Abstract
We have shown previously that the function of Ycf1p, yeast ortholog of multidrug resistance-associated protein 1 (MRP1), is regulated by yeast casein kinase 2α (Cka1p) via phosphorylation at Ser251. In this study, we explored whether casein kinase 2α (CK2α), the human homolog of Cka1p, regulates MRP1 by phosphorylation at the semiconserved site Thr249. Knockdown of CK2α in MCF7-derived cells expressing MRP1 [MRP1 CK2α(−)] resulted in increased doxorubicin sensitivity. MRP1-dependent transport of leukotriene C4 and estradiol-17β-d-glucuronide into vesicles derived from MRP1 CK2α(−) cells was decreased compared with MRP1 vesicles. Moreover, mutation of Thr249 to alanine (MRP1-T249A) also resulted in decreased MRP1-dependent transport, whereas a phosphomimicking mutation (MRP1-T249E) led to dramatic increase in MRP1-dependent transport. Studies in tissue culture confirmed these findings, showing increased intracellular doxorubicin accumulation in MRP1 CK2α(−) and MRP1-T249A cells compared with MRP1 cells. Inhibition of CK2 kinase by 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole resulted in increased doxorubicin accumulation in MRP1 cells, but not in MRP1 CK2α(−), MRP1-T249A, or MRP1-T249E cells, suggesting that CK2α regulates MRP1 function via phosphorylation of Thr249. Indeed, CK2α and MRP1 interact physically, and recombinant CK2 phosphorylates MRP1-derived peptide in vitro in a Thr249-dependent manner, whereas knockdown of CK2α results in decreased phosphorylation at MRP1-Thr249. The role of CK2 in regulating MRP1 was confirmed in other cancer cell lines where CK2 inhibition decreased MRP1-mediated efflux of doxorubicin and increased doxorubicin cytotoxicity. This study supports a model in which CK2α potentiates MRP1 function via direct phosphorylation of Thr249.
Introduction
Development of multidrug resistance is a major cause of treatment failure in cancer. Increased drug efflux from cancer cells resulting from up-regulation of one or more members of the ATP-binding cassette (ABC) transporter family is one of the drug resistance mechanisms. Within the ABC superfamily, ABCB1 (P-glycoprotein/multidrug resistance), ABCG2 (breast cancer resistance protein), and several members of the ABCC subfamily were shown to act as extrusion pumps for a broad range of therapeutic agents (Juliano and Ling, 1976; Cole et al., 1992; Grant et al., 1994; Lautier et al., 1996; Doyle et al., 1998; Munoz et al., 2007).
To date, the best-characterized member of the ABCC subfamily is multidrug resistance-associated protein 1 (MRP1/ABCC1). MRP1 is a 190-kDa membrane glycoprotein, composed of an ABC ‘core’ region and a unique N-terminal extension (NTE), a defining characteristic of ABCC subfamily (Fig. 1). MRP1 is ubiquitously expressed in normal human tissues, often as a component of blood-tissue barriers, limiting penetration of numerous cytotoxic agents to the tissue. Furthermore, MRP1 is up-regulated in a number of cancer types (e.g., leukemias, lung, breast, prostate, etc.) where it confers resistance to many chemotherapeutic agents (Nooter et al., 1995; Chen and Tiwari, 2011). Although MRP1 has been shown to transport several endogenous substrates, such as leukotriene C4 (LTC4) and estradiol-17β-d-glucuronide (E217βG), it also transports a wide range of structurally unrelated compounds, including chemotherapeutic agents such as doxorubicin and vincristine as well as numerous glutathione, glucuronide, and sulfate conjugates of various xenobiotics (Keppler et al., 1997).
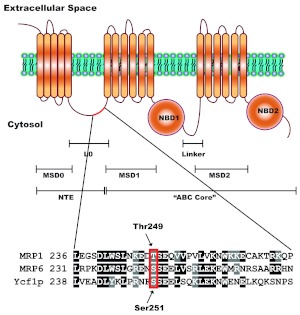
Fig. 1.
A putative CK2 phosphorylation site is semiconserved in human MRP1. The structure of MRP1 consists of a unique for ABCC subfamily NTE and a common ABC “core” region. The NTE is composed of one membrane-spanning domain (MSD0) and a cytosolic linker (L0), whereas the ABC “core” region, common to all ABC proteins, contains two membrane-spanning domains (MSD1 and MSD2) and two nucleotide-binding domains (NBD1 and NBD2). The NTE is thought to be involved in protein trafficking, stability, substrate binding, transport (Bakos et al., 2000; Fernández et al., 2002; Mason and Michaelis, 2002), and possibly in protein dimerization (Doyle et al., 1998; Yang et al., 2007, 2010). The full-length ABCC protein sequences, MRP1, MRP6, and Ycf1p were aligned using ClustalW. Alignment revealed that Ser251 of NTE, recently characterized as Cka1p regulatory site on Ycf1p (Paumi et al., 2008; Pickin et al., 2010) is semiconserved in human MRP1 as Thr249. The site is also conserved in murine MRP6 and was shown to be phosphorylated in vivo (Villén et al., 2007).
CK2 is a highly conserved serine/threonine protein kinase that forms a tetrameric complex of two catalytic (α and/or α′) and two regulatory (β) subunits; however complex formation is not required, because separate subunits are also active (Litchfield, 2003). The kinase is highly pleiotropic, with more than 300 substrates identified, and interacts with multiple signaling pathways (Meggio and Pinna, 2003; Duncan and Litchfield, 2008; Ruzzene and Pinna, 2010). CK2 kinase is considered one of the “master regulators” of the cell, playing a major role in processes related to cell growth, proliferation, death, and survival (Trembley et al., 2010). The kinase is constitutively active and ubiquitously expressed in all eukaryotes (Duncan and Litchfield, 2008). Moreover, CK2 expression is uniformly up-regulated in cancer and implicated in cellular transformation and tumorigenesis as well as multidrug resistance phenotype potentiation etc. (Yamane and Kinsella, 2005; Di Maira et al., 2007; Mishra et al., 2007; Kramerov et al., 2008; Ruzzene and Pinna, 2010). Elevation of CK2 protein expression in cancer has been associated with tumor aggressiveness and poor prognosis and thus became an attractive target for cancer therapy (Duncan and Litchfield, 2008; Pagano et al., 2010; Trembley et al., 2010).
