SUMMARY
Genomic profiling has identified a subtype of high-risk B-progenitor acute lymphoblastic leukemia (B-ALL) with alteration of IKZF1, a gene expression profile similar to BCR-ABL1-positive ALL and poor outcome (Ph-like ALL). The genetic alterations that activate kinase signaling in Ph-like ALL are poorly understood. We performed transcriptome and whole genome sequencing on 15 cases of Ph-like ALL, and identified rearrangements involving ABL1, JAK2, PDGFRB, CRLF2 and EPOR, activating mutations of IL7R and FLT3, and deletion of SH2B3, which encodes the JAK2 negative regulator LNK. Importantly, several of these alterations induce transformation that is attenuated with tyrosine kinase inhibitors, suggesting the treatment outcome of these patients may be improved with targeted therapy.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy, and relapsed B-lineage ALL remains a leading cause of cancer death in young people (Pui et al., 2008). B- ALL is characterized by recurring chromosomal abnormalities including aneuploidy, chromosomal rearrangements (e.g. ETV6-RUNX1, BCR-ABL1 and TCF3-PBX1), and rearrangements of MLL and CRLF2 (Pui et al., 2008; Mullighan et al., 2009a; Russell et al., 2009a; Harvey et al., 2010a; Yoda et al., 2010). However, leukemic cells from many patients with relapsed B-ALL lack known chromosomal alterations. Therefore, identifying the full repertoire of genetic lesions in high-risk ALL is essential to improve the treatment outcome of this disease.
Genome-wide analyses have identified genetic alterations targeting transcriptional regulators of lymphoid development (PAX5, EBF1 and IKZF1) in over 60% of B-ALL patients (Kuiper et al., 2007; Mullighan et al., 2007; Mullighan et al., 2009b). IZKF1 alteration is a hallmark of Philadelphia chromosome (Ph+) ALL with BCR-ABL1 fusion (Mullighan et al., 2008; Iacobucci et al., 2009), and is also associated with poor outcome in both BCR-ABL1-positive and negative ALL (Den Boer et al., 2009; Martinelli et al., 2009; Mullighan et al., 2009b). Notably, IKZF1 mutated, BCR-ABL1-negative cases commonly exhibit a gene expression profile similar to BCR-ABL1-positive ALL, and these cases are referred to as “Ph-like ALL” (Den Boer et al., 2009; Mullighan et al., 2009b). Ph-like ALL comprises up to 15% of pediatric B-ALL, and these patients have a higher risk of relapse compared to other BCR-ABL1-negative patients, with 5-year event-free survival rates of 63% and 86%, respectively (unpublished data from the Children’s Oncology Group). Approximately 50% of Ph-like patients harbor rearrangements of CRLF2 (CRLF2r) (Harvey et al., 2010a), with concomitant Janus kinase (JAK) mutations detected in approximately 50% of CRLF2r cases (Mullighan et al., 2009a; Russell et al., 2009a; Harvey et al., 2010a; Yoda et al., 2010). However, the genetic alterations responsible for activated kinase signaling in the remaining Ph-like cases are unknown. To identify the genetic basis of this subtype, we performed transcriptome and whole genome sequencing on tumor and matched normal material from 15 patients with Ph-like ALL.
RESULTS
Chromosomal rearrangements in Ph-like ALL
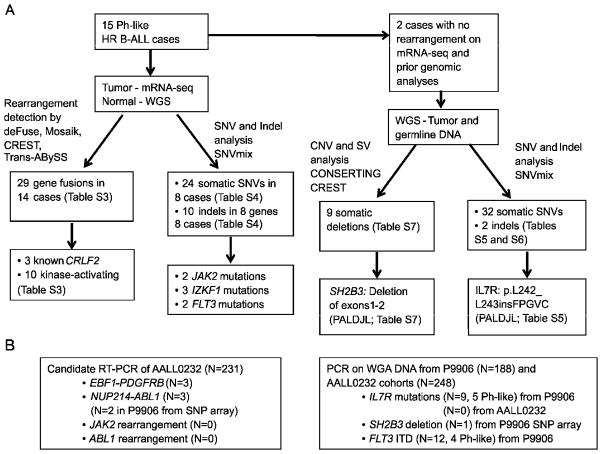
To identify genetic alterations in Ph-like ALL, we performed paired-end messenger RNA sequencing (mRNA-seq) on 15 B-ALL cases that were identified as Ph-like using prediction analysis of microarrays (PAM; Table 1 and S1) (Tibshirani et al., 2002) and Recognition of Outliers by Sampling Ends (ROSE) (Harvey et al., 2010b). Importantly, the gene expression profile of Ph-like ALL determined by limma (Linear Models for Microarray Analysis; Table S2) (Smyth, 2004) exhibited highly significant enrichment for the previously described signature of high risk, IKZF1-deleted ALL (Mullighan et al., 2009b) (data not shown). Whole genome sequencing (WGS) of tumor DNA was also performed for two cases lacking kinase-activating rearrangements on analysis of mRNA-seq data. We used multiple complementary analysis pipelines including deFuse (McPherson et al., 2011), Mosaik (Marth, 2010), CREST (Wang et al., 2011), CONSERTING (Zhang et al., 2012) and Trans-ABySS (Robertson et al., 2010) to identify rearrangements, structural variations and sequence mutations. Putative somatic sequence variants were identified by comparing tumor data to WGS data of matched normal DNA, and were validated using orthogonal sequencing methods. Overviews of methodology and findings are provided in Figures 1 and S1.
Table 1.
Chromosomal rearrangements detected in high-risk B-lineage ALL.
| Sample ID | Cohort | Rearrangement | Sex | Age (yr) | WCC × 109L | Key lesions | Karyotype |
|---|---|---|---|---|---|---|---|
| PAKTAL | P9906 | STRN3-JAK2* | Female | 12.2 | 478 | IKZF1 deletion and p.Leu117fs mutation | N/A |
| PAKKCA | P9906 | EBF1-PDGFRB* | Male | 11.7 | 236.4 | IKZF1 (IK6); EBF1 deletion; PAX5 inversion*; CDKN2A/CDKN2B deletion | 46, XY, del(6)(q13q23), del(9) (p22)[20] |
| PAKVKK | P9906 | NUP214-ABL1* | Male | 14.4 | 220.7 | IKZF1 p.Ser402fs mutation; PAX5 deletion; CDKN2A/CDKN2B deletion | N/A |
| PALIBN | P9906 | IGH@-EPOR* | Male | 14.3 | 29.9 | IKZF1 e1–5 deletion; CDKN2A/CDKN2B deletion | N/A |
| PAKYEP | P9906 | BCR-JAK2* | Male | 2.7 | 958.8 | IKZF1 (IK6); EBF1 deletion; PAX5 deletion and p.Gly24Arg mutation; CDKN2A/CDKN2B deletion | 47, XY,+2, del(2)(p23), t(3;22;9) (p12; q11.2; p24) [10]/46, XY[2] |
| PAMDRM | P9906 | IGH@-CRLF2** | Male | 7.9 | 351.3 | JAK2 p.Ile682_Arg683insGlyPro*; IKZF1 deletion e1-e6; EBF1 deletion; PAX5 p.Val319fs; CDKN2A/CDKN2B deletion | 46, XY[20] |
| PAKKXB | P9906 | IGH@-CRLF2** | Female | 14.5 | 92.7 | IKZF1 (IK6); CDKN2A/CDKN2B deletion; FLT3 p.Asn609ins23aa*** | 46, XX[21] |
| PALETF | P9906 | None | Female | 7.6 | 105.7 | EBF1 deletion; FLT3 p.Leu604ins23aa*** | 47, XX,+10[3]/46, XX,+10,−21[7]/46, XX[8] |
| PAKHZT | P9906 | IGH@-CRLF2** | Male | 13.9 | 307 | JAK2 p.Arg867Gln; CDKN2A/CDKN2B deletion | N/A |
| PALJDL | P9906 | None | Male | 3.2 | 156 | PAX5 deletion; CDKN2A/CDKN2B deletion; IL7R p.L242_L243insFPGVC mutation#; SH2B3 e1–2 deletion# | N/A |
| PANNGL | AALL0232 | PAX5-JAK2* | Female | 12.9 | 15.8 | IKZF1 deletion | 47, XX, r(7)(p12q31),+9[14]/46, XX[6] |
| PANSFD | AALL0232 | ETV6-ABL1* | Male | 5.4 | 83 | IKZF1 (IK6); PAX5 deletion; CDKN2A/CDKN2B deletion | 46, XY, ins(12;9)(p13; q34q34)[20] |
| PANEHF | AALL0232 | RCSD1-ABL1* | Male | 15.7 | 47.8 | N/A | N/A |
| SJBALL085 | Total XV | NUP214-ABL1* | Male | 16.3 | 135.6 | IKZF1 (IK6) and p.Ala79fs mutation* | 46, XY |
| SJBALL010 | Total XVI | RANBP2-ABL1* | Male | 15 | 121 | PAX5 deletion* | 46, XY, t(2;9)(q21; q34)[14]/46, XY[6] |
Chromosomal rearrangements affecting kinase and cytokine receptor signaling identified by mRNA-seq in 15 Ph-like cases. Genetic lesions disrupting B-cell development (IKZF1, EBF1, and PAX5) and JAK2 activating mutations are also shown. IK6 refers to the deletion of IKZF1 exons 4–7 (coding exons 3–6) that results in the expression of a dominant negative IKZF1 isoform that lacks the N-terminal DNA-binding zinc fingers. All cases were of B-precursor immunophenotype and did not exhibit expression of T-lineage markers. Frame shifts (fs) are designated using the short nomenclature as outlined by the Human Genome Variation Society. aa, amino acid; e, exon; ITD, internal tandem duplication; N/A, not available; WCC, white cell count (x109/L).
