Abstract
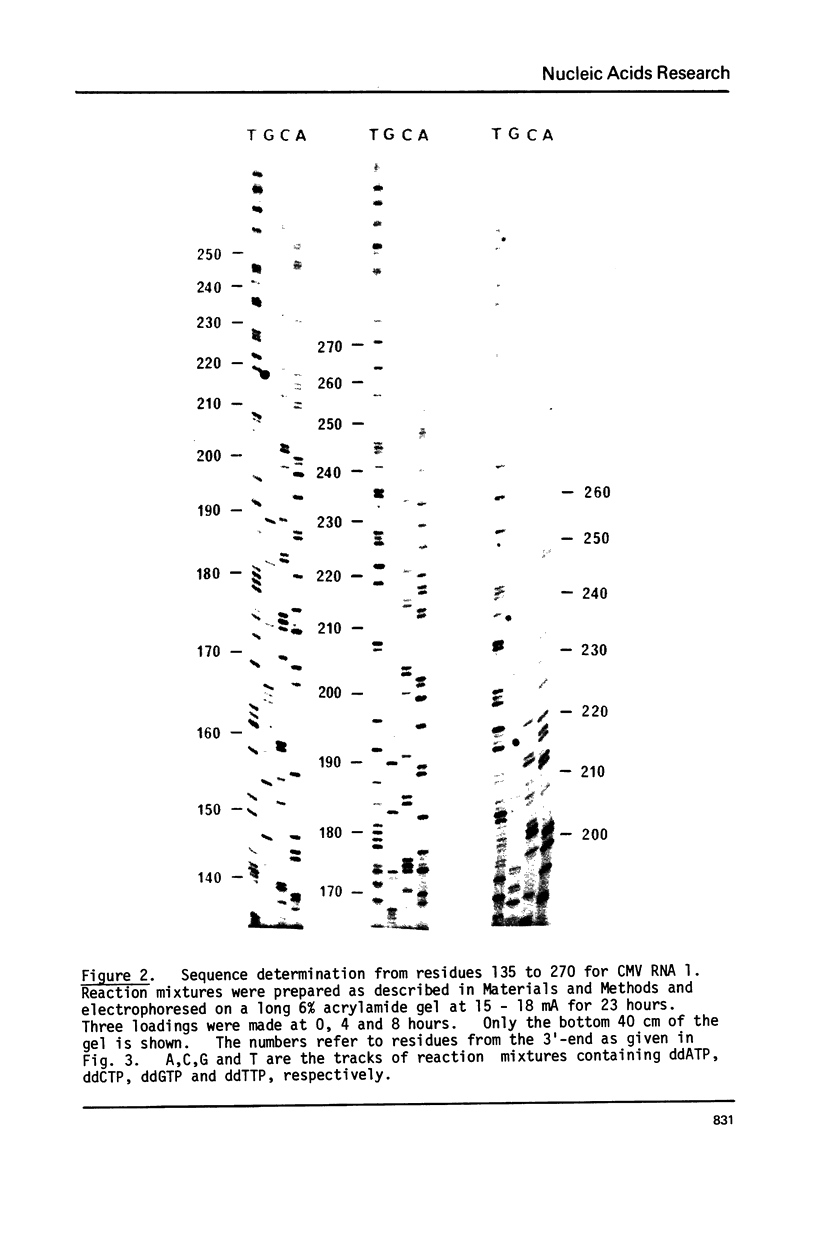
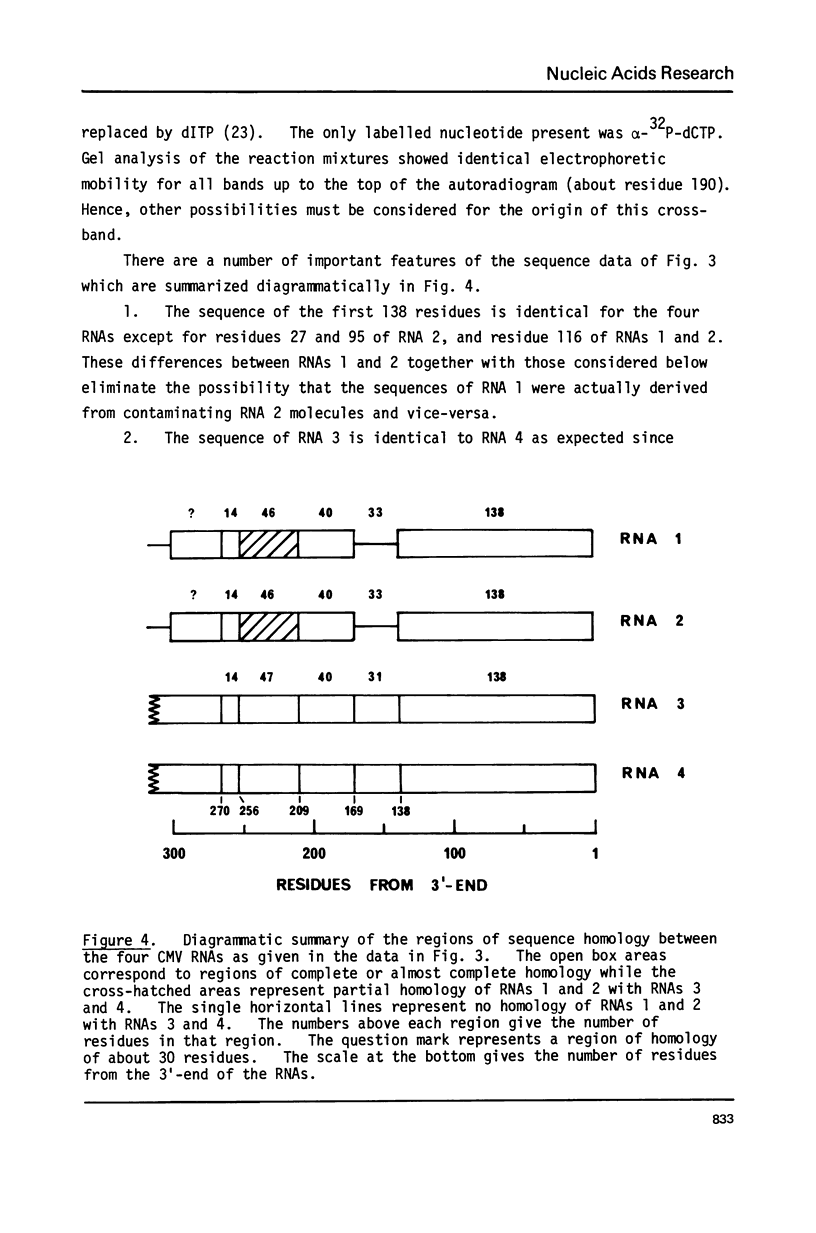
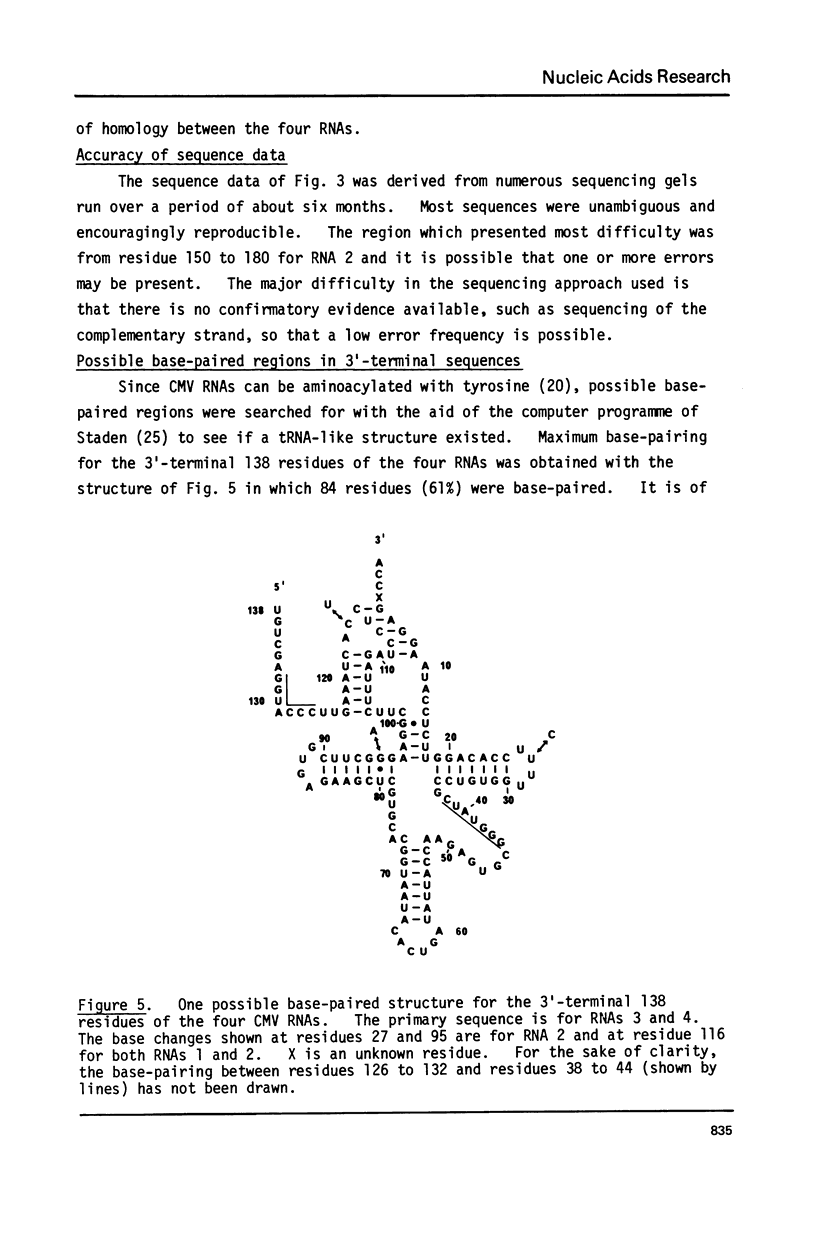
The sequences of 270 residues from the 3'-termini of the four RNAs of cucumber mosaic virus have been determined by copying the in vitro polyadenylated RNAs with reverse transcriptase using d(pT8G) as primer and the 2',3'-dideoxynucleoside 5'-triphosphates as specific chain terminators. The terminal sequences of RNAs 3 and 4 were identical; this was expected since hybridization data has shown that the sequence of RNA 4 was present at the 3'-end of RNA 3 (Gould and Symons (1978) Eur. J. Biochem. 91, 269-278). The first 138 residues of RNAs 1 and 2 were identical to those of RNAs 3 and 4 except for one residue in RNA 1 and three residues in RNA 2. From residue 139 to 270 from the 3'-terminus, RNAs 1 and 2 showed, relative to RNAs 3 and 4, a non-homologous region of 33 residues, a homologous region of 40 residues, a partially homologous region of 14 residues which probably extended to about residue 300. There were 11 residues different between RNAs 1 and 2.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Both G. W., Air G. M. Nucleotide sequence coding for the N-terminal region of the matrix protein influenza virus. Eur J Biochem. 1979 May 15;96(2):363–372. doi: 10.1111/j.1432-1033.1979.tb13048.x. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Cartwright E. M. Rapid gel sequencing of RNA by primed synthesis with reverse transcriptase. J Mol Biol. 1977 Jul;114(1):93–117. doi: 10.1016/0022-2836(77)90285-6. [DOI] [PubMed] [Google Scholar]