To date, post-translational regulation in the ABC family has been poorly characterized. Phosphorylation, among other post-translational modifications, is widely used by cells as a rapid and reversible mechanism regulating protein activity (Burnett and Kennedy, 1954; Hunter and Sefton, 1991). Several ABC transporters, including ABCB1 and (cystic fibrosis transmembrane conductance regulator (CFTR; ABCC7), were reported to be regulated by phosphorylation; however, high-throughput liquid chromatography/mass spectrometry analyses of the human phosphoproteome identified multiple phosphopeptides derived from other ABC transporters, suggesting that nearly all ABC proteins are phosphorylated to some extent (Chambers et al., 1994; Chappe et al., 2003; Stolarczyk et al., 2011). For MRP1, the majority of the phosphorylation sites identified to date by high-throughput screens are within the “linker” region/“R-like” domain (Stolarczyk et al., 2011; http://www.phosphosite.org).
A proven and tractable genetic model to study ABCC function and regulation is the MRP1 ortholog yeast cadmium factor 1 (Ycf1p) of Saccharomyces cerevisiae (Paumi et al., 2009). In an effort to identify new pathways by which the ABCC transporters are regulated, our group has carried out a number of high-throughput protein interactor studies (Paumi et al., 2008, 2009). As part of these studies, CK2α was identified as a regulator of Ycf1p function in response to salt stress. We showed that Cka1p, the yeast counterpart of human CK2α, regulates Ycf1p function via phosphorylation of Ser251 within its L0 region (Paumi et al., 2008; Pickin et al., 2010). It is noteworthy that the CK2 consensus site within Ycf1p is semiconserved in human MRP1 as Thr249 (Fig. 1). In the study described herein, we examined the role of human CK2α in the regulation of MRP1 function via putative phosphorylation at Thr249. We provide evidence that strongly suggests that CK2α regulates MRP1 function via phosphorylation of Thr249. Furthermore, we show that MRP1 is regulated by CK2 in a variety of cancer cells. Inhibition of CK2 with CK2-specific inhibitors decreases MRP1-dependent efflux of doxorubicin and increases doxorubicin cytotoxicity.
Materials and Methods
Materials.
[3H]LTC4, [3H]E217βG, and [32P]γ-ATP were purchased from PerkinElmer Life and Analytical Sciences (Waltham, MA). CK2 and ABC transporter inhibitors used in this study were purchased as indicated: cycloheximide (CHX), 2-dimethylamino-4,5,6,7-tetrabromobenzimidazole (DMAT), and 4,5,6,7-tetrabromo-benzimidazole (TBBz) from Sigma-Aldrich (St. Louis, MO); (E)-3-[[[3-[2-(7-chloro-2-quinolinyl)ethenyl]phenyl]-[[3-dimethylamino)-3-oxopropyl]thio]methyl]thio]-propanoic acid (MK571) from Cayman Chemicals (Ann Arbor, MI); and valspodar (PSC883) and fumitremorgin C from Solvo Biotechnology (Budapest, Hungary). DMEM, FBS, penicillin/streptomycin, and G418 (Geneticin) were purchased from Invitrogen (Carlsbad, CA). Puromycin and doxorubicin were from Calbiochem/Merck KGaA (Darmstadt, Germany). MTT was from AMRESCO LLC (Solon, OH). FuGENE6 Transfection Reagent, Complete EDTA-free, and PhosSTOP tablets were obtained from Roche Diagnostics (Basel, Switzerland). Phenylmethylsulfonyl fluoride and Pepstatin A were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Culture.
All cell lines were derived from parental human breast cancer cells MCF7/WT and MCF7/MRP1 (a gift from Dr. Charles Morrow, Department of Biochemistry, Wake Forest University School of Medicine, Winston-Salem, NC) and cultured in high-glucose DMEM supplemented with 10% FBS and 1× penicillin/streptomycin. Selection antibiotics (0.5 μg/ml puromycin and/or 1.5 mg/ml G418) were added to the media depending on plasmid construct used for knockdown or mutant overexpression. A549 cells were a gift from Dr. Rolf Craven (University of Kentucky, Lexington, KY) and cultured in F-12K with 20% FBS and penicillin/streptomycin. H460 and HeLa cells were a gift from Dr. Vivek Rangnekar (University of Kentucky, Lexington, KY) and were cultured in RPMI 1640 medium and DMEM with 10% FBS and 1× penicillin/streptomycin, respectively.
Site-Directed Mutagenesis and Transductions.
The retroviral expression vector pLNCX-MCS-X/S-MRP1, containing coding sequence for human MRP1, was obtained as a kind gift from Dr. Charles Morrow. MRP1 was mutated at Thr249 to Ala or Glu using QuikChange XL (Stratagene/Agilent Technologies, Santa Clara, CA). Primers were designed using the QuikChange primer design program. Mutagenesis was carried-out according to the manufacturer's instructions and yielded two new products, pLNCX-mcs-X/S-MRP1-T249A and pLNCX-mcs-X/S-MRP1-T249E. Mutagenesis and MRP1 sequence integrity were confirmed by sequencing (Eton Biosciences, Inc., San Diego, CA). Expression vectors pLNCX-mcs-X/S-MRP1-T249A and pLNCX-mcs-X/S-MRP1-T249E were transfected into PA317 packaging cell line by the calcium phosphate precipitation method (Ausubel et al., 1987). Media containing viral particles was collected from above the cells and used to transduce MCF7/WT cells. Transduced cells were grown under G418 selection, and clones were selected and analyzed for MRP1 expression via Western blotting as described previously (Paumi et al., 2003). At least two clones for each mutation were analyzed in cytotoxicity, transport, and doxorubicin accumulation assays that yielded similar results. Clones with MRP1 expression levels matching the one from MRP1 cells are described in this study.
CK2α Knockdowns.