identified by RNA-seq analysis.
Previously known (Harvey et al., 2010a);
Previously known (Zhang et al., 2011);
Identified by whole genome sequencing. See Tables S1–S9.
Figure 1.
Flow chart of methodology. (A) 15 Ph-like high-risk (HR) ALL cases were subjected to mRNA-seq, with matched normal DNA subjected to whole genome sequencing (WGS). Two cases also had WGS of tumor DNA. (B) For recurrence testing of ABL1, JAK2 and PDGFRB fusions, cases with available RNA from AALL0232 were screened by RT-PCR. The two NUP214-ABL1 cases identified in P9906 showed gain of 9q34 between NUP214 and ABL1 on SNP array analysis, and the presence of NUP214-ABL1 was confirmed by RT-PCR. Whole genome amplified (WGA) leukemic DNA was used for recurrence of IL7R and SH2B3 mutations. *FLT3 mutations were reported previously (Zhang et al., 2011). See also Figure S1.
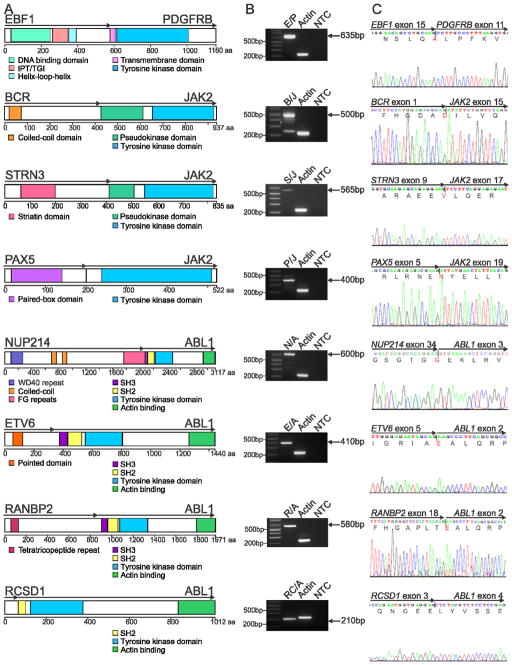
Strikingly, we identified alteration of genes encoding cytokine receptors and regulators of kinase signaling in all 15 cases studied (Table 1). Putative rearrangements were validated by reverse transcription followed by polymerase chain reaction (RT-PCR) and Sanger sequencing (Figure 2), with an average of 1.9 rearrangements identified per case, (range 0–5; Table S3 and Figure S2). The rearrangements included two cases with NUP214-ABL1, one case with insertion of the erythropoietin receptor gene (EPOR) into the immunoglobulin heavy chain locus (IGH@-EPOR), and one case each with the in-frame fusions EBF1-PDGFRB, BCR-JAK2, STRN3-JAK2, PAX5-JAK2, ETV6-ABL1, RANBP2-ABL1 and RCSD1-ABL1. These rearrangements were either cryptic on cytogenetic analysis, or the fusion partners could not be identified on examination of karyotypic data alone (Table 1). In each case multiple paired-end reads mapped to the partner genes, and split reads mapping across the fusion were identified (Figure 3A and S3A). Additional putative fusion transcripts were identified for each case (Figure S2 and Table S3); however, these commonly showed a low level of read support, did not encode an open-reading frame (SEMA6A-FEM1C, OAZ1-KLF2 and ZNF292-SYNCRIP) or involved intronic fusion break points (DOCK8-CBWD2 and TSHZ2-SLC35A1), suggesting they do not contribute to leukemogenesis. We also identified an inversion involving PAX5 and the adjacent gene ZCCHC7, resulting in a reciprocal fusion that disrupts the open reading frame of PAX5 (Figure S2H). Deletions, translocations and sequence mutations of PAX5 are detected in approximately 30% of B-ALL patients (Mullighan et al., 2007), and this inversion represents another mechanism for PAX5 inactivation.
Figure 2.
Rearrangements in Ph-like ALL. (A) Predicted domain structure of the in-frame fusions EBF1-PDGFRB (E/P), BCR-JAK2 (B/J), STRN3-JAK2 (S/J), PAX5-JAK2 (P/J), NUP214-ABL1 (N/A), ETV6-ABL1 (E/A), RANBP2-ABL1 (R/A) and RCSD1-ABL1 (RC/A) identified by mRNA-seq in eight Ph-like ALL cases. Confirmation of predicted fusions by RT-PCR (B) and Sanger sequencing (C). The two bands for BCR-JAK2 correspond to two different fusion breakpoints within JAK2 (exon 15 and 17), both of which were confirmed by Sanger sequencing. IPT/TIG, immunoglobulin-like fold, plexins, transcription factors/transcription factor immunoglobulin; FG, phenylalanine and glycine; SH, Src homology domain. See also Figure S2.
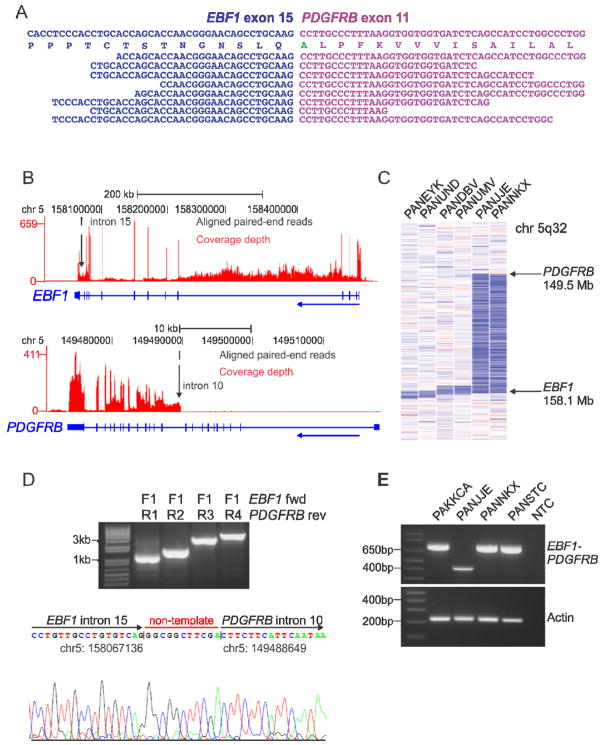
Figure 3.
