Summary
In the current investigation, we analyzed all the known small nucleolar RNAs (snoRNAs) in the deeply branching protozoan parasite Giardia lamblia for potential microRNAs (miRNAs) that might be derived from them. Two putative miRNAs have since been identified by Northern blot, primer extension, 3′-RACE and co-immunoprecipitation with Giardia Argonaute (GlAgo), and designated miR6 and miR10. Giardia Dicer (GlDcr) is capable of processing the snoRNAs into the corresponding miRNAs in vitro. Potential miR6 and miR10 binding sites in Giardia genome were predicted bioinformatically. A miR6 binding site was found at the 3′-untranslated regions (UTR) of 44 variant surface protein (vsp) genes, whereas a miR10 binding site was identified at the 3′-end of 159 vsp open-reading frames. Thirty-three of these vsp genes turned out to contain binding sites for both miR6 and miR10. A reporter mRNA tagged with the 3′ end of vsp1267, which contains the target sites for both miRNAs, was translationally repressed by both miRNAs in Giardia. Episomal expression of an N-terminal c-myc tagged VSP1267 was found significantly repressed by introducing either miR6 or miR10 into the cells and the repressive effects were additive. When the 2′-O-methyl antisense oligos (ASOs) of either miR6 or miR10 was introduced, however, there was an enhancement of tagged VSP1267 expression suggesting an inhibition of the repressive effects of endogenous miR6 or miR10 by the ASOs. Of the total 220 vsp genes in Giardia, we have now found 178 of them carrying putative binding sites for all the miRNAs that have been currently identified, suggesting that miRNAs are likely the regulators of VSP expression in Giardia.
Introduction
Giardia lamblia is a deeply branching, unicellular, flagellated parasitic protozoan that infects many mammalian species including humans. It is a major causative agent of waterborne outbreaks of diarrhea (Morrison et al., 2007). There are numerous unique features of this organism that make it a useful model for studying evolutionary eukaryotic biology (Adam, 2001). During the vegetative trophozoite stage, Giardia is binucleated with a highly plastic genome that contains few transcriptional regulatory elements like those in higher eukaryotes (Holberton and Marshall, 1995), suggesting a distinctively simplified transcriptional regulation system (Best et al., 2004; Yee et al., 2007). The untranslated regions (UTRs) in Giardia mRNAs are usually shorter than 30 nucleotides (nts) (Adam, 2001), which may restrict many aspects of gene regulation. For instance, there is no ribosome scanning in translation initiation in Giardia, making the mechanism of translational regulation more similar to that of the Archaea (Li and Wang, 2004). There are two sets of genes in Giardia whose expression is apparently regulated by mechanisms at specific stages of cell development. The encystation-specific genes in Giardia are only expressed as the trophozoites begin to differentiate into cysts (Davis-Hayman and Nash, 2002). Several transcription factors such as Myb are apparently involved in regulating the expression of encystation-specific genes (Morf et al., 2010; Su et al., 2011), but the mechanism behind this regulation is still unclear at the present time. Secondly, of a total of 220 annotated variant surface protein genes (vsps), only one is expressed on the trophozoite membrane surface at a given time (Adam, 2001), despite the fact that multiple VSP transcripts are apparently synthesized in the trophozoite (Prucca et al., 2008; Faghiri and Widmer, 2011). Regulation of VSP expression is thus most likely at the post-transcriptional level.
Recently, a micro(mi)RNA-mediated translational repression mechanism was found in Giardia (Saraiya and Wang, 2008; Li et al., 2011; Saraiya et al., 2011). miRNAs are small noncoding RNAs whose imperfect complementary pairing to the 3′-ends of target mRNAs regulates expression of more than 60% of the genes in metazoa through translational repression and mRNA destabilization (Lee et al., 1993; Bartel, 2004; Lewis et al., 2005; Filipowicz et al., 2008; Grimson et al., 2008; Friedman et al., 2009). Recent studies in measuring endogenous protein levels in response to altered miRNA expression levels indicated that a specific miRNA could modestly inhibit the production of hundreds of proteins in a cell, thus constituting a rather complex picture of post-transcriptional regulation (Baek et al., 2008; Selbach et al., 2008).
In metazoa, miRNAs are derived from long primary miRNA (pri-miRNA) precursors that are synthesized by RNA polymerase II in the nucleus (Cai et al., 2004; Kim et al., 2009). The pri-miRNAs are digested by the nuclear RNase III Drosha/Pasha (Winter et al., 2009) to release approximately 70 nt long hairpin-shaped products, the precursor miRNAs (pre-miRNAs), which are then exported to the cytoplasm by Exportin 5 (Lund et al., 2004). The pre-miRNA is subsequently cleaved by a second RNase III enzyme, Dicer, into approximately 22-nucleotide miRNA duplexes and bound to the Argonaute protein (Hutvágner et al., 2001; Knight and Bass, 2001). One of the two strands in the miRNA duplex is predominantly transferred with Argonaute to the RNA-induced silencing complex (RISC) (Schwarz et al., 2003), which mediates translation silencing, deadenylation or cleavage of the target mRNA, depending on the level of complementarity between the miRNA and the target site (Hutvágner and Zamore, 2002).
No homologue of Drosha/Pasha or Exportin 5 has been found in Giardia (Saraiya and Wang, 2008). An Argonaute homologue (GlAgo) and a Dicer homologue (GlDcr), however, were identified and found to be functional in this organism (Macrae et al., 2006; Saraiya and Wang, 2008; Li et al., 2011; Saraiya et al., 2011). The three dimensional structure of GlDcr was resolved by X-ray crystallography and found to generate small RNA products in the range of 25 to 27 nts (Macrae et al., 2006). In our recent studies, several potential miRNAs, each with an estimated size of 26 nts, were identified in Giardia (Saraiya and Wang, 2008; Li et al., 2011; Saraiya et al., 2011). One is derived from an un-annotated open reading frame (ORF) through two consecutive digestions of the transcript by GlDcr (Saraiya et al., 2011), whereas three others are derived from snoRNAs through an apparent single GlDcr digestion, thus bypassing the required actions of Drosha/Pasha and Exportin 5 (Saraiya and Wang, 2008; Li et al., 2011). The use of snoRNAs as precursors of miRNAs has since been demonstrated among a variety of metazoa (Ender et al., 2008; Politz et al., 2009; Scott et al., 2009; Taft et al., 2009) indicating that the snoRNAs are actually one of the sources of functional miRNAs preserved throughout evolution.
Tentative functional studies of the potential miRNAs in Giardia showed that the putative target sites for some of them are localized to the 3′-end of many genes including some VSP genes (Saraiya and Wang, 2008; Li et al., 2011; Saraiya et al., 2011), and two of the miRNAs are derived from snoRNAs. This observation suggests that miRNAs may play a role in regulating VSP expression in Giardia and that the snoRNAs in Giardia may provide a source of additional miRNAs, and some of them may be involved in regulating VSP expression.
There are 25 putative snoRNAs in Giardia (Niu et al., 1994; Yang et al., 2005; Luo et al., 2006). Four of them have already been found to be the precursors of miRNAs (Saraiya and Wang, 2008; Li et al., 2011). Here, we analyzed the remaining 21 snoRNAs for being potential miRNA precursors and successfully identified two of them acting as precursors of miR6 and miR10, respectively. Putative target sites for miR6 and miR10 were found among annotated ORFs, non-annotated ORFs and VSP genes. Thirty three of the VSP genes had target sites for both miR6 and miR10 arranged in the similar manner. Expression of a reporter transcript carrying the 3′ end of VSP1267, containing target sites for both miRNAs, was repressed by either miRNA. An N-terminal c-myc tagged VSP1267 was expressed in Giardia cells. The expression was repressed by the two miRNAs but enhanced by their corresponding 2′-O-methyl antisense oligos (ASO). Thus, multiple miRNAs are apparently involved in regulating VSP gene expression in Giardia, and several of them are derived from snoRNAs.
Results
Identification of small RNAs derived from 21 Giardia snoRNAs
Of the 21 snoRNAs identified in Giardia (Niu et al., 1994; Yang et al., 2005; Luo et al., 2006) but not yet been further analyzed as potential precursors for miRNAs, we found GlsR3 as a part of 5S ribosomal RNA, whereas GlsR4 and GlsR12 and GlsR18 and GlsR22 share the same sequences. There are thus only 18 distinct snoRNAs in Giardia remaining for investigation. To systematically screen for snoRNA-derived miRNAs, we performed Northern blot analysis of size-fractionated small RNA (< 200 nts) from Giardia trophozoites that could be derived from the 18 snoRNAs (Figure S1). A small RNA band of ~30 nt was detected in the Northern blots of 8 of the 18 snoRNAs; GlsR1, GlsR8, GlsR10, GlsR13, GlsR14, GlsR15, GlsR18 and GlsR24 (Figure S1).