Nonsilencing (scrambled) CK2α-specific shRNAs in GIPZ transfer vectors were purchased from Open Biosystems (Huntsville, AL). To maximize transfection efficiency, GIPZ plasmids were transfected into WT and MRP1 cells with use of FuGENE6 (Roche Diagnostics) and ArrestIn (Open Biosystems) transfection reagents. In brief, cells were plated in six-well dishes at 105 cells/well; the next day, plasmid DNA (1 μg/well) was diluted in small volume of serum-free media, mixed with transfection reagent (5 μl of reagent per 1 μg of DNA), incubated for 30 min at room temperature to allow complex formation, and added drop-wise to cells. Forty-eight hours after transfection, fresh medium with 0.5 μg/ml puromycin was added to scrambled control lines, and cells were grown until polyclonal stable lines were obtained. As for CK2α knockdowns, 48 h after transfection, cells were trypsinized, transferred to a larger dish, and grown under puromycin selection to obtain stable monoclonal cultures. Multiple stable clones were analyzed by immunoblotting for CK2α expression. Several of them with variable suppression of CK2α were further characterized by cytotoxicity and doxorubicin accumulation assays, which confirmed similar behavior (data not shown). Clones with matching levels of CK2α suppression are described in this study.
Immunocytochemistry.
Cells were plated on coverslips the day before staining. The next day, immunostaining was carried out as follows: coverslip cultures were rinsed twice with ice-cold phosphate-buffered saline (PBS) and fixed by incubation with ice-cold methanol/water (v/v) for 10 min at 4°C. The cells were washed three times with ice-cold PBS, blocked in 1% bovine serum albumin (BSA) in PBS for 30 min, and incubated for 1 h with primary antibody MRPr1 (1:40; Santa Cruz Biotechnology, Santa Cruz, CA) in 1% BSA in PBS. Then the cells were washed three times with 0.1% BSA in PBS and incubated in the dark with a secondary antibody conjugated to fluorochrome (Alexa Fluor 568; Invitrogen), washed, mounted on glass slides with ProLong antifade reagent with 4,6-diamidino-2-phenylindole (Molecular Probes/Invitrogen), and analyzed by fluorescence microscopy [Olympus BX512F (Olympus, Tokyo, Japan) equipped with fluorescence (EXFO X-cite 120 Fluorescence Illuminating System; Lumen Dynamics Group Inc., Mississauga, ON, Canada) and Nomarski interference contrast]. Images were captured with an Olympus XM10 camera and digitally documented using cellSens software (Olympus).
Cycloheximide Chase Assay.
Cells were seeded at 3 × 105 cells/well in six-well dishes in 2 ml of DMEM/10% FBS and allowed to grow overnight at 37°C and 5% CO2. The following day, cells were washed twice with ice-cold PBS; freshly prepared media containing 100 μg/ml of CHX was added, and the cells were returned to the incubator. Cell lysates were prepared at 0 (no CHX added), 8, 24, and 32 h after CHX addition. In brief, cells were washed twice with ice-cold PBS and lysed by the addition of 250 μl of ice-cold RIPA buffer, containing phenylmethylsulfonyl fluoride and protease inhibitors. Cells were incubated in the presence of RIPA buffer for 5 min to allow for complete cell lysis. Cell lysates were then placed in a new tube containing 50 μl of 6× SDS-PAGE loading buffer and frozen at −20°C. Frozen samples were later thawed and incubated at 37°C for 1 h; 70 μl of each sample was run on a 7% SDS-PAGE gel followed by Western blotting procedure as described previously (Paumi et al., 2003). MRP1 protein was probed with rat MRP1r1 antibody (1:1000). Quantification of band density was carried out using Adobe Photoshop CS4 (Adobe Systems, Mountain View, CA).
Cytotoxicity Assays.
Doxorubicin cytotoxicity was determined using MTT assay. Cells were plated in 96-well plates at 3 × 103/well for MCF7 cells and 5 × 104/well for HeLa, H460, and A549 cells. Twenty-four hours after cells were plated, fresh media containing drug or vehicle were added and incubated at 37°C under 5% CO2 for 72 h. MTT solution in amount equal to 10% of the culture volume was added to the final concentration of 0.5 μg/ml, and cells were returned to the incubator. After 3 h, media was removed, DMSO was added, and plates were shaken until all MTT crystals dissolved. Absorbance was read at 560 nm with Titertek Multiskan MCC/340 plate reader (Thermo Fisher Scientific, Waltham, MA). The data were normalized to the baseline absorbance (no drug added) and analyses were performed with use of Prism 5 software (GraphPad Software, San Diego, CA). Drug sensitivity was characterized by three summary statistics: AUC was calculated by the trapezoidal method were derived from the survival-drug concentration smooth fit curve (Moon, 1980); IC50 and IC90 were derived from four-parameter logistic nonlinear regression curve fits (Campling et al., 1991).
Inside-out Plasma Membrane Vesicles Preparation and Transport Assay.
Inside-out vesicles were prepared by nitrogen cavitation as described previously (Loe et al., 1996; Paumi et al., 2001). In brief, frozen cell pellets (4 × 108 cells) were thawed on ice in 7 ml of the homogenization mixture. Cells were disrupted by nitrogen cavitation at 1250 psi with constant stirring for 20 min at 4°C. The homogenate was centrifuged at 1700 rpm in a centrifuge (Beckman Coulter, Fullerton, CA) at 4°C for 15 min. The supernatant was overlaid on a 3-ml sucrose cushion [35% (w/v) in 10 mM Tris, pH 7.5, and 1 mM EDTA]. After centrifugation at 35,000 rpm for 2 h at 4°C (SW41 rotor; Beckman Coulter), the opaque interface was collected, diluted into five parts TS buffer (10 mM Tris, pH 7.5, and 250 mM sucrose), and centrifuged at 35,000 rpm for 40 min at 4°C (SW41 rotor). The pellet was suspended in 1 ml of 50 mM Tris, pH 7.5, and 250 mM sucrose, gently dispersed by 10 to 15 passages through a 27-gauge needle, and stored in aliquots at −80°C.