mRNA-seq data, recurrence screening and genomic mapping of the EBF1-PDGFRB fusion. (A) Split reads mapping across the EBF1-PDGFRB fusion point for case PAKKCA. Amino acid substitution from wild-type PDGFRB (Ser>Ala), is highlighted in green. (B) Coverage depth for all mRNA-seq reads at the EBF1 and PDGFRB locus in case PAKKCA, showing expression across the EBF1 locus and increased expression of PDGFRB at intron 10 (arrowed). The vertical height of the red bar indicates the number of reads covering the site. (C) SNP 6.0 microarray log2 ratio DNA copy number heatmap showing deletion (blue) between EBF1 and PDGFRB for two EBF1-PDGFRB cases (PANJJE and PANNKX), and four non-rearranged cases with focal EBF1 deletions (left). (D) Genomic mapping of the EBF1-PDGFRB rearrangement breakpoint by PCR (top) and sequencing (bottom), showing juxtaposition of EBF1 intron 15 (chr5:158067136) to PDGFRB intron 10 (chr5:149488649), with the addition of non-template nucleotides between the breakpoints. (E) RT-PCR confirmation of EBF1-PDGFRB fusion in four high-risk B-ALL cases with exon 14 (bottom band) or exon 15 (top band) of EBF1 fused to exon 11 of PDGFRB. See also Figure S3.
CRLF2 is over-expressed in up to 7% of B-ALL, including over 50% of ALL cases in children with Down syndrome, and occurs via multiple mechanisms involving either a cryptic translocation that juxtaposes CRLF2 to the regulatory elements of the immunoglobulin heavy chain locus (IGH@-CRLF2) (Mullighan et al., 2009a; Russell et al., 2009a), or an interstitial deletion of the pseudoautosomal dominant region (PAR1) centromeric to CRLF2 resulting in the P2RY8-CRLF2 rearrangement (Mullighan et al., 2009a). Less frequently, the point mutation affecting codon 232 (p.Phe232Cys) has also been identified (Chapiro et al., 2010). Mutations activating JAK are present in approximately 50% of CRLF2r cases (Mullighan et al., 2009a; Russell et al., 2009a; Harvey et al., 2010a; Hertzberg et al., 2010; Yoda et al., 2010); however, the nature of kinase-activating mutations in CRLF2r cases lacking JAK mutations is unknown. Three cases in the discovery cohort were known to have the IGH@-CRLF2 translocation, and one of these harbored a known JAK2 mutation (p.Arg867Gln) (Mullighan et al., 2009c). Two additional CRLF2r cases lacking known JAK mutations were sequenced, one of which harbored a FLT3 internal tandem duplication (ITD; p.Asn609ins23aa) (Zhang et al., 2011), and the other harbored a complex JAK2 mutation (p.Ile682_Arg683insGlyPro in case PAMDRM) that was not identified by previous Sanger sequencing (Mullighan et al., 2009c). No additional kinase-activating lesions were identified in the CRLF2r cases. A full listing of somatic single nucleotide variants (SNVs) and insertions/deletions identified by mRNA-seq are provided in Table S4.
Case PAKKCA harbored the previously unknown EBF1-PDGFRB fusion that was present in the predominant leukemic clone, as confirmed by fluorescence in-situ hybridization (FISH) (Figure S3). mRNA-seq coverage analysis for this case showed a sharp increase in read depth at intron 10 of PDGFRB that corresponds to the genomic breakpoint (Figure 3B). Both genes are located on chromosome 5q, and analysis of DNA copy number data revealed a deletion between the two breakpoints (Figure 3C). Genomic PCR identified the breakpoint 0.5 kb downstream of EBF1 exon 15 and 2.3 kb upstream of PDGFRB exon 11 in the index case (Figure 3D). Several copy number alterations and rearrangements in B-ALL arise from aberrant recombination activating gene (RAG) activity (Mullighan et al., 2007; Mullighan et al., 2009a); however, analysis of the sequences adjacent to the genomic breakpoints of EBF1 and PDGFRB showed no evidence of RAG-mediated activity in this case.
The NUP214-ABL1 rearrangement has not previously been reported in B-ALL but is present in 5% of T-lineage ALL, and commonly accompanies episomal amplification of 9q34 (Graux et al., 2004). Notably, both NUP214-ABL1 cases had pre-B-ALL immunophenotype with no expression of T-lineage markers, and in contrast to T-ALL, did not show high level episomal amplification by FISH analysis (data not shown). Instead, we observed gain of only one copy of DNA between the two partner genes at 9q34 (Figure S2I). The ABL1 breakpoints correspond to those observed in NUP214-ABL1 T-ALL (De Braekeleer et al., 2011) and Ph+ chronic myeloid leukemia or B-ALL (Melo, 1996), which retain the SH2, SH3 and kinase domains of ABL1.
Case PAKYEP harbored the BCR-JAK2 fusion, which has previously been identified in myeloid leukemia (Griesinger et al., 2005; Cirmena et al., 2008), but not in B-ALL. Visualization of mRNA-seq split-reads using Bambino (Edmonson et al., 2011) identified two BCR-JAK2 fusion transcripts in this case involving exon 1 of BCR fused to either exon 15 or 17 of JAK2, both of which were validated by RT-PCR and sequencing (Figure S4A). Using Bambino, we also mapped the genomic breakpoint at intron 1 of BCR located within the minor breakpoint cluster region, to intron 14 of JAK2 (Figure S4B). Notably, all JAK2 fusions identified in this study are in-frame and disrupt the pseudokinase domain of JAK2, which is thought to relieve auto-inhibition of the kinase domain, thus resulting in a constitutively active fusion protein.
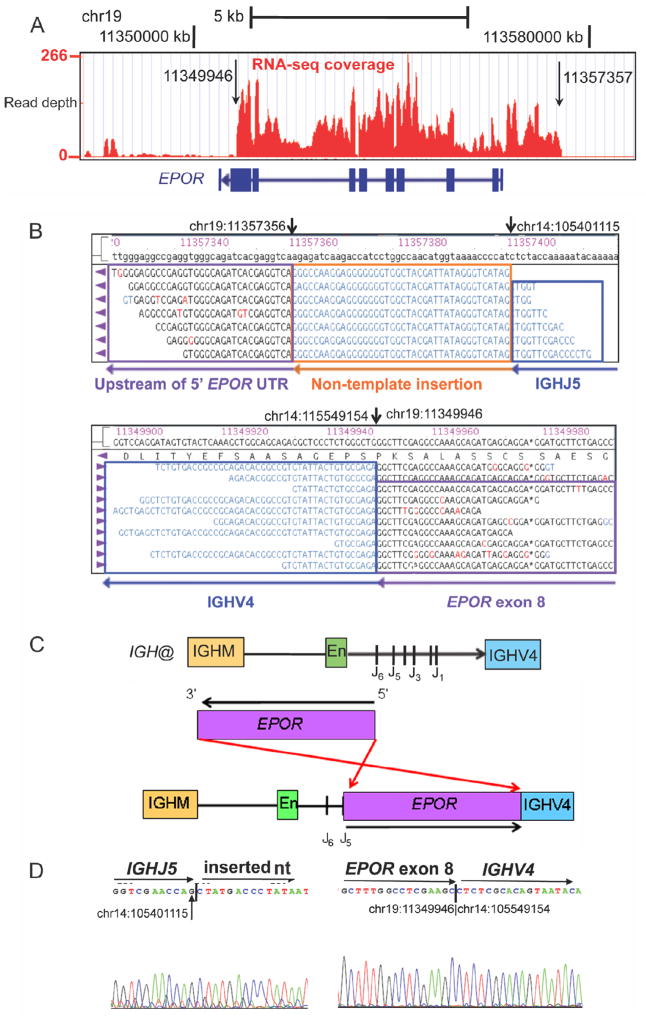
The IGH@-EPOR rearrangement arising from a reciprocal t(14;19)(q32; p13) translocation has been documented in B-cell precursor ALL (Russell et al., 2009b). However, FISH for the t(14;19) rearrangement in case PALIBN was negative. Detailed analysis of mRNA-seq data and genomic mapping demonstrated that the rearrangement involved a 7.5 kb insertion of EPOR into the immunoglobulin heavy chain locus downstream of the IgH enhancer domain with similar cytogenetic breakpoints as the previously identified translocation, thus identifying another mechanism of IGH@-EPOR rearrangement (Figure 4).