The 8 snoRNAs that showed the possibility of being processed into a ~30 nt small RNA were further analyzed on the Northerns with smaller probes corresponding to the 5′-portion, middle portion or 3′-portion of the snoRNA to determine the potential location of the small RNA in the snoRNA. The results showed that the small RNAs are likely generated from the 3′-portion of GlsR1 and GlsR14, from the middle portion of GlsR13 and from the 5′-portion of GlsR10 (Figure S2). There were, however, also some ambiguities in the data from GlsR8, GlsR15, GlsR18 and GlsR24, where the small RNA band hybridized apparently with more than one portion of the probing snoRNA antisense sequence (Figure S2). We attributed these ambiguities to either overlapping sequences between the probes or the presence of more than one small RNA species derived from different parts of the snoRNA. These small RNAs underwent thus further primer extension and 3′-RACE analysis for sequence determination.
Size-fractioned RNAs (<200 nts) from Giardia trophozoites were used as templates for primer extension reactions. An in vitro synthesized snoRNA was used as a control template in order to distinguish nonspecific stops caused by secondary structures from the real stops. The relative location of each small RNA in the corresponding snoRNA was estimated based on the previous Northern blot data and a primer was synthesized to hybridize with the 3′ end of each presumed small RNA (Figure S2). This included the 3′-ends of GlsR1, GlsR8, GlsR14, GlsR15, GlsR18 and GlsR24, the middle portion of GlsR13 and the 5′-ends of GlsR8 and GlsR10. A snoRNA sequencing reaction was run in parallel with the primer extension reaction using the same primer to identify the precise stopping site of each primer extension product. The results showed that there were small RNA bands of ~26 nt from GlsR1, GlsR8, GlsR10, GlsR13 and GlsR18 (Figure S3A). But apparently no small RNA was derived from GlsR14, GlsR15 or GlsR24 (Figure S3B), which had since been eliminated from further study. A total of 9 small RNAs had been thus identified from the 5 snoRNAs by primer extension and were designated snoRNA-derived small RNA (sdRNA) 1 to 9 (Table 1).
Table 1. The miRNAs derived from 8 snoRNAs in Giardia.
| snoRNA | snoRNA lengtha |
Northern blot of overlapped portions |
Primer Extension |
Small RNA |
Position of small RNA in snoRNAc |
chromosomal localization |
Sequences of small RNAs (after 3′-RACE) |
Small RNA length |
Co-IP with GlAgo |
Designation | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′-portion | Mid-portionb | 3′-portion | ||||||||||
| GlsR1 | 85 nt (77 nt) | − | N/D | + | 3′-end | sdR1 | 54~81 | GLCHR03 (+) 412207..412234 |
GACGCGTGACGAAGTTTGTCGTATTCTG | 28 nt | + | miR6 |
| GlsR8 | 70 nt (67 nt) | + | N/D | + | 5′-end | sdR2 | 4~25 | GLCHR03 (−) 136711..136732 |
AGATGAAGAGAGATAAATCAGC | 22 nt | − | − |
| 3′-end | sdR3 | 43~70 | GLCHR03 (−) 136666..136693 |
TGAGGAAGAAACCGCCTTTCGTCTGACC | 28 nt | + | miR10 | |||||
| GlsR10 | 68 nt (65 nt) | + | N/D | − | 5′-end | sdR4 | 2~28 | GLCHR05 (+) 3181814..3181840 |
GAATGATGAGACGTGTTCCTCTCTCCT | 27 nt | − | − |
| GlsR13 | 100 nt (100 nt) |
+ | + | − | Mid-portion | sdR5 | 15~38 | GLCHR05 (−) 4314800..4314823 |
GATATGATGATTGGGAGCGACCTA | 24 nt | − | − |
| sdR6 | 14~38 | GLCHR05 (−) 4314800..4314824 |
AGATATGATGATTGGGAGCGACCTA | 25 nt | − | − | ||||||
| sdR7 | 13~35 | GLCHR05 (−) 4314803..4314825 |
GAGATATGATGATTGGGAGCGAC | 23 nt | − | − | ||||||
| GlsR14 | 69 nt (73 nt) | − | N/D | + | 3′-end | − | − | − | − | − | − | − |
| GlsR15 | 87 nt (61 nt) | − | N/D | + | 3′-end | − | − | − | − | − | − | − |
| GlsR18 | 109 nt (101 nt) |
− | + | + | 3′-end | sdR8 | 65~90 | GLCHR01 (+) 149540..149565 |
GTGGGGAGCGGATCCCGTCCATCCTC | 26 nt | − | − |
| sdR9 | 62~87 | GLCHR01 (+) 149537..149562 |
TAGGTGGGGAGCGGATCCCGTCCATC | 26 nt | − | |||||||
| GlsR24 | 125 nt (116 nt) |
+ | + | + | 3′-end | − | − | − | − | − | − | − |
The sequences and the lengths of Giardia snoRNAs used for probing Northern blot are based on Yang et al., 2005. After primer extension and 3′-RACE, the revised sequences of Giardia snoRNAs listed here were further determined (Figure S3). The numbers in brackets were the revised length of each snoRNA in the present study.
N/D: Not Determined.
The numbers showed the start and the end positions of each small RNA in the snoRNA. The snoRNA sequences designated here were by the previously reported sequences (Yang et al., 2005).
The precise 5′ end of each sdRNA and its precursor snoRNA was determined from the sequencing ladder (Figure S3A [red arrows] and Table 1). A 3′ RACE was performed to determine the 3′ ends of the snoRNAs and the corresponding sdRNAs (Figure S3A [blue arrows]). The precise sequence and the location of each sdRNA in the precursor snoRNA are shown in Figure S3A. With the exception of GlsR13, the sequences of all the snoRNAs studied turned out to require minor sequence refinements from the originally published data (Niu et al., 1994; Yang et al., 2005; Luo et al., 2006). These newly revised sequences of snoRNAs are also presented in Figure S3.
Co-immunoprecipitation of the sdRNAs with GlAgo
In order to verify if the 9 sdRNAs could be miRNAs, we immunoprecipitated tagged GlAgo from transfected Giardia to see if the protein could bring down any of the 9 sdRNAs (Saraiya et al., 2011). An N-terminal 2×HA-tagged GlAgo (HA-GlAgo) was over-expressed in Giardia and effectively immunoprecipitated from the cell lysate with anti-HA beads (Saraiya et al., 2011). Analysis of the TRIzol extracted RNA showed the presence of a clearly defined 26~30 nt small RNA band (Saraiya et al., 2011). We have previously shown by RT-qPCR that this band contains all the miRNAs (miR2, miR3, miR4 and miR5) we have previously identified in Giardia (Saraiya et al., 2011). This small RNA sample was used in the current study as template for RT-qPCR to amplify each of the 9 sdRNAs identified in the primer extension. The results, summarized in Figure S4A, indicate that only 3 out of the 9 sdRNAs show significant CT value differences between the GlAgo pull-down sample and the control. They are sdR1, sdR3 and sdR8 from GlsR1, GlsR8 and GlsR18, respectively. When the three RT-qPCR samples were further examined in gel electrophoresis, however, only sdR1 and sdR3 were shown in recognizable discrete bands with the anticipated size (Figures S4B and S4C). They were regarded as potential miRNAs and tentatively designated as miR6 and miR10.
miR6 and miR10 are processed by GlDcr from their respective precursor snoRNAs
The precursors of miR6 and miR10, GlsR1 and GlsR8, are box C/D snoRNAs (Yang et al., 2005). Their secondary structures were analyzed by MFOLD (Figure S5A and S5B), and each was found to fold into a hairpin structure with the corresponding miRNA sequence localized at the 3′-end in one of the two arms of the stem. This is the typical structure of a miRNA precursor that can be processed by Dicer to generate the mature miRNA duplex (Hutvágner et al., 2001; Knight and Bass, 2001). To test this possibility, N terminally tagged GlDcr (3×c-myc GlDcr) was pulled down by anti-c-myc beads and used in an in vitro dicing assay (Li et al., 2011). Full-length GlsR1 and GlsR8 were transcribed in vitro and each incubated with the GlDcr beads at 37°C for 16 hrs and 24 hrs. The products were probed on a Northern blot with the antisense sequence of miR6 and miR10, respectively. The results indicated that both GlsR1 and GlsR8 can be processed by the GlDcr beads to a small RNA band corresponding to the 28 nt size of miR6 and miR10, respectively (Figure S5C and S5D). During the GlDcr digestion, levels of the snoRNAs were significantly reduced, while the levels of the two small RNAs increased with incubation time, suggesting that GlDcr is responsible of converting the two snoRNAs to the two corresponding miRNAs. The intermediary products from GlDcr digestion of GlsR1 could not be clearly identified due to apparent band diffusion. But multiple intermediary RNA bands larger than miR10 were identified from GlDcr-beads digested GlsR8 (Figure S5D, asterisks). The sizes of these bands correspond to those of the fragments predicted from the secondary structure of GlsR8 after a digestion of the loop regions (shown in Figure S5B, arrows). The same sized fragments were also found in the previous primer extension of GlsR8 (Figure S3A, asterisks), supporting the possibility that the fragments are the intermediate products of GlDcr digestion of GlsR8.