The MRP1-dependent uptake of 3H-labeled conjugate by vesicles was determined using an adaptation of the membrane rapid filtration method (Keppler et al., 1998; Paumi et al., 2003). In brief, 50-μl reaction mixtures contained 50 mM Tris, pH 7.5, 10 mM MgCl, and 250 mM sucrose, 4 mM ATP or 5′-adenylyl (β,γ-methylene)diphosphonate (nonhydrolyzable ATP control), and various concentrations of [3H]LTC4 or [3H]E217βG. Mixtures were warmed to 37°C, and reactions were initiated by addition of membrane vesicles (5 μg/50-μl reaction). After a 1-min incubation, the reactions were terminated by the addition of 1 ml of ice-cold TS buffer. Samples were immediately filtered with vacuum through 25 mm hydrophilic membrane filters (GVWP; Millipore Corp., Billerica, MA), and the retained vesicles were washed twice with 1 ml of ice-cold TS buffer before liquid scintillation counting.
Doxorubicin Accumulation Assay.
Cells were plated in complete DMEM, without selection antibiotics, at 3 × 105 cells/well in 24-well dishes the day before assay to be roughly 90% confluent at the time of the assay. The next day, cells were incubated with 50 μM doxorubicin for 1 h, followed by a 30-min efflux step without doxorubicin. Where indicated, cells were pretreated with inhibitor for 30 min to 1 h before addition of doxorubicin. Next, cells were washed with PBS, trypsinized, transferred to 1.7-ml Eppendorf tubes, spun down, and lysed in 700 μl of RIPA buffer. Lysate (200 μl; in duplicate or triplicate) was then transferred to Optilux 96-well Microplates (Falcon; BD Biosciences Discovery Labware, Bedford, MA) and, for all MCF7-derived cell lines, fluorescence was read by SpectraMax M2 (Molecular Devices, Sunnyvale, CA), with excitation at 536 nm, emission at 648 nm, and cutoff at 630 nm (to exclude green fluorescent protein-derived fluorescence from GIPZ plasmid). For all other cell lines, we used Synergy 2 Multi-Mode Microplate Reader (BioTek Instruments, Inc.) with excitation filter 460/40 and emission 560/15. After fluorescence reading, 25 μl of lysate from each well was taken for protein measurement by bicinchoninic acid assay (Thermo Fisher Scientific).
Synthesis of Synthetic Peptides.
All peptides were assembled by 9-fluorenylmethoxycarbonyl solid-phase peptide synthesis on a TETRAS peptide synthesizer (CreoSalus, Louisville, KY). In brief, the peptides were synthesized using Wang resins substituted with the first amino acid. Amino acid deprotection and coupling then followed standard methods to extend the peptide sequence as designed. Biotin was incorporated at the N terminus via standard coupling with the exposed carboxylic acid functionality. Upon completion of assembly, the resin was washed with dimethylformamide, MeOH, and CH2Cl2 and then dried. The peptide was cleaved from the resin by gentle mixing of the resin with a mixture of 6% phenol, 4% double-distilled water, and 90% trifluoroacetic acid for 5 h, and the resin was filtered away. The peptides were precipitated by the addition of ice-cold diethyl ether and dried under vacuum. The crude peptide was purified by reversed-phase high-performance liquid chromatography (double-distilled water/acetonitrile and 0.5% trifluoroacetic acid), and the mass of each of the peptides was confirmed by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry. Peptide synthesis yielded three peptides for use in the recombinant human CK2α kinase assays. The three peptides synthesized were biotin-Ahx-RRRADDSDDDDDK (Control), biotin-Ahx- LNKEDTSEQVV (MRP1-Thr249 peptide, Thr249 in bold), and biotin-Ahx-LNKEDASEQVV (MRP1-T249A peptide, Ala249 in bold). Electrospray mass spectrometry confirmed the mass of each peptide.
In Vitro Phosphorylation Assay.
Kinase assays were carried out using a modification of the Sigma-Aldrich biotin substrate-tagged method. In brief, radiometric assays were carried out in 50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 0.5 mM dithiothreitol, 100 ng/ml BSA, 100 μM ATP, and 2 μl of [32P]γ-ATP (10 Ci/mmol) (PerkinElmer Life and Analytical Sciences), and 200 mM NaCl using 250 μM biotinylated synthetic peptide as the substrate in a 100-μl reaction volume. Reactions were initiated by the addition of recombinant human CK2α (Assay Designs, Ann Arbor, MI) and carried out for 10 min at 30°C. Reactions were stopped by the addition of a 25-μl reaction aliquot to 4 μl of 0.5M EDTA and 4 μl of 250 mg/ml avidin (Thermo Fisher Scientific) and allowed to incubate at room temperature for 5 min. Terminated reaction aliquots were added to a 30-kDa spin column (Pall Scientific, East Hills, NY) spun at 14,000 rpm for 5 min and subsequently washed two times with 100 μl of wash solution (0.5 M phosphate and 0.5 M NaCl, pH 8.5). Spin columns were submerged directly into scintillation vials containing 12 ml of scintillation cocktail and counts per minute of phosphorylated peptide were determined via a scintillation counter (PerkinElmer Life and Analytical Sciences). All experiments were normalized to a zero time point and performed in triplicate. Results are reported as nanomoles of phosphorylated peptide formed per 5 min of reaction per milligram of recombinant protein.
Synthesis and Purification of the Thr249 Phosphospecific Antibody.
The synthesis and purification of the MRP1-Thr249-P phosphospecific antibody was performed by Open Biosystems. In brief, two small peptides containing the MRP1-Thr249 CK2 consensus site in a nonphosphorylated and a nonhydrolyzable form [CWSLNKEDTSEQVVP and CWSLNKED(pT)SEQVVP, respectively] were synthesized using standard techniques and conjugated to keyhole limpet hemocyanin. Two rabbits were injected with the phosphorylated version of the synthetic peptide over a 90-day period (Open Biosystems 90-day protocol). Serum was collected at three separate times (days 0, 35, and 90) and shipped. The specificity of each aliquot was tested by immunoprecipitation and Western blotting for MRP1. One rabbit was found to produce MRP1-specific antibodies. Serum containing MRP1-specific antibody was then subjected to negative affinity chromatography according to the Open Biosystems standard protocol.