Figure 4.
Schematic of the IGH@-EPOR rearrangement. (A) Plot of read depth obtained from mRNA-seq data showing increased read depth across the EPOR locus. The arrows correspond to the genomic breakpoints identified by genomic PCR and sequencing. (B) Bambino viewer of mRNA-seq reads showing IGHJ5 with 40bp of inserted sequence joined to ~1.3 kb upstream of EPOR 5′ untranslated region (UTR) on chromosome 19. Bottom view shows split reads spanning exon 8 of EPOR adjacent to IGHV4. (C) IGH@ contains an IGHM domain, enhancer (En), J domains (J1–J6) and the downstream IGHV4 gene segment. The EPOR locus is inverted so the 5′ end is adjacent to the J5 domain and the 3′ end is within the IGHV4 gene segment. (D) Sanger sequencing confirming the rearrangement at IGHJ5 (chr14:105401115) to non-template sequence (left), and exon 8 of EPOR (chr19:11349946) to IGHV4 (chr14:105549154). See also Figure S4.
Sequence mutations and deletions in Ph-like ALL
WGS of tumor and normal DNA was performed on two Ph-like cases for which a kinase-activating rearrangement was not identified by mRNA-seq. Case PALJDL harbored two alterations predicted to activate tyrosine kinase signaling, the first being an in-frame insertion in the transmembrane domain of the interleukin 7 receptor, IL7R (p.Leu242_Leu243insFPGVC; Figure 5A). Using the mRNA-seq mutant allele read counts, we estimated the IL7R mutation to be expressed in approximately 93.4% of cells in the sample sequenced. Similar activating mutations in IL7R have recently been described in pediatric B and T-lineage ALL (Shochat et al., 2011; Zenatti et al., 2011; Zhang et al., 2012). Interestingly, case PALJDL also harbored a focal homozygous deletion removing the first two exons of SH2B3 that was not evident by single nucleotide polymorphism (SNP) array analysis, with a concomitant absence of SH2B3 expression by mRNA-seq analysis (Figure 5B–C). By comparing the coverage in the region of homozygous deletion (1.15x) to that of the undeleted region downstream on the same chromosome (30.86x) we estimate this deletion to be in at least 96% of cells in the sample sequenced. SH2B3 encodes the protein LNK, which is a negative regulator of JAK2 signaling (Tong et al., 2005), and inactivating mutations within exon 2 have been identified in JAK2 p.Val617Phe-negative myeloproliferative neoplasms (MPN) (Oh et al., 2010; Pardanani et al., 2010) and early T-cell precursor ALL (Zhang et al., 2012).
Figure 5.
IL7R activating mutations and SH2B3 deletion. (A) Protein domain structure of IL7R with location of sequence mutations identified in the transmembrane domain of nine B-ALL cases. (B) Copy number variant analysis of PALDJL comparing tumor and matched non-tumor DNA at the SH2B3 locus. (C) Transcript read depth data from mRNA-seq analysis comparing three cases with normal expression of SH2B3 to PALJDL. The vertical height of the red bar indicates the number of reads covering the site. (D) Sanger sequencing confirming the deletion from chr12:110325422 (first arrow) to chr12:110361365 (second arrow). See also Figure S5.
Case PALETF was found to harbor an in-frame ITD within the FLT3 juxtamembrane domain (p.Leu604ins23aa; Table S4). FLT3 ITDs and increased expression of wild-type FLT3 are also present in high-risk acute myeloid and lymphoblastic leukemia (Schnittger et al., 2002; Armstrong et al., 2003; Paietta et al., 2004; Zhang et al., 2012). Similar to PDGFRB and JAK2 rearrangements, FLT3 mutations facilitate leukemic transformation by inducing constitutive kinase activation and signaling through the Ras and JAK/STAT5 pathways (Mizuki et al., 2000). Additional SNVs and structural variations identified by WGS of PALJDL and PALETF are provided in Tables S5–S7 and Figure S5.
Recurrence of genetic alterations in Ph-like B-ALL
We next performed recurrence screening of extended cohorts of high-risk B-ALL to determine the frequency of these genetic alterations (Figure 1). RT-PCR for the EBF1-PDGFRB, BCR-JAK2, STRN3-JAK2, PAX5-JAK2, NUP214-ABL1, ETV6-ABL1, RANBP2-ABL1 and RCSD1-ABL1 fusions was performed for 231 cases from a separate consecutively recruited cohort of high-risk B-progenitor ALL obtained from the Children’s Oncology Group (COG) AALL0232 study. Screening for these fusions in the COG P9906 discovery cohort was not possible due to lack of RNA for many cases. We investigated the presence of IL7R and SH2B3 variants in both the P9906 and AALL0232 cohorts by Sanger sequencing of tumor DNA and SNP array analysis of tumor and matched non-tumor DNA. CRLF2 rearrangements, JAK mutations and amplification between NUP214 and ABL1 were examined in both P9906 and AALL0232 cohorts by SNP array analysis, genomic PCR and sequencing, and FISH for cases with 9q34 amplification. We also performed RT-PCR for the fusions and genomic sequencing for IL7R in other hematopoietic malignancies including 23 MPN cases lacking JAK2 or MPL mutations, 25 chronic myelomonocytic leukemia (CMML) cases and 44 childhood acute myeloid leukemia (AML) cases, including 34 that lacked recurring chromosomal rearrangements.
Forty of 231 cases (14%) of the AALL0232 cohort were identified as Ph-like (Table S8). Twenty-five cases (8.8%) had high CRLF2 expression, 19 of which were Ph-like and 6 non-Ph-like. JAK mutations were present in 10 cases with high CRLF2 expression, all of which were Ph-like (Table S9). The EBF1-PDGFRB fusion was detected in three additional Ph-like patients (8% of Ph-like ALL), in which exon 15 (N=2) or exon 14 (N=1) of EBF1 was fused to exon 11 of PDGFRB (Figure 3E). Each of the EBF1-PDGFRB cases showed an increase in PDGFRB expression by gene expression profiling and two of these patients (PANJJE and PANNKX) had an interstitial deletion between the partner gene breakpoints (Figure 3C).
No additional cases with the ABL1 or JAK2 rearrangements identified in the discovery cohort were observed in the AALL0232 cohort. Analysis of SNP array data identified two cases in P9906 (PAMBWU and PALFBA, one Ph-like) with a single copy gain of DNA between NUP214 and ABL1 (Figure 2I). The presence of the NUP214-ABL1 rearrangement was confirmed by RT-PCR and Sanger sequencing (Figure 2J), indicating that this fusion is also recurrent in B-ALL. No ABL1, JAK2 or PDGFRB rearrangements were identified in the MPN, CMML and AML cohorts, and have not been detected in other childhood B-ALL subtypes studied by WGS and mRNA-seq (Downing et al., 2012), indicating these genetic lesions are highly enriched in the Ph-like subtype.
Mutations within the transmembrane domain of IL7R were found in eight additional cases from P9906, five of which were Ph-like (12.5% of Ph-like ALL) (Figure 5A). An additional Ph-like case from the AALL0232 cohort (PANKMB) had a focal homozygous deletion removing exons 1–2 of SH2B3 that was identified using the higher resolution SNP 6.0 microarray, and subsequently confirmed by PCR (data not shown). Interestingly, this case harbors a P2RY8-CRLF2 rearrangement, but lacks a JAK mutation, suggesting that removal of JAK2 regulation by LNK augments JAK signaling in this case. No additional somatic SH2B3 mutations or deletions were identified in this study. Sanger sequencing of FLT3 in the P9906 cohort reported mutations in 12 cases, 4 of which were Ph-like (Table S9) (Zhang et al., 2011).