- Catterall J. F., O'Malley B. W., Robertson M. A., Staden R., Tanaka Y., Brownlee G. G. Nucleotide sequence homology at 12 intron--exon junctions in the chick ovalbumin gene. Nature. 1978 Oct 12;275(5680):510–513. doi: 10.1038/275510a0. [DOI] [PubMed] [Google Scholar]
- Dasgupta R., Kaesberg P. Sequence of an oligonucleotide derived from the 3' end of each of the four brome mosaic viral RNAs. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4900–4904. doi: 10.1073/pnas.74.11.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Vournakis J. N., Donis-Keller H., Chaconas G., Dougall D. K., Kafatos F. C. End labeling of enzymatically decapped mRNA. Nucleic Acids Res. 1977 Dec;4(12):4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould A. R., Palukaitis P., Symons R. H., Mossop D. W. Characterization of a satellite RNA associated with cucumber mosaic virus. Virology. 1978 Feb;84(2):443–455. doi: 10.1016/0042-6822(78)90261-1. [DOI] [PubMed] [Google Scholar]
- Gould A. R., Symons R. H. Alfalfa mosaic virus RNA. Determination of the sequence homology between the four RNA species and a comparison with the four RNA species of cucumber mosaic virus. Eur J Biochem. 1978 Nov 2;91(1):269–278. doi: 10.1111/j.1432-1033.1978.tb20962.x. [DOI] [PubMed] [Google Scholar]