Immunoprecipitations.
For immunoprecipitations with MRP1-Thr249-P phosphospecific antibody, cells were lysed by brief sonication, and membrane fractions were prepared in the presence of protease and phosphatase inhibitors. Membranes were resuspended in 20 mM Tris-HCl and 0.5% n-dodecyl-β-d-maltopyranoside, and standard immunoprecipitation procedures were followed. In brief coimmunoprecipitation, cells were lysed by addition of lysis buffer (20 mM Tris-HCl, 0.5% n-dodecyl-β-d-maltopyranoside, and 1× protease inhibitor cocktail) and incubated while rocking for 10 min at 4°C, Lejjft on ice for 1 h, and subsequently spun down at 500g for 15 min, and the supernatant was retained. Protein concentration was measured by bicinchoninic acid assay (Thermo Fisher Scientific). One milligram of membrane or total protein lysate was incubated overnight while rocking at 4°C with primary antibodies, MRP1-Thr249-P (phosphospecific antibody synthesized by Open Biosystems), anti-MRP1 (QCRL-1; Santa Cruz Biotechnology), or anti-CK2α (Santa Cruz Biotechnology). The next day, Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) was added, and reactions were incubated overnight. Immunoprecipitates were washed three times in lysis buffer, then 2× Laemmli sample buffer (Bio-Rad Laboratories, Hercules, CA) was added, and preparations were incubated for 1 h at 37°C followed by SDS-PAGE electrophoresis. Proteins were transferred by semidry transfer (Bio-Rad Laboratories) to nitrocellulose membrane, blocked in 3% BSA in Tris-buffered saline/Tween 20 (for phosphospecific antibody) or with 5% nonfat dry milk in Tris-buffered saline/Tween 20, and incubated with primary antibody to detect the protein of interest.
Results
Suppression of CK2α Protein Expression Results in Decreased MRP1 Transport Activity.
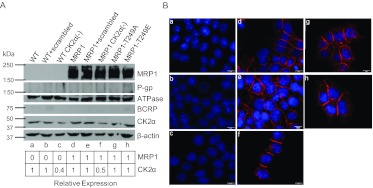
We chose MCF7 cells as our study model because they lack ABCB1 and ABCG2 transporter expression, which was crucial for study of MRP1 function, because substrate specificity of these three transporters greatly overlaps. We generated a number of stable MCF7-derived cell lines, and those with matched CK2α and MRP1 expression were selected for further analysis. This was critical for cross–cell-line comparisons. To determine whether MRP1 is regulated by CK2α, we measured the effect of reduced CK2α expression on MRP1-mediated cellular resistance to doxorubicin and on MRP1-dependent in vitro transport. We reasoned that if CK2α regulates MRP1 function, then decreasing cellular CK2α activity via shRNA-mediated silencing of CK2α protein should result in a change in MRP1 function. MRP1-overexpressing (MRP1) and wild-type (WT) MCF7 cells were transfected with scrambled or CK2α-specific shRNAs, and stable clones with CK2α expression reduced by half were obtained (Fig. 2A, lanes b, c, e, and f). Immunofluorescence staining was performed to assess whether shRNA delivery or CK2α knockdown altered cellular localization of MRP1; however, no difference in localization of MRP1 protein was observed compared with MRP1 cell line (Fig. 2B, compare e, f, and d).
Fig. 2.
Expression of relevant proteins in MCF7-derived cell lines used in this study. Cell lines depicted in A and B are designated as follows: a, WT cells. b, WT scrambled. c, WT CK2α(−). d, MRP1. e, MRP1 scrambled. f, MRP1 CK2α(−). g, MRP1-T249A. h, MRP1-T249E. A, cell lysates (100 μg) were subjected to electrophoresis on 9% SDS-PAGE gel and immunoblotted with antibodies specific for human MRP1 (mouse monoclonal QCRL-1, 1:200; Santa Cruz Biotechnology), Na+/K+-ATPase α-1 (mouse monoclonal Na+/K+ ATPase, 1:20,000; Millipore Corp.), ABCB1 (mouse C219, 1 μg/ml; Calbiochem), ABCG2 (BXP-21; Santa Cruz Biotechnology), CK2α [goat CK2α (C-18), 1:500; Santa Cruz Biotechnology], β-actin (mouse β-actin, 1:1000; Abcam Inc., Cambridge, MA). Densitometric analysis was performed with use of Adobe Photoshop CS4, and expression of MRP1 and CK2α was normalized to β-actin. B, immunofluorescence staining of all MCF7-derived cell lines showing no visible changes in cellular localization of overexpressed MRP1 protein compared with control (MRP1): red, MRP1 signal (MRPr1 antibody, Alexa 568); blue, 4,6-diamidino-2-phenylindole nuclear stain.
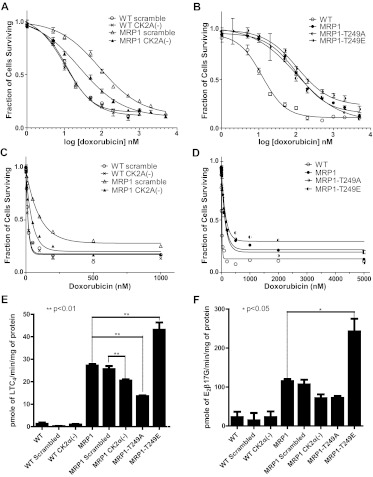
The effect of CK2α on MRP1-mediated resistance to doxorubicin was measured by MTT assay, and AUC, IC50, and IC90 values were extrapolated from the cell survival plots (Fig. 3, A and C; Table 1). Knockdown of CK2α expression had no effect on cellular viability of WT cells [Fig. 3, A and C; Table 1; WT scrambled versus WT CK2α(−)], whereas it significantly decreased resistance to doxorubicin in MRP1-expressing cells [Fig. 3, A and C; Table 1; MRP1 scrambled versus MRP1 CK2α(−)].
Fig. 3.