Rearrangements are transforming and sensitive to tyrosine kinase inhibitors
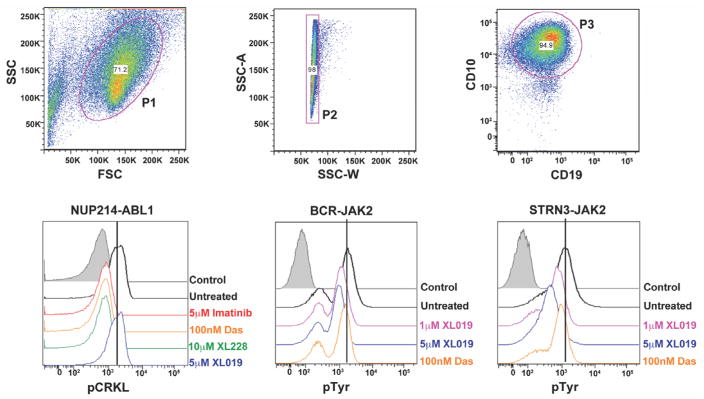
Recent phosphoflow cytometry studies have shown that B-ALL leukemic cells harboring CRLF2 rearrangements (with or without concomitant JAK mutations) have enhanced signaling through oncogenic pathways that can be targeted with JAK or PI3K inhibitors (Tasian et al., 2012). To determine if the genetic alterations we identified in Ph-like ALL activate kinase signaling and respond to TKIs, we performed flow cytometric phosphosignaling analysis on four primary leukemic samples (two cases with the NUP214-ABL1 fusion, one case with the BCR-JAK2 fusion, and one case with the STRN3-JAK2 fusion). All cases demonstrated activation of downstream signaling pathways, with phosphorylation of the ABL1 substrate CRKL in the NUP214-ABL1 cases and tyrosine phosphorylation in the cases with BCR-JAK2 and STRN3-JAK2 fusions (Figure 6). Importantly, this basal level of phosphorylation was reduced with imatinib, dasatinib and XL228 in samples harboring the ABL1 fusion, and the JAK2 inhibitor, XL019, in the JAK2-rearranged samples (Figure 6). Notably, XL019 had no effect on CRKL phosphorylation in ABL1-positive cases, however we did observe slight inhibition of tyrosine phosphorylation with dasatinib in the JAK2-cases. Five non Ph-like B-ALL cases were also assessed by phosphoflow and showed minimal activation of signaling pathways compared to Ph-like ALL, with no response to ABL1 or JAK2 inhibitors (Figure S6).
Figure 6.
Phosphosignaling analysis of primary leukemic blasts harboring NUP214-ABL1, BCR-JAK2 and STRN3-JAK2 rearrangements. Cells were untreated or treated with indicated tyrosine kinase inhibitors for 1 hour and levels of phosphorylated CRKL or tyrosine were assessed by phosphoflow cytometric analysis. Viable (P1) and single cells (P2) were gated by expression of CD10 and CD19 (P3). Das, dasatinib. Control is secondary antibody alone. See also Figure S6.
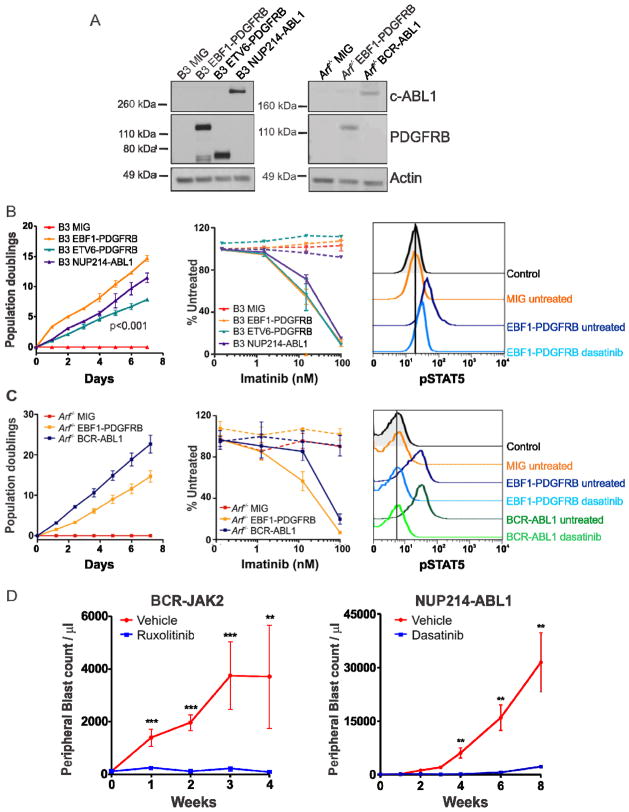
To evaluate the transforming potential of the EBF1-PDGFRB fusion, we assessed the ability of murine Ba/F3 and Arf−/− pre-B cells (Williams et al., 2006) expressing EBF1-PDGFRB to proliferate in the absence of exogenous cytokines. EBF1-PDGFRB expression (Figure 7A) conferred growth factor independence and resulted in significantly faster proliferation compared to Ba/F3 cells expressing the most common PDGFRB rearrangement, ETV6-PDGFRB (Figure 7B–C). Importantly, cytokine-independent proliferation was inhibited by imatinib (Figure 7B–C), the ABL1 and Src inhibitor dasatinib, and the multi-kinase inhibitor dovitinib (Figure S7A). Accordingly, imatinib treatment reduced phosphorylation of the PDGFRB receptor, with no change in total PDGFRB expression (Figure S7B). Several oncogenic pathways were constitutively activated by EBF1-PDGFRB in pre-B cells, demonstrated by elevated levels of pSTAT5, pAKT and pERK1/2. Notably, this signaling was also inhibited with dasatinib (Figure 7B–C and S7C). In addition, we also have evidence of a patient with EBF1-PDGFRB+ B-ALL refractory to induction chemotherapy entering remission with the addition of imatinib (data not shown).
Figure 7.
Kinase-activating fusions induce growth factor-independence and show response to tyrosine kinase inhibitors. (A) Immunoblot of c-ABL1 and PDGFRB in Ba/F3 (B3) and Arf−/− pre-B cells expressing empty vector (MIG), EBF1-PDGFRB, ETV6-PDGFRB or NUP214-ABL1. Transduced Ba/F3 (B) or Arf−/− pre-B cells (C) were grown in the absence of cytokine and cell number was recorded as indicated (left). Ba/F3 or Arf−/− pre-B cells were grown in increasing concentrations of imatinib (middle). No cytokine (solid line) or cytokine (dotted line). Error bars represent mean ± SD of three independent experiments. Cells were untreated or treated with dasatinib for 1 hour and levels of phosphorylated STAT5 were assessed by phosphoflow cytometric analysis (right). (D) Xenograft model of BCR-JAK2 and NUP214-ABL1. Mice were randomized to receive vehicle (40% dimethyl acetamide, 60% propylene glycol; N=5), ruxolitinib (30 mg/kg/day; N=7) or dasatinib (20 mg/kg/day; N=5). Error bars represent mean ± SEM. **, p<0.001; ***, p<0.0001. See also Figure S7.
We next investigated the therapeutic efficacy of the JAK2 inhibitor, ruxolitinib, in a xenograft model of BCR-JAK2-rearranged ALL (case PAKYEP). In this model cryopreserved BCR-JAK2+ cells were injected into NOD. Cg-Prkdcscid l2rgtm1Wjl/Szj (NSG) mice, and continuous infusion of ruxolitinib or vehicle was commenced once engraftment exceeded 5% of peripheral blood leukocytes (determined by measuring human CD19+/45+ cells). The presence of the fusion in xenografted cells was confirmed by RT-PCR (data not shown). We observed a striking decrease in leukemic burden after 4 weeks of ruxolitinib treatment compared to vehicle-treated controls, as measured by reduced peripheral blood (p<0.001; Figure 7D) and spleen blast counts (data not shown). Furthermore, a xenograft model of NUP214-ABL1 ALL responded to dasatinib up to 8 weeks of treatment (Figure 7D), confirming that cells expressing NUP214-ABL1 are sensitive to TKIs (Quintas-Cardama et al., 2008; Deenik et al., 2009). In addition, ruxolitinib significantly decreased peripheral blood and spleen blast counts in a xenograft model of case PALJDL, which harbors both an IL7R activating mutation and a somatic SH2B3 (LNK) deletion (data not shown). Together, these data indicate that EBF1-PDGFRB, BCR-JAK2, NUP214-ABL1 fusions and sequence mutations in IL7R/SH2B3 are transforming, and represent excellent candidates for therapy with currently available TKIs.