Predicting potential target sites for the two putative miRNAs in Giardia using RNAhybrid
We used the RNAhybrid (ver2.2) program to predict the potential target sites of the two putative miRNAs in Giardia genome (no G:U wobbles, ΔG < −20 kcal/mol). The program has been used successfully to predict multiple potential binding sites of miRNAs in the 3′ UTRs in Drosophila (Rehmsmeier et al., 2004). It is similar to the miRanda program that we used in our previous studies (Saraiya and Wang, 2008; Saraiya et al., 2011; Li et al., 2011). Both methods depend on seed sequence complementation and free energy of binding between miRNA and mRNA for the prediction (Rehmsmeier et al., 2004; John et al., 2005). We tested both programs in predicting target sites in the current study, and the outcomes showed a 95% agreement.
Since the 3′ UTRs in Giardia genome are relatively short and a growing number of mRNAs in other organisms have now been found targeted by miRNAs within the ORFs rather than the 3′-UTRs (Forman et al., 2008; Tay et al., 2008; Chi et al., 2009), we set the domain of search for a target site to 150 nts with 100 nts upstream and 50 nts downstream from the stop codon of an ORF. A total of 5,901 ORFs in the GiardiaDB (version 2.2) (Aurrecoechea et al., 2009) were examined. The results showed 105 genes bearing potential target sites for miR6, of which 40 encoded hypothetical proteins. The other 65 annotated genes contained 44 genes encoding the VSPs (Table S2). The putative target sites in these 44 VSP genes have highly similar sequences and locations with the miR6 seed sequence located at the 3′-UTR of the transcripts immediately upstream from the polyadenylation site. Putative target sites for miR10 were identified in 260 genes with 56 for hypothetical proteins and 204 for annotated proteins, within which, 159 were VSPs (Table S3). The target sites in the 159 VSP mRNAs also share similar sequences and localizations inside the ORFs. None of the rest of the annotated genes carrying potential target sites for miR6 or miR10 appears to be related to VSP or to belong to any particular gene family, except for three high cysteine membrane proteins each carrying a potential target site for miR10 but not for miR6 (Tables S2 and S3).
Among a total of 220 annotated vsp genes in GiardiaDB (Aurrecoechea et al., 2009), a common C-terminal CRGKA sequence was identified in 159 of the encoded VSP proteins. They were recently designated as VSPs each assigned with a number for identification by Adam et al. (Adam et al., 2010). We adopted this system along with the previously designated names of VSPs in our current study (Table S4). For the rest of the VSPs with C-terminal sequences other than CRGKA (totaling 61), we took the liberty of assigning them as VSP-292 to VSP-352 by following the gene ID numbers to facilitate the subsequent analysis (Table S4).
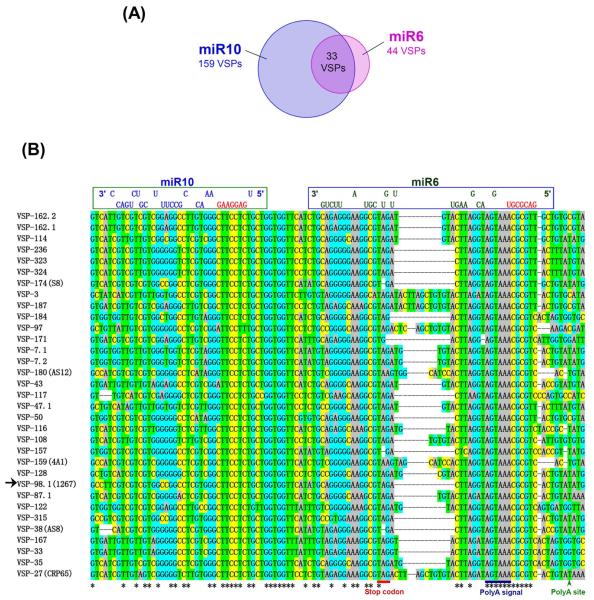
Between the two sets of vsp mRNAs carrying putative target sites for miR6 and miR10, 33 of them contain target sites for both miR6 and miR10 in a highly similar spatial arrangement (Figure 1A). The two sites do not overlap and maintain a ~16 nt gap in between. The sequences and localizations of these 33 dual target sites are extremely similar to one another and are aligned in Figure 1B for comparison.
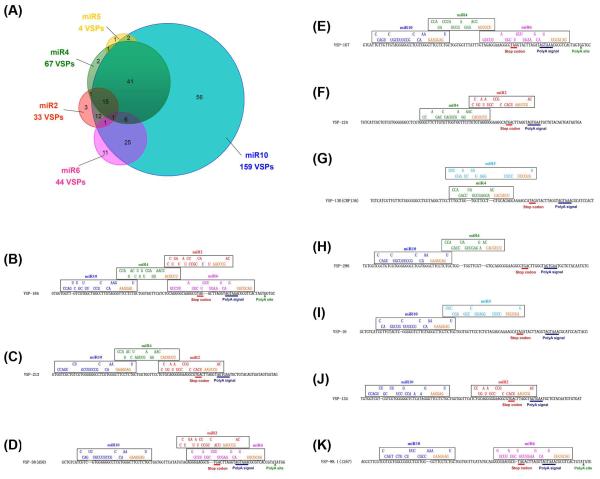
Figure 1. The vsp genes carrying putative target sites for both miR6 and miR10.
(A) Fortyfour vsp genes were predicted to have the miR6 binding site, whereas 159 vsp genes have putative target site for miR10. There are 33 vsp transcripts carrying the dual target sites for both miRNAs. (B) The sequence alignment of the 3′-ends of 33 vsp genes bearing the dual target sites for miR6 and miR10. The target site for miR10 is within the open reading frame near the stop codon and the target site for miR6 is largely in the 3′-UTR upstream from the poly-adenylation site. There is a ~16 nt linker between the two target sites. Potential binding of miR6 and miR10 to the target sites is demonstrated on the top. Sequences in red are the “seed sequences” of miR6 and miR10. The arrow shows vsp1267 chosen for further studies.
Two additional Giardia assemblages B and E have their genomic sequences made available at the present time. miR6 and miR10 are well conserved in assemblage E isolate P15. There are 156 annotated vsp genes in this isolate (Aurrecoechea et al., 2009) with 45 carrying the miR6 target site at the 3′ UTRs and 122 vsp genes showing the miR10 target site at the 3′-ends of the ORF, whereas 41 of them have the target sites for both miR6 and miR10 with a physical arrangement equivalent to that observed in the WB isolate (Figure 1B). The pattern of miR6 and miR10 binding to the 3′ ends of vsp genes in isolate P15 may thus highly resemble what we found in the WB isolate. For the assemblage B isolate GS, miR6 has a mutation in the seed sequence and two additional ones downstream, whereas miR10 is more conserved with only one mutation at the 3′ end. There are 39 annotated vsp genes in the GS isolate (Aurrecoechea et al., 2009), among which 17 carry the miR10 targeting site at the 3′-ends of the ORFs similar to that in isolate WB. However, there is no apparent target site in the 39 vsp genes for the mutated miR6 in the genome of GS isolate. Thus, the binding of miR6 and miR10 to the 3′ ends of vsp genes is apparently well conserved in P-15 isolate, but only the function of miR10 is preserved in GS isolate, suggesting that miRNAs may play similar, albeit somewhat different, roles in regulating vsp expression in the three Giardia isolates.
Functional studies of miR6 and miR10 on a reporter transcript carrying dual target sites
Among higher eukaryotes, two consecutive miRNA target sites within 40 nts (but not closer than 8 nts) have been shown to enable the two miRNAs to act cooperatively (Bartel, 2009). Cooperative miRNA function provides a mechanism by which repression can become more sensitive to small changes in miRNA expression levels. It greatly enhances the regulatory effect and utility of combinatorial miRNA expression.