Knockdown of CK2α and MRP1-Thr249 mutations alter MRP1 function in tissue culture and in vitro transport assays. Doxorubicin cytotoxicity profiles were determined by MTT assay as described under Materials and Methods. Points represent mean values ± 95% confidence intervals of three or more separate experiments performed with repeated measures in 96-well plates. The data were normalized to the baseline absorbance. Four-parameter logistic nonlinear regression model fit and fraction of cell survival-doxorubicin concentration data plots were generated with use of GraphPad Prism 5. Summary statistics derived from these data are listed in Table 1. A, four-parameter logistic nonlinear regression curve fit for WT scrambled, WT CK2α (−), MRP1 scrambled, MRP1 CK2α(−). B, four-parameter logistic nonlinear regression curve fit for WT, MRP1, MRP1-T249A, and MRP1-T249E. C, fraction of cell survival-doxorubicin concentration data plot for WT scrambled, WT CK2α (−), MRP1 scrambled, and MRP1 CK2α(−). D, fraction of cell survival-doxorubicin concentration data plot for WT, MRP1, MRP1-T249A, and MRP1-T249E. In vitro transport into inside-out vesicles prepared from presented cell lines was measured for two 3H-labeled MRP1 substrates, LTC4 (E) and E217βG (F). Transport experiments were performed in triplicate or greater, and the bar graph shows mean values ± S.D. Statistical analysis was performed using one-way ANOVA followed by Bonferroni post test.
TABLE 1.
Doxorubicin sensitivity of MCF7-derived cell lines used in this study
AUC was derived from fraction cell survival-doxorubicin concentration plots in Fig. 3, C and D. IC50 and IC90 values derived from four-parameter logistic nonlinear regression curve fit presented in Fig. 3, A and B.
| Cell Line | AUC | IC50 | 95% CI | IC90 | 95% CI |
|---|---|---|---|---|---|
| nM | nM | nM | |||
| WT scrambled | 162a | 11.7 | 10.90–12.52 | 2.16 | 1.81–2.58 |
| WT CK2A(−) | 155a | 10.6 | 9.459–11.90 | 1.46 | 1.08–1.98 |
| MRP1 scramble | 360a | 88.6b | 79.23–98.97 | 5.45b | 3.99–7.46 |
| MRP1 CK2A(−) | 229a | 24.9b,c | 21.55–28.73 | 1.31c | 0.9036–1.885 |
| WT | 564 | 12.2 | 10.98–13.47 | 2.45 | 1.9–3.16 |
| MRP1 | 1114 | 105.3d | 97.85–113.3 | 12.93d | 10.68–15.67 |
| MRP1-T249A | 961 | 101.7d | 87.53–118.2 | 5.58d,e | 3.76–8.28 |
| MRP1-T249E | 1514 | 107.7d | 94.61–122.6 | 11.67d | 0.39–16.22 |
CI, confidence interval.
AUC calculated only for data points between 0 and 1 μM to allow for comparison between the lines.
Significantly different from WT scrambled control.
Significantly different from MRP1 scrambled.
Significantly different from WT control.
Significantly different from MRP1.
To determine whether the decrease in MRP1-mediated resistance to doxorubicin in MRP1 CK2α(−) cells was due to a decrease in MRP1 function, in vitro transport assays were performed on WT scrambled, WT CK2α(−), MRP1 scrambled, and MRP1 CK2α(−) derived inside-out vesicles. ATP-dependent transport of LTC4 and E217βG was measured as described under Materials and Methods. Transport of LTC4 into MRP1 CK2α(−) vesicles was significantly reduced compared with MRP1 and MRP1 scrambled control (Fig. 3E). Transport of E217G also shows a trend to be reduced; however, it did not reach statistical significance (Fig. 3F).
MRP1 Phosphorylation at Thr249 Positively Regulates Transport Activity in Vitro.
To determine whether phosphorylation of Thr249 affects MRP1 transporter function, two site-specific mutants were generated: MRP1-T249A, with alanine blocking phosphorylation at Thr249; and MRP1-T249E mutant, mimicking threonine phosphorylation. WT cells were retrovirally transduced with each mutant vector, and stable clones of both MRP1-T249A and MRP1-T249E were selected to have expression comparable with MRP1 cells (Fig. 2A, lanes g and h versus lane d). None of the cell lines expressed ABCB1 or ABCG2. Immunofluorescence staining confirmed that mutations had no visible effect on MRP1 cellular localization, and both mutants localized predominantly to the plasma membrane, similar to wild-type MRP1 (Fig. 2B, d, g, and h). In addition, we performed cycloheximide chase assay to address whether mutation of Thr249 to Ala or Glu affects MRP1 protein stability. We have found that the stability of both MRP1-T249A and MRP1-T249E does not differ from that of the wild-type MRP1 protein, suggesting that the mutations at Thr249 do not alter protein folding (Supplemental Fig. 1).
The resistance of MRP1-, MRP1-T249A-, and MRP1-T249E-expressing cells to doxorubicin and in vitro transport assays were carried out as described above for the CK2α knockdowns. Overexpression of MRP1 resulted in approximately 10 fold increase in cellular resistance to doxorubicin (Fig. 3B; Table 1; WT versus MRP1), consistent with previous reports (Paumi et al., 2003). Although cell survival plots for MRP1-T249A and MRP1-T249E do not seem to differ from those for the MRP1 cell line, a modest increase in sensitivity can be seen for MRP1-T249A cells, and a trend to confer more resistance can be seen for MRP1-T249E cells (Fig. 3, B and D; Table 1). In vitro transport assays (Fig. 3, E and F) revealed that MRP1-T249A dependent transport of both LTC4 and E217βG was decreased by approximately half, and MRP1-T249E dependent transport of LTC4 and E217βG was roughly double that of MRP1. These results support our hypothesis that phosphorylation at Thr249 positively regulates MRP1 transport activity.
Inhibition of CK2α Results in Decreased Doxorubicin Efflux from Mrp1 Expressing Cells in Cell Culture.