DISCUSSION
Ph-like ALL represents approximately 10% of childhood B-ALL and 15% of high-risk B-ALL, and is three to four times more common than Ph+ ALL. Among a large cohort of patients with high-risk B-ALL treated on COG AALL0232, the Ph-like phenotype is associated with older age (12.4 vs. 9.5 years, p<0.0001), and significantly inferior 5 year event-free survival compared to non Ph-like patients (unpublished data). Using next-generation sequencing, we have shown that rearrangements and sequence mutations activating tyrosine kinase and cytokine receptor signaling are a hallmark of Ph-like ALL. Moreover, each of the cases studied harbored genomic lesions affecting lymphoid transcription factors (most commonly deletions and/or mutations of IKZF1), suggesting that perturbation of these two pathways cooperate to induce B-lineage ALL and drive the Ph-like gene expression profile.
Chromosomal rearrangements resulting in activated tyrosine kinase signaling are recognized as driver lesions in a number of hematopoietic malignancies, the prototype being BCR-ABL1 in CML (Melo, 1996) and Ph+ B-ALL (De Braekeleer et al., 2011). Here we report three new fusions in B-ALL, (EBF1-PDGFRB, STRN3-JAK2 and RANBP2-ABL1), and several that have been reported in very few patients including IGH@-EPOR (Russell et al., 2009b), PAX5-JAK2 (Nebral et al., 2009), ETV6-ABL1 and RCSD1-ABL1 (De Braekeleer et al., 2011).
Rearrangements involving the PDGFRB receptor are present at low frequency in Ph-negative myeloid neoplasms (Golub et al., 1994; Cross and Reiter, 2008), The identification of a PDGFRB fusion is of clinical importance, as patients with chronic myeloproliferative disease and activating PDGFRB rearrangements show complete hematologic and molecular responses to imatinib treatment (Apperley et al., 2002). For EBF1-PDGFRB, the coding region of EBF1 is juxtaposed to the C-terminal region of PDGFRB, preserving the transmembrane and kinase domains. It is predicted that the EBF1 helix-loop-helix domain mediates homo-dimerization (Hagman et al., 1995) and facilitates constitutive activation of PDGFRB, as is observed with ETV6-PDGFRB (Carroll et al., 1996). Furthermore, EBF1 is a transcription factor that plays a major role in regulating B-cell differentiation (Hagman and Lukin, 2006), and deletions that abolish normal EBF1 function have been reported in B-lineage ALL (Mullighan et al., 2007). The fusion of EBF1 to PDGFRB is also likely to impair the normal function of EBF1, and represents a mechanism resulting in PDGFRB overexpression.
We also identified RANBP2 as a fusion partner for ABL1. RANBP2 (or NUP358) localizes to the cytoplasmic side of the nuclear pore complex via interaction with NUP88, and forms a sub-complex with NUP214 (Bernad et al., 2004). The structural features of RANBP2 retained in the fusion protein include the leucine zipper, which is predicted to mediate homo-dimerization of RANBP2-ABL1, as observed with RANBP2-ALK in atypical myeloproliferative leukemia (Rottgers et al., 2010). Furthermore, localization of NUP214-ABL1 to the nuclear pore complex and interaction with additional nuclear pore proteins is required for ABL1 kinase activity of this fusion protein (De Keersmaecker et al., 2008). Thus, we hypothesize that RANBP2-ABL1 may be activated in a similar manner.
While a diverse range of kinase lesions are present in Ph-like ALL, activation of ABL1 and/or JAK/STAT signaling pathways is a common mechanism for transformation. The dramatic improvement in outcome observed in Ph+ B-ALL patients treated with chemotherapy and imatinib (Schultz et al., 2009), and our demonstration that Ph-like leukemic cells are sensitive to currently available TKIs provide a strong rationale to test chemotherapy plus TKI treatment in Ph-like ALL patients. At present, next-generation sequencing is not widely available in diagnostic laboratories. However, our results indicate that flow cytometric phosphosignaling analysis can identify Ph-like cases with activation of kinase pathways, and in conjunction with flow-cytometric detection of CRLF2 overexpression (Mullighan et al., 2009a), may be implemented as a routine diagnostic test. In addition, the gene expression profile of Ph-like ALL can be used to design targeted low density gene expression arrays suitable for diagnostic use. Although the majority of Ph-like patients do not harbor known recurring chromosomal rearrangements, initial screening may be performed on all ALL cases. Patients identified as Ph-like can then undergo additional testing for known genetic lesions associated with this subtype, and be directed to treatment that combines chemotherapy with ABL1, PDGFRB or JAK inhibitors. It is important to note that rare non Ph-like patients that harbor kinase alterations (e.g NUP214-ABL1) may also benefit from the addition of TKI therapy.
In summary, this study illustrates how the use of genomic analysis can identify rationale therapeutic targets that drive tailored treatment, and provides a model that can be applied to a wide range of cancer subtypes to benefit patients with high-risk disease.
EXPERIMENTAL PROCEDURES
Patients and Samples
Ten Ph-like ALL cases from the COG P9906 high-risk B-ALL study (Bowman et al., 2011), three cases enrolled on the high-risk COG AALL0232 study (ClinicalTrials.gov Identifier NCT00075725) and two cases treated on the St Jude Children’s Research Hospital Total XV (Pui et al., 2009) and Total XVI protocols (ClinicalTrials.gov Identifier NCT00137111 and NCT00549848, respectively) were selected for mRNA-seq based on a similar gene expression profile to BCR-ABL1 ALL, as determined by ROSE clustering (Harvey et al., 2010b), PAM (Tibshirani et al., 2002), and the availability of suitable genomic material. All samples were obtained with patient or parent/guardian provided informed consent under protocols approved by the Institutional Review Board at each COG institution and St Jude Children’s Research Hospital. Details on case selection and recurrence are outlined in the Supplemental Experimental Procedures.
mRNA-seq and whole genome sequencing
mRNA-seq was performed using a method similar to that previously described (Morin et al., 2010). For WGS, Illumina paired-end whole genome shotgun libraries were prepared from 1 μg of genomic DNA as described (Shah et al., 2009). Sequencing was performed on the Illumina Genome Analyzer GAIIx or HiSeq 2000 platforms. Methods for library preparation, sequencing and detection of rearrangements, DNA copy number alterations and sequence variations are provided in the Supplemental Experimental Procedures.
RT-PCR, genomic mapping and sequencing
Putative rearrangements identified by mRNA-seq were validated by RT-PCR and Sanger sequencing. Leukemic cell RNA was reverse-transcribed using Superscript III (Life Technologies) and fusion products amplified with Phusion HF polymerase (New England Biolabs). Genomic mapping of the EBF1-PDGFRB and BCR-JAK2 rearrangement breakpoints was performed using whole genome amplified (Qiagen, Germany) leukemic cell DNA.
Retroviral constructs, infection and cell proliferation assays
The full-length EBF1-PDGFRB fusion was amplified from leukemic cell cDNA, cloned into pGEM-T-Easy (Promega), then subcloned into the MSCV-IRES-GFP retroviral vector. Retroviral supernatants containing MSCV-EBF1-PDGFRB-IRES-GFP, MSCV-ETV6-PDGFRB-IRES-GFP (Carroll et al., 1996), MSCV-NUP214-ABL1-IRES-GFP (De Keersmaecker et al., 2008) or MSCV-BCR-ABL1-IRES-GFP (p185) (Williams et al., 2006) were produced using the ecotropic Phoenix packaging cell line and used to infect murine hematopoietic progenitor Ba/F3 or primary Arf−/− pre-B cells (Williams et al., 2006). To evaluate factor-independent growth, cells were washed three times, seeded in triplicate without cytokine and cell number was recorded daily using a Vicell cell counter (Beckman Coulter). Proliferation rates of each cell line were compared using a linear mixed-effect model with order-1 autoregressive covariance structure for longitudinal data in the SAS package (SAS Inc, Cary). Drug sensitivity was assessed using the CellTiter-Blue Cell Viability Assay (Promega) according to manufacturer’s instructions, and IC50 was determined using nonlinear regression (GraphPad Prism). Each experiment was performed three times.