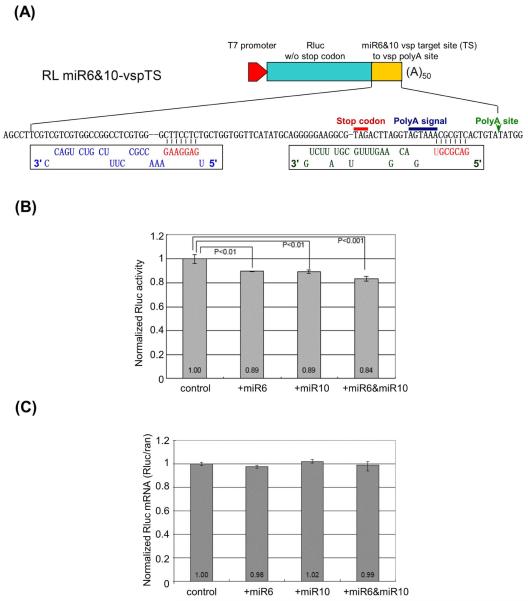
In order to verify whether miR6 and miR10 would act cooperatively or interfere with each other on the expression of the 33 vsp mRNAs carrying the non-overlapping dual target sites, we examined first whether the two putative miRNAs act as bona fide miRNAs in repressing translation of mRNAs in Giardia. One of the 33 vsps carrying the dual target site, vsp1267 (GiardiaDB gene ID: GL50803_112208) (VSP-98.1 by Adam et al., 2010), was chosen for further investigation (see Figure 1B, arrow). We constructed a plasmid, RL miR6&10-vspTS, where the original stop codon in an RLuc gene was removed and replaced with a 90 nt sequence from vsp1267 consisting of the dual target sites starting from the miR10 target site to the site of poly-adenylation (Figure 2A). The in vitro transcribed chimeric RLuc mRNA from this construct was transfected into Giardia trophozoites with chemically synthesized miR6 or miR10 and incubated at 37°C for 4 hours (Figure 2B). The RLuc activity was reduced by 11% in the presence of either miRNA. However, when both miR6 and miR10 were introduced, the RLuc expression was further reduced by ~16%, suggesting a coordinated action between miR6 and miR10 (Figure 2B). The extent of reduced expression in the study was relatively small, but it was repeated many times and statistical analysis of the data indicated that they are statistically significant (Figure 2B).
Figure 2. miR6 and miR10 inhibit the expression of an RLuc mRNA carrying the dual target sites.
(A) Diagram of the RL miR6&10-vspTS reporter constructs. The original stop codon of RLuc mRNA was replaced by a 90-nt-long sequence (yellow), which starts from the miR10 potential binding site to the poly(A) site in vsp1267 transcript. (B) Normalized Luciferase activity in RL miR6&10-vspTS transcript transfected Giardia trophozoites. The control was set at 1. The introduction of exogenous miR6 or miR10 (1 μg) represses RLuc activity by 11%. When both miR6 and miR10 were introduced, the RLuc expression was reduced by15%. The results and standard deviations were derived from three independent transfection experiments. The p-values indicated were calculated by two-tailed Student’s t-test. (C) RT-qPCR results showed the mRNA levels of the RL miR6&10-vspTS remained unchanged when exogenous miR6 or/and miR10 were introduced into the cells, suggesting that miR6 and miR10 act likely by translational repression. The results and standard deviations were from three independent transfection experiments.
Total RNAs were extracted with TRIzol from the Giardia trophozoites 4 hrs after the transfections described above. RT-qPCR was performed on the RNA samples for an estimation of the levels of chimeric RLuc mRNA. The results, presented in Figure 2C, indicate that the level of the mRNA was not significantly affected by the introduction of miR6, miR10 or both. Thus, the inhibitory effects of the two miRNAs are not by degrading the mRNA but rather likely by translational repression.
Expression of N-terminal 3×c-myc tagged VSP1267 in Giardia
To see if the miRNAs repress the expression of a VSP mRNA carrying the dual target sites, an N-terminal 3×c-myc tagged VSP1267 (myc-VSP1267) was cloned into a tetracycline (Tet)-inducible expression vector and the construct was transfected into Giardia trophozoites to be expressed (Figure S6A) (Sun et al., 2005).
The expression of myc-vsp1267 mRNA after Tet induction was confirmed by RT-PCR (Figure S6B). The expression of Tet-induced myc-VSP1267 protein was also demonstrated by a Western blot stained with an anti-c-myc antibody (Figure S6C). To localize the myc-VSP1267 protein expressed in Giardia trophozoites, a Mem-PER Eukaryotic Membrane Protein Extraction Reagent Kit (Thermo Scientific) was used to extract the membrane proteins from the cell lysate. The separated membrane and cytoplasmic proteins were compared on a Western blot and the results show that myc-VSP1267 can be found only in the membrane fraction of Tet-induced cells (Figure S6D). This is an interesting indication that myc-VSP1267 may be expressed on the membrane surface of Giardia trophozoites, though it could be also associated with the nuclear envelope endoplasmic reticulum cisternae during synthesis. The induced cells were then stained with an anti-c-myc-FITC antibody in an immuno-fluorescence assay that showed expression of myc-VSP1267 uniformly in and around the cells (Figure S6E), suggesting that this N-terminal c-myc tagged VSP may be expressed on the membrane surface of nuclei, endoplasmic reticulum as well as Giardia trophozoites.
miR6 and miR10 repress the expression of myc-VSP1267 in Giardia
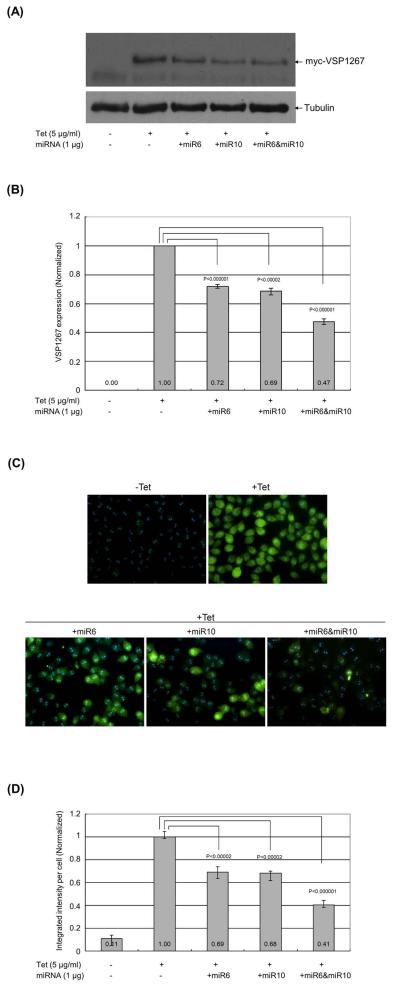
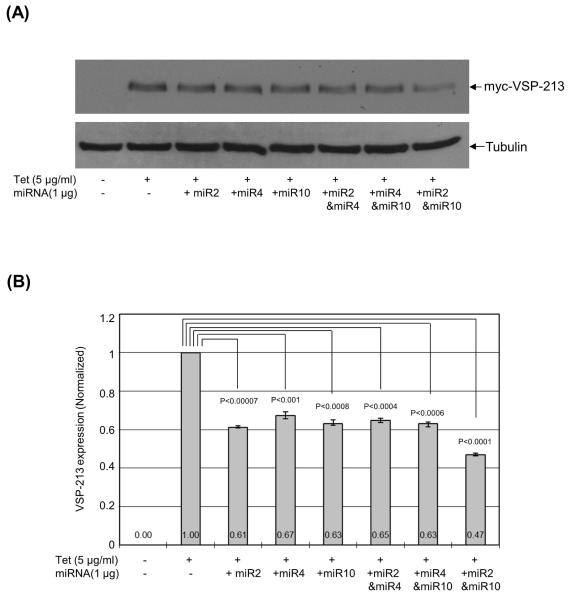
To study the potential effect of miR6 or/and miR10 on the expression of myc-VSP1267 in transfected Giardia trophozoites, the chemically synthesized miRNAs were introduced into the transfected trophozoites expressing myc-VSP1267. After a 16 hr incubation at 37°C, the cells were lysed and examined on Western blot. The results, shown in Figure 3A, indicate that miR6 repressed the expression of myc-VSP1267 by ~28% and miR10 by ~31% (Figure 3B). When both miR6 and miR10 were introduced into the cell, the expression of myc-VSP1267 was repressed by ~53% (Figure 3B). The data thus provided evidence that the two miRNAs are capable of each significantly repressing the expression of a VSP gene carrying the dual target sites in Giardia, and that the actions of the two miRNAs are cooperative.
Figure 3. miR6 and miR10 can repress the expression of myc-VSP1267 in Giardia.
(A) Expression of myc-VSP1267 after introducing miR6 or/and miR10 was examined in Western blot. Synthetic miR6, miR10 or miR6+miR10 was introduced into the trophozoites, which express Tet-inducible myc-VSP1267. The protein level of myc-VSP1267 was monitored after a 16 hr incubation at 37°C. Five independent experiments were carried out. (B) The quantitative data from (A) were shown with the expression of myc-VSP1267 without miRNAs set at 1. The p-values indicated were calculated by two-tailed Student’s t-test. (C) Immuno-fluorescence assay of myc-VSP1267-expressing trophozoites after introducing miR6 or/and miR10. The same cell samples used in Western blot analysis were used for immunostaining of myc-VSP1267. The cell images were taken and the mean integrated intensity of fluorescence per cell in each image was measured using the CellProfiler program (Lamprecht et al., 2007). For each sample, at least five images were taken. (D) The quantitative integrated intensity of fluorescence per cell was analyzed. The p-values indicated were calculated by two-tailed Student’s t-test.