- Gould A. R., Symons R. H. Determination of the sequence homology between the four RNA species of cucumber mosaic virus by hybridization analysis with complementary DNA. Nucleic Acids Res. 1977 Nov;4(11):3787–3802. doi: 10.1093/nar/4.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habili N., Francki R. I. Comparative studies on tomato aspermy and cucumber mosaic viruses. I. Physical and chemical properties. Virology. 1974 Feb;57(2):392–401. doi: 10.1016/0042-6822(74)90179-2. [DOI] [PubMed] [Google Scholar]
- Hamlyn P. H., Browniee G. G., Cheng C. C., Gait M. J., Milstein C. Complete sequence of constant and 3' noncoding regions of an immunoglobulin mRNA using the dideoxynucleotide method of RNA sequencing. Cell. 1978 Nov;15(3):1067–1075. doi: 10.1016/0092-8674(78)90290-8. [DOI] [PubMed] [Google Scholar]
- Hidaka S., Shimotohno K., Miura K., Takanami Y., Kubo S. Nucleotide sequence near the 5'-terminal of cucumber mosaic virus RNA No. 5 segment. FEBS Lett. 1979 Feb 1;98(1):115–118. doi: 10.1016/0014-5793(79)80165-9. [DOI] [PubMed] [Google Scholar]
- Kohl R. J., Hall T. C. Aminoacylation of RNA from several viruses: amino acid specificity and differential activity of plant, yeast and bacterial synthetases. J Gen Virol. 1974 Nov;25(2):257–261. doi: 10.1099/0022-1317-25-2-257. [DOI] [PubMed] [Google Scholar]