Phosphoflow analysis and immunoblotting
To assess signaling within leukemic samples and cell lines, intracellular phosphoflow cytometric analysis were performed as previously described (Kotecha et al., 2008). Briefly, cryopreserved patient samples were thawed, or cells in culture were harvested at 1×106 cells per tube and treated with the TKIs imatinib (Novartis), dasatinib (Bristol Myers Squibb), XL228 or XL019 (Exelixis) for one hour. Cells were fixed, permeabilized and stained with either anti-phosphotyrosine-4G10 (Upstate), anti-pAKT (S573), -pCRKL (Y207), -pERK1/2 (T202/Y204), or -pSTAT5 (Y694; Cell Signaling Technology) then Alexa Fluor 647 conjugated anti-rabbit or Pacific Blue conjugated anti-mouse IgG secondary antibodies (Life Technologies). Cellular fluorescence data were collected on an LSR II flow cytometer (BD Biosciences) using DIVA software (BD Biosciences), and analyzed with FlowJo (Tree Star). For immunoblotting, cells were lysed in RIPA buffer, subjected to SDS-PAGE and probed with anti-phosphotyrosine-4G10 (Upstate), anti-c-ABL, anti-PDGFRB and anti-Actin (Santa Cruz).
Xenograft models
Xenograft models of case PAKYEP (BCR-JAK2) and PAKVKK (NUP214-ABL1) were established as previously described with modifications (Teachey et al., 2006). Primary leukemia cells from bone marrow were intravenously injected into the tail vein of NSG mice (107 cells/mouse). Following engraftment (>5% human CD19+/45+ blasts in peripheral blood), BCR-JAK2 mice were randomized to receive ruxolitinib (30 mg/kg/day; Incyte) or vehicle (40% dimethyl acetamide, 60% propylene glycol) by continuous subcutaneous infusion using implanted mini-osmotic pumps (Alzet). For NUP214-ABL1 mice, dasatinib (20 mg/kg; Bristol Myers Squibb) or vehicle (10% citric acid in 80mM sodium citrate) was given 5 days a week by oral gavage. Disease burden was assessed weekly by flow cytometric determination of human CD19+/45+ blast count in peripheral blood, using CountBright beads (Invitrogen). Deaths within 72 hours of pump placement were considered secondary to anesthesia or surgery, and these mice were censored at the time of death. All experiments were conducted on protocols approved by the Institutional Animal Care and Use Committee and Institutional Review Board of The Children’s Hospital of Philadelphia.
Supplementary Material
SIGNIFICANCE.
Ph-like ALL patients comprise up to 15% of childhood ALL, exhibit a high risk of relapse and have a poor outcome. Using next-generation sequencing, we have shown that genetic alterations activating kinase or cytokine receptor signaling are a hallmark of this subtype, and that a number of these lesions are sensitive to tyrosine kinase inhibitors (TKIs). Thus, our findings support screening at diagnosis to identify Ph-like ALL patients that may benefit from the addition of TKI treatment to current chemotherapeutic regimens. Furthermore, this study illustrates how genomic analysis can be used to drive tailored therapy for cancer patients.
Acknowledgments
We thank M. Tomasson and J. Cools for providing the MSCV-ETV6-PDGFRB-IRES-GFP and MSCV-NUP214-ABL1-IRES-GFP constructs, respectively; D. Pei and C. Cheng for statistical analyses of cell line proliferation data; Garry Nolan for providing the Phoenix cell line (http://www.stanford.edu/group/nolan/retroviral_systems/phx.html); Beckman Coulter Genomics for Sanger Sequencing, and the Flow Cytometry Core Facility, Tissue Resources Core Facility and Clinical Application of Core Technology (Affymetrix) Laboratory of the Hartwell Center for Bioinformatics and Biotechnology of St Jude Children’s Research Hospital. The correlative biology studies described in this manuscript were funded by grants from the NIH, and philanthropic funds of the Children’s Oncology Group, and not a commercial entity. The sequencing was part-funded with Federal funds from the NCI and NIH under Contract No. N01-C0-12400 as part of the Therapeutically Applicable Research to Generate Effective Treatments initiative. This work was supported by funds provided as a supplement to the Children’s Oncology Group Chair’s award (CA098543, G.R.); grants to the COG including U10 CA98543 (COG Chair’s grant), U10 CA98413 (COG Statistical Center), and U24 CA114766 (COG Specimen Banking); a National Cancer Institute Strategic Partnering to Evaluate Cancer Signatures Program award CA114762 (W.L.C., I-M.C., R.C.H., C.L.W.); NIH Cancer Center Core Grant CA21765 (J.R.D., C.G.M., W.E.E., C.-H.P., S.J.); St Jude Children’s Research Hospital - Washington University Pediatric Cancer Genome Project; a Stand Up To Cancer Innovative Research Grant (C.G.M.); and the American Lebanese Syrian Associated Charities of St Jude Children’s Research Hospital. K.G.R. is supported by a National Health and Medical Research Council (Australia) Overseas Training Fellowship and a Haematology Society of Australasia and New Zealand Novartis New Investigator Scholarship. R.D.M. is a Vanier Scholar (CIHR) and holds a MSFHR senior graduate studentship. M.A.M. is a UBC Canada Research Chair in Genome Science and a Michael Smith Senior Research Scholar. S.P.H. is the Ergen Family Chair in Pediatric Cancer. C.G.M. is a Pew Scholar in the Biomedical Sciences and a St. Baldrick’s Scholar. S.P.H. is a member of the Bristol Myers Squibb pediatric dasatinib advisory board.
Footnotes
Supplemental Information includes seven figures, nine tables and Supplemental Experimental Procedures.
