al. 2001. Direct phosphorylation of NF-κB p105 by the IκB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 276: 22215– 22222 [DOI] [PubMed] [Google Scholar]
- 36. Sanjabi S, Hoffmann A, Liou H-C, Baltimore D, Smale ST. 2000. Selective requirement for c-Rel during IL-12 p40 gene induction in macrophages. Proc. Natl. Acad. Sci. U. S. A. 97: 12705– 12710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Savinova OV, Hoffmann A, Ghosh G. 2009. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol. Cell 34: 591– 602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sha WC, Lious H-C, Tuomanen EI, Baltimore D. 1995. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell 80: 321– 330 [DOI] [PubMed] [Google Scholar]
- 39. Smale ST. 2010. Selective transcription in response to an inflammatory stimulus. Cell 140: 833– 844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sriskantharajah S, et al. 2009. Proteolysis of NF-kB1 p105 is essential for T cell antigen receptor-induced proliferation. Nat. Immunol. 10: 38– 47 [DOI] [PubMed] [Google Scholar]
- 41. Trinchieri G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3: 133– 146 [DOI] [PubMed] [Google Scholar]
- 42. Vallabhapurapu S, Karin M. 2009. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27: 693– 733 [DOI] [PubMed] [Google Scholar]
- 43. Verstrepen L, Carpentier I, Verhelst K, Beyaert R. 2009. ABINs: A20 binding inhibitors of NF-κB and apoptosis signaling. Biochem. Pharm. 78: 105– 114 [DOI] [PubMed] [Google Scholar]
- 44. Warren MK, Vogel SN. 1985. Bone marrow-derived macrophages: development and regulation of differentiation markers by colony-stimulating factor and interferons. J. Immunol. 134: 982– 989 [PubMed] [Google Scholar]
- 45. Waterfield M, Jin W, Reiley W, Zhang MY, Sun S-C. 2004. IKβ kinase is an essential component of the TPL-2 signaling pathway. Mol. Cell. Biol. 24: 6040– 6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Waterfield MR, Zhang M, Norman LP, Sun S-C. 2003. NF-κB1/p105 regulates lipopolysaccharide-stimulated MAP kinase signaling by governing the stability and function of the TPL-2 kinase. Mol. Cell 11: 685– 694 [DOI] [PubMed] [Google Scholar]
- 47. Yamamoto M, et al. 2004. Regulation of Toll / IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430: 218– 222 [DOI] [PubMed] [Google Scholar]