- Lane L. C. The bromoviruses. Adv Virus Res. 1974;19:151–220. doi: 10.1016/s0065-3527(08)60660-0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Lot H., Marchoux G., Marrou J., Kaper J. M., West C. K., van Vloten-Doting L., Hull R. Evidence for three functional RNA species in several strains of cucumber mosaic virus. J Gen Virol. 1974 Jan;22(1):81–93. doi: 10.1099/0022-1317-22-1-81. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Turnbull N. T. Analysis of the 3'-terminal nucleotide sequence of vesicular stomatitis virus N protein mRNA. Nucleic Acids Res. 1978 Nov;5(11):4007–4024. doi: 10.1093/nar/5.11.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W., Symons R. H. Cucumber mosaic virus contains a functionally divided genome. Virology. 1973 Jun;53(2):487–492. doi: 10.1016/0042-6822(73)90232-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel P. F., Wells R. D. The synthesis and purification of (gamma-32P)-adenosine triphosphate with high specific activity. J Biol Chem. 1973 Dec 10;248(23):8319–8321. [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons R. H. The rapid, simple and improved preparation of high specific activity alpha-[32P]dATP and alpha-[32P]ATP. Nucleic Acids Res. 1977 Dec;4(12):4347–4355. doi: 10.1093/nar/4.12.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmern D., Kaesberg P. 3'-terminal nucleotide sequence of encephalomyocarditis virus RNA determined by reverse transcriptase and chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4257–4261. doi: 10.1073/pnas.75.9.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]