ACCESSION NUMBERS
The sequence data and SNP microarray data have been deposited in the database of genotypes and phenotypes (dbGAP, http://www.ncbi.nlm.nih.gov/gap) database under the accession number phs000218.v1.p1. The gene expression data for COG P9906 has been deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), accession GSE11877. The gene expression data without metadata for COG AALL0232 is deposited at the National Cancer Institute caArray site (https://array.nci.nih.gov/caarray/project/EXP-578). The EBF1-PDGFRB sequence has been deposited into the NCBI Genbank database (http://www.ncbi.nlm.nih.gov/genbank) database (Accession JN003579).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apperley JF, Gardembas M, Melo JV, Russell-Jones R, Bain BJ, Baxter EJ, Chase A, Chessells JM, Colombat M, Dearden CE, et al. Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med. 2002;347:481–487. doi: 10.1056/NEJMoa020150. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Bernad R, van der Velde H, Fornerod M, Pickersgill H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman WP, Larsen EL, Devidas M, Linda SB, Blach L, Carroll AJ, Carroll WL, Pullen DJ, Shuster J, Willman CL, et al. Augmented therapy improves outcome for pediatric high risk acute lymphocytic leukemia: results of Children’s Oncology Group trial P9906. Pediatr Blood Cancer. 2011;57:569–577. doi: 10.1002/pbc.22944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M, Tomasson MH, Barker GF, Golub TR, Gilliland DG. The TEL/platelet-derived growth factor beta receptor (PDGF beta R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF beta R kinase-dependent signaling pathways. Proc Natl Acad Sci U S A. 1996;93:14845–14850. doi: 10.1073/pnas.93.25.14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapiro E, Russell L, Lainey E, Kaltenbach S, Ragu C, Della-Valle V, Hanssens K, Macintyre EA, Radford-Weiss I, Delabesse E, et al. Activating mutation in the TSLPR gene in B-cell precursor lymphoblastic leukemia. Leukemia. 2010;24:642–645. doi: 10.1038/leu.2009.231. [DOI] [PubMed] [Google Scholar]
- Cirmena G, Aliano S, Fugazza G, Bruzzone R, Garuti A, Bocciardi R, Bacigalupo A, Ravazzolo R, Ballestrero A, Sessarego M. A BCR-JAK2 fusion gene as the result of a t(9;22)(p24; q11) in a patient with acute myeloid leukemia. Cancer Genet Cytogenet. 2008;183:105–108. doi: 10.1016/j.cancergencyto.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Cross NC, Reiter A. Fibroblast growth factor receptor and platelet-derived growth factor receptor abnormalities in eosinophilic myeloproliferative disorders. Acta Haematol. 2008;119:199–206. doi: 10.1159/000140631. [DOI] [PubMed] [Google Scholar]
- De Braekeleer E, Douet-Guilbert N, Rowe D, Bown N, Morel F, Berthou C, Ferec C, De Braekeleer M. ABL1 fusion genes in hematological malignancies: a review. Eur J Haematol. 2011;86:361–371. doi: 10.1111/j.1600-0609.2011.01586.x. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker K, Rocnik JL, Bernad R, Lee BH, Leeman D, Gielen O, Verachtert H, Folens C, Munck S, Marynen P, et al. Kinase activation and transformation by NUP214-ABL1 is dependent on the context of the nuclear pore. Mol Cell. 2008;31:134–142. doi: 10.1016/j.molcel.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Deenik W, Beverloo HB, van der Poel-van de Luytgaarde SC, Wattel MM, van Esser JW, Valk PJ, Cornelissen JJ. Rapid complete cytogenetic remission after upfront dasatinib monotherapy in a patient with a NUP214-ABL1-positive T-cell acute lymphoblastic leukemia. Leukemia. 2009;23:627–629. doi: 10.1038/leu.2008.318. [DOI] [PubMed] [Google Scholar]
- Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, Ley TJ, Evans WE. The pediatric cancer genome project. Nat Genet. 2012;44:619–622. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonson MN, Zhang J, Yan C, Finney RP, Meerzaman DM, Buetow KH. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics. 2011;27:865–866. doi: 10.1093/bioinformatics/btr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, Vermeesch JR, Stul M, Dutta B, Boeckx N, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–1089. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- Griesinger F, Hennig H, Hillmer F, Podleschny M, Steffens R, Pies A, Wormann B, Haase D, Bohlander SK. A BCR-JAK2 fusion gene as the result of a t(9;22)(p24; q11.2) translocation in a patient with a clinically typical chronic myeloid leukemia. Genes Chromosomes Cancer. 2005;44:329–333. doi: 10.1002/gcc.20235. [DOI] [PubMed] [Google Scholar]
- Hagman J, Gutch MJ, Lin H, Grosschedl R. EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 1995;14:2907–2916. doi: 10.1002/j.1460-2075.1995.tb07290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol. 2006;18:127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, Kang H, Liu W, Dobbin KK, Smith MA, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010a;115:5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010b;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg L, Vendramini E, Ganmore I, Cazzaniga G, Schmitz M, Chalker J, Shiloh R, Iacobucci I, Shochat C, Zeligson S, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–1017. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S, Astolfi A, Chiaretti S, Vitale A, Messa F, et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP) Blood. 2009;114:2159–2167. doi: 10.1182/blood-2008-08-173963. [DOI] [PubMed] [Google Scholar]
- Kotecha N, Flores NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S, Diaz-Flores E, Coram M, Shannon KM, Nolan GP, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14:335–343. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, Hoogerbrugge PM. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- Marth G. MOSAIK assembler. 2010 http://bioinformatics.bc.edu/marthlab/Mosaik.
- Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, Soverini S, Vitale A, Chiaretti S, Cimino G, et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27:5202–5207. doi: 10.1200/JCO.2008.21.6408. [DOI] [PubMed] [Google Scholar]
- McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–2384. [PubMed] [Google Scholar]
- Mizuki M, Fenski R, Halfter H, Matsumura I, Schmidt R, Muller C, Gruning W, Kratz-Albers K, Serve S, Steur C, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, Ma J, Coustan-Smith E, Harvey RC, Willman CL, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009a;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009b;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009c;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, Strehl S. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:134–143. doi: 10.1038/leu.2008.306. [DOI] [PubMed] [Google Scholar]
- Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD, Jr, Merker JD, Zehnder JL, Nolan GP, Gotlib J. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116:988–992. doi: 10.1182/blood-2010-02-270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta E, Ferrando AA, Neuberg D, Bennett JM, Racevskis J, Lazarus H, Dewald G, Rowe JM, Wiernik PH, Tallman MS, et al. Activating FLT3 mutations in CD117/KIT(+) T-cell acute lymphoblastic leukemias. Blood. 2004;104:558–560. doi: 10.1182/blood-2004-01-0168. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Lasho T, Finke C, Oh ST, Gotlib J, Tefferi A. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia. 2010;24:1713–1718. doi: 10.1038/leu.2010.163. [DOI] [PubMed] [Google Scholar]
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, Ribeiro RC, Rubnitz JE, Raimondi SC, Onciu M, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360:2730–2741. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A, Tong W, Manshouri T, Vega F, Lennon PA, Cools J, Gilliland DG, Lee F, Cortes J, Kantarjian H, et al. Activity of tyrosine kinase inhibitors against human NUP214-ABL1-positive T cell malignancies. Leukemia. 2008;22:1117–1124. doi: 10.1038/leu.2008.80. [DOI] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- Rottgers S, Gombert M, Teigler-Schlegel A, Busch K, Gamerdinger U, Slany R, Harbott J, Borkhardt A. ALK fusion genes in children with atypical myeloproliferative leukemia. Leukemia. 2010;24:1197–1200. doi: 10.1038/leu.2010.18. [DOI] [PubMed] [Google Scholar]
- Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, Chandrasekaran T, Chapiro E, Gesk S, Griffiths M, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009a;114:2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- Russell LJ, De Castro DG, Griffiths M, Telford N, Bernard O, Panzer-Grumayer R, Heidenreich O, Moorman AV, Harrison CJ. A novel translocation, t(14;19)(q32; p13), involving IGH@ and the cytokine receptor for erythropoietin. Leukemia. 2009b;23:614–617. doi: 10.1038/leu.2008.250. [DOI] [PubMed] [Google Scholar]
- Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, Loffler H, Sauerland CM, Serve H, Buchner T, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- Schultz KR, Bowman WP, Aledo A, Slayton WB, Sather H, Devidas M, Wang C, Davies SM, Gaynon PS, Trigg M, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol. 2009;27:5175–5181. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, Te Kronnie G, Cario G, Cazzaniga G, Kulozik AE, Stanulla M, et al. Gain-of-function mutations in interleukin-7 receptor-{alpha} (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med. 2011;208:901–908. doi: 10.1084/jem.20110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Tasian SK, Doral MY, Borowitz MJ, Wood BL, Chen I-M, Harvey RC, Gastier-Foster JM, Willman CL, Hunger SP, Mullighan CG, Loh ML. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012 doi: 10.1182/blood-2011-12-389932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teachey DT, Obzut DA, Cooperman J, Fang J, Carroll M, Choi JK, Houghton PJ, Brown VI, Grupp SA. The mTOR inhibitor CCI-779 induces apoptosis and inhibits growth in preclinical models of primary adult human ALL. Blood. 2006;107:1149–1155. doi: 10.1182/blood-2005-05-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W, Zhang J, Lodish HF. Lnk inhibits erythropoiesis and Epo-dependent JAK2 activation and downstream signaling pathways. Blood. 2005;105:4604–4612. doi: 10.1182/blood-2004-10-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, Ma J, Rusch MC, Chen K, Harris CC, Ding L, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda A, Yoda Y, Chiaretti S, Bar-Natan M, Mani K, Rodig SJ, West N, Xiao Y, Brown JR, Mitsiades C, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2010;107:252–257. doi: 10.1073/pnas.0911726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, Tritapoe J, Hixon JA, Silveira AB, Cardoso BA, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43:932–939. doi: 10.1038/ng.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, Buetow KH, Carroll WL, Chen IM, Devidas M, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2011;118:3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.