The cells used in the experiment were stained with anti-c-myc antibody conjugated with FITC and examined in an immuno-fluorescence assay (Figure 3C). Images of approximately 50 cells were randomly taken in each experiment, and the integrated intensity of fluorescence in each image was measured using the CellProfiler program (Lamprecht et al., 2007). This total intensity was then divided by the number of cells in the image estimated with a phase contrast microscope, which resulted in the integrated fluorescence intensity per cell. These data, presented in Figure 3D, demonstrate that the fluorescence intensity is repressed by 31% by miR6, 32% by miR10 and 59% by miR6+miR10. The close agreement between the immunofluorescence data and those from the Western blot (Figure 3B) further confirms the cooperative repressive effect of the two miRNAs.
The functions of endogenous miR6 and miR10 are inhibited by introducing their 2′-O-methylated antisense oligos (ASO) into Giardia
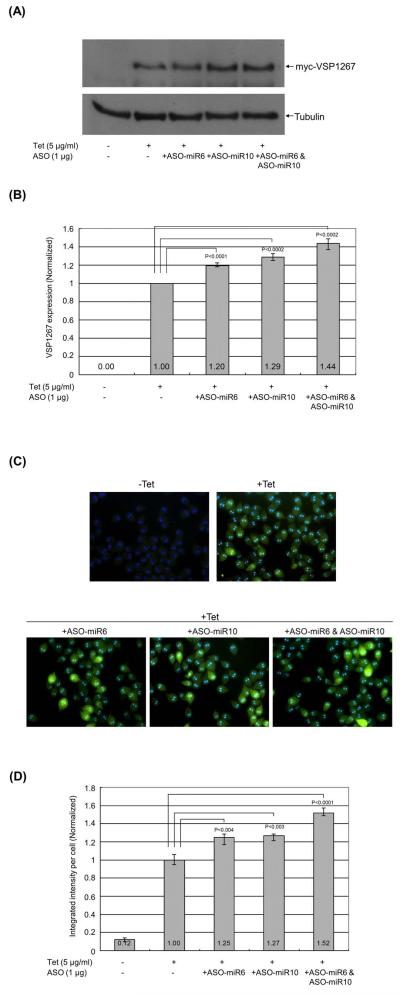
One of the most conclusive ways of demonstrating whether a miRNA represses the expression of a mRNA by hybridizing to the target site is by introducing its 2′-O-methylated antisense oligo (ASO) into the cell. The 2′-O-methylated antisense sequences are known to bind to the corresponding miRNAs in RISCs with very high affinity, effectively out-competing the target mRNA and relieving the inhibitory effect of the miRNA (Horwich and Zamore, 2008). ASOs of the two miRNAs under investigation were synthesized (ASO-miR6 and ASO-miR10), and introduced into the Giardia cells expressing myc-VSP1267 to see; (1) if there are endogenous miR6 and miR10 functions in Giardia; and (2) if these functions could be inhibited by the corresponding ASOs. One μg of ASO-miR6 or/and ASO-miR10 was introduced into the cells and the expression of myc-VSP1267 was examined by Western blot (Figure 4A). The expression of myc-VSP1267 was enhanced by 20% in the presence of ASO-miR6, 29% in the presence of ASO-miR10 and 44% in the presence of both ASO-miR6 and ASO-miR10 (Figure 4B). In the corresponding immuno-fluorescence assay (Figure 4C), there was an increased fluorescence intensity of 25% by ASO-miR6, 27% by ASO-miR10 and 52% by ASO-miR6+ASO-miR10 (Figure 4D). These closely related data from Western blot and immunofluorescence once again indicate strongly that there are in Giardia trophozoites endogenous miR6 and miR10 functions that jointly repress the expression of VSP1267 by binding to their respective target sites at the 3′-end of the mRNA.
Figure 4. The ASOs of miR6 and miR10 can suppress the endogenously repressed myc-VSP1267 expression.
(A) The expression of myc-VSP1267 was detected by Western blot after introducing ASO-miR6 or/and ASO-miR10 into the trophozoites. The level of myc-VSP1267 protein was monitored by Western blot after a 16 hr incubation at 37°C. Five independent experiments were carried out. (B) The quantitative data on the expression of myc-VSP1267 without ASOs was set at 1. The p-values indicated were calculated by two-tailed Student’s t-test. (C) The immuno-fluorescence assay of Giardia trophozoites expressing myc-VSP1267 after introducing ASO-miR6 or/and ASO-miR10. The same cell samples used in Western blot analysis were used for immunostaining of myc-VSP1267. The cell images were taken and the mean integrated intensity of fluorescence per cell in each image was measured using the CellProfiler program (Lamprecht et al., 2007). For each sample, at least five images were taken. (D) The quantitative integrated intensity of fluorescence per cell was analyzed. The p-values indicated were calculated by two-tailed Student’s t-test.
Repression of VSP-213 expression by miR2, miR4 and miR10 in Giardia
Another vsp gene (GiardiaDB gene ID: GL50803_114122) (VSP-213 by Adam et al., 2010), which was found in our previous study, carries the overlapping target sites for miR2 and miR4 (Saraiya et al., 2011). Our data from the present study showed that there is an additional miR10 binding site at the 3′-end of the ORF of this vsp transcript. It is 13 nt upstream from the miR2 target site. But its seed sequence overlaps with the 3′ end of miR4 binding site (see Figure 6C below). One would thus anticipate that miR10 would repress the expression of VSP213 and act cooperatively with miR2 but mutually exclusively with miR4.
Figure 6. Five miRNAs are involved in regulating the expression of 178 vsp genes.
(A) A scheme of miRNA-mediated regulation of VSP expression in Giardia by the 5 identified miRNAs is shown. More than half of the VSP mRNAs (105) carry multiple miRNA target sites without redundancy. (B-K) Bindings of multiple miRNAs to various vsp genes. Each vsp gene shown here has the target site(s) for different miRNAs arranged in different manners, suggesting the complexity and diversity of regulation of VSP expression by miRNAs. (B) VSP-184 has the target sites for 4 of the 5 miRNAs at the 3′ end. (C-E) VSP-213, VSP-38 (AS8) and VSP-167 have target sites for 3 of the 5 miRNAs at the 3′ end. (F-K) VSP-124, VSP-130 (CRP136), VSP-296, VSP-16, VSP-134 and VSP-98.1 (VSP1267) are targeted by 2 of the 5 miRNAs with the corresponding target sites positioned in different ways.
To verify this expectation, a construct for expressing an N-terminal 3×c-myc tagged VSP-213 (myc-VSP-213) was introduced into Giardia trophozoites. The expression was induced with Tet and detectable only in the membrane fraction in Western (Figure S7). When miR2, miR4 or miR10 was introduced into the cell, Western results from three independent transfection experiments indicated that miR2 repressed the expression of myc-VSP-213 by ~39%, miR4 by ~33% and miR10 by ~37% (Figures 5A and 5B). The introduction of miR2+miR4 or miR4+miR10, resulted in a repression of ~35% and ~37% respectively (Figures 5A and 5B), indicating a lack of cooperative effect. However, a combination of miR2 and miR10 caused more than half (~53%) reduction of myc-VSP-213 expression (Figures 5A and 5B). It is thus clearly demonstrated that the physical arrangement of multiple miRNA target sites at the 3′-end of a VSP mRNA determines whether the net repressive effect from multiple miRNA actions is additive or mutually exclusive.
Figure 5. Repression of myc-VSP-213 expression by miR2, miR4 and miR10.
(A) Expression of myc-VSP213 in transfected Giardia was monitored by Western analysis after introduction of the miRNAs into the trophozoites for a 16-hr incubation at 37°C. Three independent experiments were performed. (B) The data were analyzed quantitatively with the no miRNA control value set at 1. The p-values indicated were calculated by two-tailed Student’s t-test.
Discussion
The pathway using snoRNAs as precursors of miRNAs in Giardia
We have now analyzed all the snoRNAs identified in Giardia (Niu et al., 1994; Yang et al., 2005; Luo et al., 2006) and shown that 5 of them are precursors of functional miRNAs. They are GlsR17 as pre-miR2 (Saraiya and Wang, 2008), GlsR16 as pre-miR3 (Saraiya and Wang, 2008), GlsR2 as pre-miR5 (Li et al., 2011), GlsR1 as pre-miR6 and GlsR8 as pre-miR10. One common feature among these 5 snoRNAs is that they are all box C/D snoRNAs with readily identifiable C and D boxes. Another similar aspect is that all the mature miRNAs are derived from a 26 to 28 nt stretch at the 3′-ends of the snoRNAs, which is located in the stem region of the MFOLD predicted hairpin structures. The stem region in a hairpin structure constitutes the miRNA duplex released by Dicer digestion (Hutvágner et al., 2001; Knight and Bass, 2001), which has been observed by us in the GlDcr digestions of the snoRNAs (Saraiya and Wang, 2008; Li et al., 2011; Saraiya et al., 2011). The size of 26-28 nts among the mature miRNAs also agrees well with the predicted product size based on the crystal structure of GlDcr (Macrae et al., 2006). These data provide evidence that a biogenetic pathway of snoRNA-derived miRNA exists in Giardia. It may constitute an ancient means of gene regulation when Drosha/Pasha and Exportin 5 have not yet become involved in miRNA biogenesis (Matera et al., 2007; Brameier et al., 2010; Ono et al., 2011; Scott and Ono, 2011).