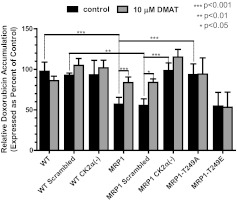
As a complementary analysis to the work described above, we assessed MRP1-mediated drug efflux from cells pretreated with doxorubicin by measuring intracellular accumulation of the drug. Consistent with cytotoxicity and transport data described earlier, CK2α knockdown in MRP1-expressing cells resulted in increased doxorubicin accumulation in these cells compared with MRP1 scrambled control [Fig. 4, compare black bars for MRP1 CK2α(−) versus MRP1 scrambled, p < 0.001]; no difference upon knockdown of CK2α was observed in WT cells [Fig. 4, WT scrambled versus WT CK2α(−)]. Compared with wild-type MRP1 cells, we observed increased doxorubicin accumulation in MRP1-T249A mutant cell line (Fig. 4, MRP1 versus MRP1 T249A; p < 0.001) and no change in MRP1-T249E mutant cells (Fig. 4, MRP1 versus MRP1 T249E).
Fig. 4.
CK2α knockdown, inhibition of CK2, and mutation of the putative CK2 phosphorylation site at Thr249 to alanine results in decreased MRP1-mediated doxorubicin efflux. Doxorubicin accumulation assays were performed as described under Materials and Methods. Where indicated, cells were pretreated with 10 μM DMAT, a potent CK2α inhibitor, before the addition of doxorubicin (gray bars). The bars represent mean value ± S.D. of three or more separate experiments with treatments performed in triplicate or greater. Statistical analysis was performed using ANOVA followed by Bonferroni post test.
The same assay was employed as a tool to determine the effect of CK2 kinase inhibition on MRP1 function. We reasoned that if the reduced resistance to doxorubicin cytotoxicity and decreased transport of LTC4 and E217βG in MRP1 CK2α(−) is the result of decreased MRP1 function, then treatment of the MRP1 CK2α(−) cells with a CK2 inhibitor (DMAT) should have no effect on the MRP1-dependent decreased doxorubicin accumulation associated with MRP1 overexpression [Fig. 4, DMAT treatment (gray bars versus black) on MRP1 CK2α(−) cells]. Furthermore, if CK2α regulates MRP1 function via Thr249, as we have proposed, then the doxorubicin efflux from MRP1-T249A and MRP1-T249E cells should not be affected by the addition of the CK2 inhibitor (Fig. 4, DMAT treatments on MRP1-T249A and MRP1-T249E). Our studies revealed this to be the case. However treatment of MRP1 and MRP1 scrambled cells with DMAT increased doxorubicin accumulation to levels similar to that of the WT, WT scrambled, WT CK2α(−), MRP1 CK2α(−), and MRP1-T249A cells (Fig. 4). Thus, through two independent techniques, we have shown that CK2 activates MRP1 through Thr249.
CK2α Regulates MRP1 Function via the Direct Phosphorylation of Thr249.
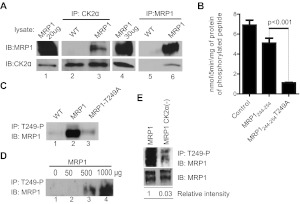
The findings described above show that MRP1 and CK2α proteins interact functionally and suggest that CK2α regulates MRP1 function via the phosphorylation of Thr249. In this light, we further hypothesized that CK2α directly phosphorylates MRP1 at Thr249, without intermediate proteins and/or pathways. Direct interaction of kinase and substrate, even if transient, is necessary for phosphorylation. Hence, to determine whether MRP1 and CK2α proteins indeed interact physically within cells, we carried out coimmunoprecipitation studies. As described under Materials and Methods, lysates from WT and MRP1 cells were subjected to coimmunoprecipitation with an antibody against either CK2α or MRP1, and immunoprecipitated protein complexes were further characterized by Western blotting. Both CK2α and MRP1 proteins were found in coimmunoprecipitations (coIPs) carried out against MRP1 and CK2α (Fig. 5A, lane 3 and 6), providing evidence for direct interaction of the two proteins.
Fig. 5.
A, MRP1 and CK2α proteins interact physically. Protein lysates prepared from WT and MRP1 cells were subjected to CoIP with an antibody against CK2α (goat CK2α; lanes 2 and 3) or an antibody against MRP1 (mouse QCRL-1; lanes 5 and 6). Protein A/G PLUS Agarose was added to precipitate the immunocomplex and proteins were analyzed by immunoblotting (IB). Small aliquot of MRP1 lysate was loaded as a positive control for immunoblotting (lanes 1 and 4). Reactions were carried-out in triplicate and blots shown here are representative of the series. B, phosphorylation of MRP1-Thr249 consensus site by recombinant CK2α is dependent on Thr249. The ability of recombinant human CK2α to phosphorylate MRP1-Thr249 consensus site was assessed by in vitro kinase assay as described under Materials and Methods. Synthetic peptides biotin-Ahx-RRRADDSDDDDDK (Control), biotin-Ahx- LNKEDTSEQVV (MRP1244–255), and biotin-Ahx- LNKEDASEQVV (MRP1244- 254T249A) were incubated in the presence of recombinant human CK2α and 32P-labeled ATP. All experiments were normalized to a zero time point and performed in triplicate, and results are reported as nanomoles of phosphorylated peptide formed per 5 min of reaction per milligram of recombinant protein. Statistical analysis was performed using Student's t test (control peptide served as a positive control to test for activity of the recombinant CK2α protein and therefore was not taken into consideration for statistical analysis). C, MRP1-Thr249-P antibody pulls down MRP1 protein from MRP1 cells but not from MRP1-T249A line. Custom-made rabbit antibody against synthetic peptide corresponding to MRP1-Thr249-P consensus site [WSLNKEDT(p)SEQVVP] was used to immunoprecipitate (IP) MRP1 protein from membrane fractions prepared from WT, MRP1, and MRP1-T249A cells, followed by IB with MRP1 antibody (QCRL-1). D, test of linearity carried out by immunoprecipitation with fixed amount of MRP1-Thr249-P antibody on increasing amount of MRP1 membrane fraction. E, knockdown of CK2α results in decreased MRP1 phosphorylation at Thr249. Membrane fractions prepared from MRP1 and MRP1 CK2α(−) cells were subjected to immunoprecipitation with rabbit MRP1-Thr249-P antibody (top). Relative phosphorylation (phosphorylated MRP1/ total MRP1) was determined by densitometric analysis with use of Adobe Photoshop CS4.