The GlDcr protein localizes to the cytoplasm of Giardia trophozoites (Prucca et al., 2008), which would require an export of the snoRNAs from the nucleolus (Saraiya and Wang, 2008) to the cytoplasm for processing. Mammalian snoRNAs are assembled and matured in snoRNPs associated with a variety of proteins including the phosphorylated export adaptor PHAX, the cap binding complex Ran and the exportin CRM1, suggesting a cytoplasmic phase during the maturation of snoRNP (Maxwell and Fournier, 1995; Filipowicz and Pogacić, 2002; Watkins et al., 2004). When U8 and U22 snoRNAs were injected into the cytoplasm of Xenopus oocytes, the snoRNAs were imported into the nuclei suggesting a mechanism for nuclear import of snoRNAs (Peculis, 2001). Additionally, the U8 pre-snoRNPs had a distinct distribution in the nucleoplasm and cytoplasm with an association with both nuclear import and export factors during maturation (Watkins et al., 2007). snoRNAs thus appear to have a cytoplasmic phase during their maturation. Ran and CRM1 homologues have been identified in the Giardia genome database (Chen et al., 1994). It is thus likely that Giardia snoRNAs can be exported to the cytoplasm by the exportin CRM1 complex during their biogenesis and subject to GlDcr degradation to produce miRNAs in the cytoplasm.
Since the identification of a snoRNA-derived miRNA in mammalian cells by Ender et al. and our simultaneous finding that miR2 was derived from GlsR17 in Giardia (Saraiya and Wang, 2008), more evidence has since been accumulated that snoRNA-derived miRNAs are a conserved feature in eukaryotes (Scott et al., 2009; Taft et al., 2009; Brameier et al., 2010; Li et al., 2011; Ono et al., 2011). Additionally, snoRNA and miRNA complexes could potentially cross talk with each other due to the presence of common protein components in their complexes (Scott and Ono, 2011). For instance, in human cells, the box C/D snoRNP component fibrillarin has been identified in the AGO2 complexes (Höck et al., 2007). Another box C/D snoRNP core protein, NOP56, was found in the AGO1 complexes (Hutvagner and Simard, 2008). Small RNA fragments derived from snoRNAs were associated with AGO7 in Arabidopsis and Ago1 in Schizosaccharomyces pombe (Taft et al., 2009). In humans, several box H/ACA snoRNA-derived fragments were identified in AGO complexes (Ender et al., 2008) and, more recently, several processed snoRNAs were found incorporated into RISC complexes (Brameier et al., 2010). Many miRNA precursors and mature miRNAs have also been detected in the nucleolus, whereas some of the miRNA precursors display sequence, structural and functional characteristics of snoRNAs (Scott et al., 2009; Ono et al., 2011). Thus, during evolution, the pathway of miRNA biogenesis without involving Drosha/Pasha and Exportin 5 seems to have been preserved and the use of snoRNAs as miRNA precursors has apparently continued in spite of the emergence of the more complicated canonical pathway.
Among the Giardia snoRNAs (Niu et al., 1994; Yang et al., 2005; Luo et al., 2006), only GlsR1 showed an rRNA modification at the snoRNA-targeted site (Yang et al., 2005). Eleven others each carries a single antisense element of rRNAs, but showed no corresponding rRNA modification (Yang et al., 2005). The rest of the 9 snoRNAs do not contain any antisense element of rRNAs, and could be classified as orphan snoRNAs. This suggests that most, if not all, of the snoRNAs may not function as guides of rRNA modifications but may perform some other functions such as being the precursors of miRNAs in Giardia (Saraiya and Wang, 2008; Li et al., 2011). Our present investigation also pointed out that 4 snoRNAs generate 7 sdRNAs other than miRNAs in Giardia (see Figure S4A and Table 1). They could play biological functions other than that of miRNAs in Giardia. snoRNA-derived small RNAs do not always function in translation regulation (Scott and Ono, 2011). A large family of box C/D snoRNAs in humans, the HBII-52s, has been found processed into smaller RNAs and shown to regulate splicing of several different transcripts (Kishore and Stamm, 2006). It is possible that some of the non-miRNA sdRNAs in Giardia could be involved in mRNA splicing, even though the Giardia genome is known to contain a relatively limited number of introns (Nixon et al., 2002; Morrison et al., 2007).
miRNA regulation of VSP expression
With a total of 220 annotated vsp genes identified in the Giardia genome (Aurrecoechea et al., 2009), only a single member of the VSP family is expressed on the membrane surface of a trophozoite at a given time (Adam, 2001). But the expression can randomly switch from one VSP to another at a rate of about once every 6 to 13 generations in an apparent effort in avoiding host immunity (Adam, 2001). Unlike the other eukaryotic pathogens, such as Trypanosoma brucei (Pays, 2005) and Plasmodium falciparum (Kyes et al., 2007), a sub-telomeric localization for vsp genes is relatively uncommon in Giardia, and is clearly not required for expression (Adam et al., 2010). Previous studies have suggested that the VSP switch occurs by an epigenetic mechanism involving histone acetylation of the promoter region (Kulakova et al., 2006). But the outcome from nuclear run-on and mRNA microarray of Giardia trophozoites indicated that VSP regulation should be at the post-transcriptional level (Prucca et al., 2008; Faghiri and Widmer, 2011). Prucca et al. proposed a vsp gene silencing mechanism by RNA interference (RNAi) (Prucca et al., 2008). Unfortunately, no RNAi activity has ever been demonstrated in Giardia trophozoites, and there is no evidence that GlDcr can process the long double-stranded (ds) RNA proposed by Prucca et al. (Prucca et al., 2008). Furthermore, the only Argonaute homologue in Giardia, GlAgo, has been found without slicer activity (Li et al., 2011; Saraiya et al., 2011).
Our technical success in expressing an N-terminally tagged VSP in the transfected Giardia trophozoites has enabled us to monitor the effects of miR2, miR4, miR6 and miR10 directly on VSP expression in Giardia trophozoites. The results confirmed that each of the miRNAs, known to repress the translation of an RLuc transcript tagged with their respective target sites in Giardia (Saraiya et al., 2008; Saraiya et al., 2011; Figure 2), can also repress the expression of a vsp gene carrying the corresponding target site (Figures 3 and 5). Furthermore, while expression of the reporter was repressed by miR 2, 4, 6 and 10 by 15% (Saraiya et al., 2011), 15% (Saraiya et al., 2011), 11% (Figure 2B) and 11% (Figure 2B) respectively, expression of the corresponding VSPs under the same experimental conditions was repressed by 39% (Figure 5B), 33% (Figure 5B), 28% (Figure 3B) and 31% (Figure 3B). This preferential inhibition of VSP expression suggests authenticity of VSP expression as the bona fide target of action of these miRNAs. The significant enhancement of VSP1267 expression by the ASO of either miR6 or miR10 (Figure 4) further supports the notion that both endogenous miR6 and miR10 are functional and repress VSP1267 expression in Giardia trophozoites. Quantative PCR of the reporter mRNA carrying the target sites for miR6 and miR10 showed no effect of the two miRNAs on mRNA levels (Figure 2C), further indicating that miR6 and miR10 act by repressing translation of the transcript rather than promoting its degradation.
The coordinated effects of different miRNAs on VSP expression
In higher eukaryotes, most biological processes are influenced by miRNAs (Bartel, 2009). Each miRNA may have hundreds of evolutionarily conserved targets (Baek et al., 2008; Chi et al., 2009), whereas multiple miRNAs have been found to modulate the expression of a single gene by directly binding to the 3′-UTR of the mRNA (Wu et al., 2010).
We have thus far characterized 5 miRNAs in Giardia involved in regulating VSP expression. Within this limited number of miRNAs, miR4 turns out incapable of acting cooperatively with miR2 or miR10 in repressing the expression of VSP-213 due to the partially overlapping target sites. miR10, on the other hand, is capable of acting cooperatively with miR6 and miR2 in repressing the expression of VSP1267 and VSP-213, because its target site locates upstream from those for miR6 and miR2 by 16 and 13 nts, respectively. Similar arrangements of dual miRNA target sites have been found among many other vsp genes with 17 of them carrying the dual sites for miR2 and miR4 (Saraiya et al., 2011), 63 for miR4 and miR10, 29 for miR2 and miR10, and 33 for miR6 and miR10 (Table S4). Expressions of these vsp genes are thus dependent not only on the presence of these miRNA target sites, but also on their physical arrangements in determining the outcome from multiple miRNA actions. An example of 10 randomly chosen vsp genes carrying various spatial arrangements of target sites for the 5 miRNAs is presented in Figure 6.
A summation of the analytical data on the number of putative target sites for the 5 identified miRNAs at the 3′ends of 220 annotated vsp genes is now presented in Figure 6A and Table S4. Of the 220 genes, 178 of them carry at least one putative target site for one of the miRNAs. Among the 178 genes, 105 have two or more target sites for multiple miRNAs, which are, interestingly, never redundant. The single target site identified in the other 73 genes is mostly for binding to miR10, which could be attributed to the relatively high number of vsp genes (159) carrying the miR10 target site. As for the remainder 42 vsp genes, which have not been found to possess any potential target site for the 5 miRNAs, it is likely that with increasing numbers of miRNAs identified in Giardia in the foreseeable future, all the vsp genes may turn out to each carry multiple target sites for the actions from multiple miRNAs. The translational repression on each vsp transcript accumulated from the actions of multiple miRNAs could thus completely inhibit the expression of most of the VSPs leaving but one of them to be effectively expressed on the surface of cell membrane by a yet un-identified mechanism. Regulation of VSP expression at any given moment may be dependent on the individual availability of each miRNA involved in the controlling machinery. One could imagine an enrichment of certain miRNAs and depletion of some other miRNAs once every 6 to 13 generations during the growth of Giardia trophozoites that would allow the selective disappearance of an existing VSP and the emergence of a new VSP. The detailed mechanisms behind this potential gene regulation would be an extremely interesting, albeit complex, subject for further investigation.
Experimental procedures
Oligonucleotides
All the DNA and RNA primers used in this study were synthesized by IDT. The names and sequences of these primers are listed in Table S1.
Cell culture and transfection
Giardia lamblia (WB clone C6, ATCC50803) trophozoites were grown anaerobically in plastic culture tubes at 37°C in the modified TYI-S-33 medium supplemented with antibiotics, as described previously (Keister, 1983). Transfections of Giardia trophozoites were carried out using electroporation. Cells at mid-to-late logarithmic phase were harvested by chilling the culture tubes on ice for 10 min and collected by centrifugation (1,000 g at 4°C for 10 min). The cells were washed twice in phosphate buffered saline (PBS), once in electroporation buffer (Cytomix buffer: 10 mM K2HPO4-KH2PO4 (pH 7.6), 25 mM HEPES (free acid), 120 mM KCl, 0.15 mM CaCl2, 2 mM EGTA, 5 mM MgCl2, 2 mM ATP, 4 mM glutathione), and then suspended to a final concentration of 2.5 × 107 cells/ml in Cytomix buffer. An aliquot of the concentrated cell suspension (400 μl, containing 107 cells) was transferred to a 0.2-cm-gap electroporation cuvette (Bio-Rad) and placed on ice. Capped mRNA (4 μg), plasmid DNA (50 μg), yeast tRNA (125 μg), synthetic 5′-phosphate-miRNA (1 μg) or synthetic 2′-O-methylated antisense oligo of miRNA (1 μg) was added to the cell suspension. The cells were immediately subjected to electroporation using a Bio-Rad Gene Pulser Xcell (Voltage: 450 V, Capacitance: 500 mF, Resistance: ∞). The electroporated cells were incubated on ice for 10 min, added to pre-warmed culture medium, and incubated at 37°C.
RNA isolation, Northern blot analysis and primer extension
Total RNA was isolated from Giardia trophozoites using TRIzol (Invitrogen). Size-fractioned RNAs (<200 nts) were isolated using the High Pure miRNA Isolation kit (Roche). The very small RNAs (25~30 nts) were purified by gel fractionation from the size-fractioned RNAs (<200 nts). In brief, size-fractionated RNAs (<200 nts) were separated in a 12% denaturing polyacrylamide gel with 8 M Urea in 1×TBE (Tris-Borate-EDTA buffer). The region containing small RNAs of 25~30 nts was excised, eluted off the gel using 300 μl 1×TE buffer with 0.3 M NaAc (pH5.3) and 1U/μl SUPERase·In (Ambion), and incubated at 37°C with shaking for at least 4 hrs. It was then followed by ethanol precipitation.
For Northern blot, MAXIscript Kit (Ambion) was used to incorporate α-32P-UTP (Perkin Elmer) into the RNA probes. Fifteen micrograms of size-fractionated RNAs (<200 nts) were separated in a 12% denaturing polyacrylamide gel, capillary blotted onto a Hybond-N membrane (Amersham) and followed by UV light irradiation. Blots were hybridized with the radiolabeled probes overnight at 42°C in a solution containing 50% formamide, 0.5% SDS, 5×SSC (150 mM NaCl, 15 mM sodium citrate), 5×Denhardt’s solution, 100 μg/ml denatured salmon sperm DNA, washed twice with 2×SSC and 0.1% SDS for 15 min at room temperature followed by two washes with 0.1×SSC and 0.1% SDS for 15 min at 42°C. The hybridization signal was monitored with a PhosphorImager screen and scanned with a GE Storm 860 (Amersham).
For primer extension assays, Primer Extension System-AMV Reverse Transcriptase (Promega) was used. A cDNA sequencing ladder was obtained using fmol DNA Cycle Sequencing System (Promega). Primers (see Table S1) used for cDNA sequencing and primer extension was PAGE-purified and γ-32P-ATP (Perkin Elmer) end-labeled using T4 polynucleotide kinase (NEB). The cDNA thus synthesized was analyzed by electrophoresis in 8% polyacrymide/8 M urea gel along with the cDNA sequencing ladder. The gel was exposed to a PhosphorImager and scanned with GE Storm 860 (Amersham).
3′-RACE of snoRNAs and sdRNAs
Giardia size-fractioned RNAs (<200 nts) and the very small RNAs (25~30 nts) were polyadenylated using E. coli poly-(A) polymerase (NEB) for the 3′-RACE of snoRNAs and sdRNAs, respectively. Reverse transcription was carried out using degenerated poly(dT) primer. A 20 nt universal primer and a snoRNA or small RNA specific primer starting from and complementary to the 5′ end of the snoRNA or sdRNA defined by the previous primer extension were used for amplification. The PCR product was cloned into a pGEM-T Easy vector using the pGEM-T Easy kit (Promega). Mutiple E. coli colonies containing the inserts were collected and the plasmid DNA was isolated and sequenced. The most abundant sequences were taken to represent the correct 3′ end of each snoRNA and sdRNA.
GlAgo immunoprecipitation and detection of snoRNA-derived small RNAs by RT-qPCR
The N-terminal HA tagged GlAgo and the associated RNAs were immunoprecipitated as previously described (Saraiya et al., 2011). RT-qPCR of sdRNAs was performed (Li et al., 2011; Saraiya et al., 2011). Briefly, the extracted ~26-30 nt RNA band co-immunoprecipitated with GlAgo was extracted from the gel and reverse transcribed using the SuperScript III RT (Invitrogen) with the RT primer. The cDNA was then amplified using iQ Supermix (Bio-Rad), with a forward primer, a reverse primer and a TaqMan probe.
In vitro dicing assay
N terminal tagged GlDcr (3×c-myc GlDcr) was over-expressed in Giardia and pulled down by anti-c-myc beads as described (Li et al., 2011). The in vitro dicing assay was carried out using 2 μl of the beads, which were incubated at 37°C with 500 ng of in vitro transcribed GlsR1 and GlsR8 in the presence of 3 mM MgCl2, 30 mM NaCl and 100 mM Hepes, pH 7.5. The final volume of each reaction was 10 μl. Reactions were stopped by adding 10 μl of formamide gel loading buffer. RNA fragments were resolved in denaturing polyacrylamide (12%) gel electrophoresis. The digested products were transferred onto a Hybond-N membrane (Amersham). The anti-miR6 or anti-miR10 sequence was γ-32P-ATP (Perkin Elmer) end-labeled and used as the probe in a Northern.
Cloning of the Rluc-reporter and expression of the in vitro transcript in miRNA assay
We have previously constructed a plasmid pRL, which carries an RLuc reporter gene under the control of a T7 promoter and a multiple cloning site followed by A50 at the 3′ end (Saraiya and Wang, 2008). The original stop codon was removed by PCR amplification using the primer set RL-seq and VSP1267TS-R1. The PCR product was gel purified and amplified again using the primer set RL-seq and VSP1267TS-R2 and eventually cloned into pGEM-T Easy (Promega). The clones containing the inserts were sequenced and sub-cloned into pRL using the BsrGI and XbaI restriction sites. The plasmid DNA was amplified using the primer set T7RL-F and RL-R in PCR. The purified PCR product was used as template for making capped RL miR6&10-vspTS mRNA. The latter was transfected into Giardia trophozoites and a miRNA assay was performed as previously described (Li et al., 2011).
3 × Myc-VSP1267 and 3 × Myc-VSP-213 cloning and expression
Genes encoding VSP1267 and VSP-213 were PCR amplified from Giardia genomic DNA using the AatII-VSP-F primer and SalI-VSP-R primer. The product was cloned into pGEM-T Easy (Promega), sequenced and sub-cloned into the pNlop4-myc vector, a derivative of the pNlop3-GTetR vector kindly provided to us by Dr. Zac Cande of UC Berkeley (Sun et al., 2005), using AatII and SalI restriction sites. The plasmid (pNlop4-VSP1267 or pNlop4-VSP-213) was transfected into Giardia trophozoites as described above and the transfectants were selected with 200 μg/ml G418. The selected cells were incubated with 5 μg/ml of tetracycline at 37°C for 24 hrs to induce expression of the VSPs, which were assayed by Western blot.
Western blot
The tetracycline-induced cells were treated with miRNAs or ASOs, cooled on ice, pelleted and washed once with cold PBS. For total protein assay, the cells were lysed with the lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 1×Halt protease inhibitor cocktail (Thermo Scientific)). After a 30 min incubation at 4°C, the cell lysate was cleared by centrifugation at 16,000 g at 4°C for 20 min. Mem-PER Eukaryotic Membrane Protein Extraction Reagent Kit (Thermo Scientific) was used to extract the membrane proteins from the cell lysate. The separated membrane and cytoplasmic proteins were then purified and concentrated using Pierce SDS-PAGE Sample Prep Kit (Thermo Scientific). The concentration of protein in each sample was quantified using the Bradford method (Bio-Rad). For SDS-PAGE separation, 25 μg of protein from each sample was used. Protein was transferred to a PVDF membrane (Bio-Rad) for detection with the anti-c-myc antibody (Invitrogen). It was subsequently stripped using Restore Western Blot Stripping Buffer (Thermo Scientific) and re-blotted with anti-Tubulin antibody (Sigma) for loading control. Protein bands were quantified using the BioRad Quantity One software package.
Immunofluorescence assay
For detecting myc-VSP expression, the harvested Giardia cells were re-suspended in 200 μl of modified TYI-S-33 culture medium, placed on a cover slip pretreated with 0.1% poly-L-lysine, and incubated at 37°C for 30 min to allow the cells to adhere. The cells were then fixed in 4% paraformaldehyde at room temperature for 30 min, washed with PBS three times for 5 min each and blocked in 5% BSA at room temperature for 60 min. The anti-myc-FITC antibody (Invitrogen) was 1:500 diluted in 1% BSA and incubated with the fixed cells at room temperature for 60 min in a dark box. The cells were then washed three times with PBS for 5 min each. The cover slip was placed face down on clean glass slides with 1 drop of Vectashield (Vector Labs) mounting media with DAPI (6-diamidino-2-phenylindole) and sealed with paraffin wax. The cells were examined using a Nikon TE2000E motorized inverted microscope equipped with 60×bright field and epifluorescence optics. Images were acquired with the NIS-Elements Advanced Research software (Nikon) and analyzed with CellProfiler (Lamprecht et al., 2007).
Supplementary Material
Acknowledgements
We thank Professor Zacheus Cande of UC Berkeley for the pNlop4 vector. This work was supported by the National Institutes of Health [R01 AI-30475].
References
- Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam RD, Nigam A, Seshadri V, Martens CA, Farneth GA, Morrison HG, et al. The Giardia lamblia vsp gene repertoire: characteristics, genomic organization, and evolution. BMC Genomics. 2010;11:424. doi: 10.1186/1471-2164-11-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, et al. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis. Nucleic Acids Res. 2009;37:D526–530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs, genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs, target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best AA, Morrison HG, McArthur AG, Sogin ML, Olsen GJ. Evolution of eukaryotic transcription, insights from the genome of Giardia lamblia. Genome Res. 2004;14:1537–1547. doi: 10.1101/gr.2256604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions, expanding the range of regulatory RNAs. Nucleic Acids Res. 2010;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen B,R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Chern Y, Ong SJ, Tai JH. Molecular cloning and characterization of a ras-related gene of ran/tc4/spi1 subfamily in Giardia lamblia. J Biol Chem. 1994;269:17297–17304. [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Hayman SR, Nash TE. Genetic manipulation of Giardia lamblia. Mol Biochem Parasitol. 2002;122:1–7. doi: 10.1016/s0166-6851(02)00063-4. [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Faghiri Z, Widmer G. A comparison of the Giardia lamblia trophozoite and cyst transcriptome using microarrays. BMC Microbiol. 2011;11:91. doi: 10.1186/1471-2180-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Pogaciy V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of posttranscriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel D. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, et al. Proteomic and functional analysis of Argonaute-containing mRNA protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holberton DV, Marshall J. Analysis of consensus sequence patterns in Giardia cytoskeleton gene promoters. Nucleic Acids Res. 1995;23:2945–2953. doi: 10.1093/nar/23.15.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Zamore PD. Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat Protoc. 2008;3:1537–1549. doi: 10.1038/nprot.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. miRanda algorithm: Human MicroRNA targets. PLoS Biol. 2005;3:e264. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakova L, Singer SM, Conrad J, Nash TE. Epigenetic mechanisms are involved in the control of Giardia lamblia antigenic variation. Mol Microbiol. 2006;61:1533–1542. doi: 10.1111/j.1365-2958.2006.05345.x. [DOI] [PubMed] [Google Scholar]
- Kyes SA, Kraemer SM, Smith JD. Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family. Eukaryot Cell. 2007;6:1511–1520. doi: 10.1128/EC.00173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71–75. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li L, Wang CC. Capped mRNA with a single nucleotide leader is optimally translated in a primitive eukaryote, Giardia lamblia. J Biol Chem. 2004;279:14656–14664. doi: 10.1074/jbc.M309879200. [DOI] [PubMed] [Google Scholar]
- Li W, Saraiya AA, Wang CC. Gene regulation in Giardia lambia is mediated by microRNA derived from small nucleolar RNA. PLoS Negl Trop Dis. 2011;5:e1338. doi: 10.1371/journal.pntd.0001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Luo J, Zhou H, Chen CJ, Li Y, Chen YQ, Qu LH. Identification and evolutionary implication of four novel box H/ACA snoRNAs from Giardia lamblia. Chin Sci Bull. 2006;51:2451–2456. [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Morf L, Spycher C, Rehrauer H, Fournier CA, Morrison HG, Hehl AB. The transcriptional response to encystation stimuli in Giardia lamblia is restricted to a small set of genes. Eukaryot Cell. 2010;9:1566–1576. doi: 10.1128/EC.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Niu XH, Hartshorne T, He XY, Agabian N. Characterization of putative small nuclear RNAs from Giardia lamblia. Mol. Biochem Parasitol. 1994;66:49–57. doi: 10.1016/0166-6851(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Nixon JE, Wang A, Morrison HG, McArthur AG, Sogin ML, Loftus BJ, Samuelson J. A spliceosomal intron in Giardia lamblia. Proc Natl Acad Sci USA. 2002;99:3701–3705. doi: 10.1073/pnas.042700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Peculis BA. snoRNA nuclear import and potential for cotranscriptional function in pre-rRNA processing. RNA. 2001;7:207–219. doi: 10.1017/s1355838201001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prucca CG, Slavin I, Quiroga R, Elías EV, Rivero FD, Saura A, et al. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456:750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Li W, Wang CC. A newly identified microRNA that regulates variant surface protein gene expression in Giardia lamblia. RNA. 2011;17:2152–2164. doi: 10.1261/rna.028118.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA Precursors with Box H/ACA snoRNA Features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Su LH, Pan YJ, Huang YC, Cho CC, Chen CW, Huang SW, et al. A novel E2F-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J Biol Chem. 2011;286:34101–34120. doi: 10.1074/jbc.M111.280206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CH, Su LH, Gillin FD. Influence of 5′ sequences on expression of the Tet repressor in Giardia lamblia. Mol Biochem Parasitol. 2005;142:1–11. doi: 10.1016/j.molbiopara.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Lührmann R. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol Cell Biol. 2007;27:7018–7027. doi: 10.1128/MCB.00516-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Lührmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- Yang CY, Zhou H, Luo J, Qu LH. Identification of 20 snoRNA-like RNAs from the primitive eukaryote, Giardia lamblia. Biochem Biophys Res Commun. 2005;328:1224–1231. doi: 10.1016/j.bbrc.2005.01.077. [DOI] [PubMed] [Google Scholar]
- Yee J, Tang A, Lau WL, Ritter H, Delport D, Page M, et al. Core histone genes of Giardia intestinalis: genomic organization, promoter structure, and expression. BMC Mol Biol. 2007;8:26. doi: 10.1186/1471-2199-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.