To determine whether CK2α kinase can phosphorylate Thr249 on MRP1, we performed in vitro kinase assays with human recombinant CK2α and three peptide substrates: a control peptide to measure general CK2 activity and two peptides derived from the CK2 consensus site in MRP1, the first containing Thr249, the second with Thr249 substituted by alanine. Peptide substrates were used instead of full-length proteind because MRP1 is a membrane protein with 17 membrane spans and high molecular weight; as such, a purification method of the full-length protein has yet to be described. Human recombinant CK2α phosphorylated both the control and the MRP1244–255 peptides, whereas the phosphorylation of the MRP1244–255-T249A was reduced 6-fold compared with the MRP1244–255 peptide (Fig. 5B). Summarizing, these results suggest that CK2α can phosphorylate MRP1 at Thr249.
Next, a phosphospecific antibody to Thr249 (MRP1-Thr249-P) was produced in a rabbit as described under Materials and Methods. Upon purification of the phosphoantibody by negative affinity chromatography, we tested its specificity by using it to immunoprecipitate the antigen from WT, MRP1, and MRP1-T249A membrane lysates (Fig. 5C). We reasoned that if MRP1 was phosphorylated at Thr249, as our data suggested, then MRP1 should be immunoprecipitated from MRP1 cells only, not from WT or MRP1-T249A. As anticipated, a strong MRP1 signal was detected in the immunoprecipitate from MRP1 cells and no signal or a very weak signal was detected in WT or MRP1-T249A lysates (Fig. 5C). To rule out the possibility that MRP1 signal is an artifact or due to unspecific background, we carried out a linearity experiment immunoprecipitating the antigen with fixed amount of phosphoantibody and increasing the amount of input MRP1 membrane fraction. In agreement with the hypothesis that the MRP1-Thr249-P is specific for phosphorylated Thr249 of MRP1 with increase in lysate input, we observed increase in MRP1 signal (Fig. 5D).
Finally, to determine whether CK2α is regulating MRP1 function via phosphorylation of Thr249, we used the same method to assess the phosphorylation status of Thr249 in MRP1 and MRP1 CK2α(−) cells. As shown in Fig. 5E, MRP1 phosphorylation at Thr249 was greatly diminished upon CK2α knockdown.
Inhibition of CK2 in MRP1-Expressing Cancer Cell Lines Results in Decreased MRP1-Mediated Doxorubicin Efflux and Increased Sensitivity to Doxorubicin.
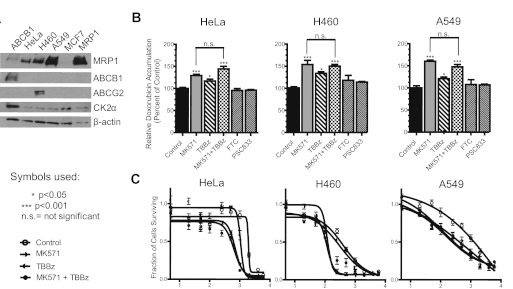
To validate our findings from MCF7 system overexpressing MRP1, we carried out doxorubicin accumulation assays in cells endogenously expressing MRP1 protein. To validate our model, a small panel of MRP1-expressing cell lines was selected: A549 non–small-cell lung cancer and HeLa cervical cancer cells. We considered the expression of other ABC proteins that transport doxorubicin, [e.g., ABCB1 and a mutant form of ABCG2 (R482T/G)]. Immunoblots in Fig. 6A show that all cell lines express MRP1 and CK2α but not ABCB1. ABCG2 was detected only in H460 (Fig. 6A). Doxorubicin accumulation assays were performed on all cell lines, untreated or pretreated with MK571 (MRP1 inhibitor), TBBz (CK2 inhibitor), fumitremorgin C (FTC; ABCG2 inhibitor), and PSC883 (ABCB1 inhibitor), where indicated (Fig. 6B). In this study, TBBz was substituted for DMAT as the CK2 inhibitor because DMAT recently went under patent and is no longer available for purchase. TBBz, the parental compound from which DMAT was derived, inhibits CK2 to a similar extent except that the IC50 is approximately 5-fold higher (Pagano et al., 2004). Therefore, in these studies, cells were treated with 50 μM TBBz as a replacement for 10 μM DMAT.
Fig. 6.
Inhibition of CK2 in HeLa, H460, and A549 cancer cell lines results in decreased MRP1-dependent doxorubicin efflux. A, Western blot showing expression of MRP1, ABCB1, ABCGw2, and CK2α proteins in HeLa, H460, A549, and MCF7 (WT) and MCF7/MRP1 (MRP1) cell lines. In lane 1, ABCB1-expressing cell line was loaded as a positive control for immunoblotting of ABCB1. B, doxorubicin accumulation assays were performed as described under Materials and Methods. Where indicated, cells were pretreated with 50 μM MK571, 40 μM TBBz, 10 μM FTC, or 1 μM PSC883 for 1 h before the addition of doxorubicin. Experiments were performed in triplicate, and results were presented as mean values ± S.D. Statistical analysis was performed using one-way ANOVA followed by Bonferroni post-test. C, cytotoxicity profiles of doxorubicin were determined by MTT assay as described under Materials and Methods. Points represent mean values ± 95% confidence intervals of two or three separate experiments performed with repeated measures in 96-well plates. The data were normalized to the baseline absorbance, and four-parameter logistic nonlinear regression curves were generated with use of GraphPad Prism 5. Summary statistics derived from these data are listed in Table 2.
As anticipated, inhibition of MRP1 with MK571 resulted in significant increase in doxorubicin accumulation in all three cell lines, consistent with high expression of MRP1 protein. Compared with untreated and MK571-treated cells, TBBz pretreatments alone resulted in an intermediate but significant increase in doxorubicin accumulation in cells. We reasoned that if TBBz-mediated increase in doxorubicin accumulation in these cells were dependent upon MRP1, then cotreatment of the cells with TBBz and MK571 would not increase doxorubicin accumulation beyond what was obtained with MK571 alone. The results depicted in Fig. 6B confirmed that this is indeed the case for HeLa, H460, and A549 cells. It is important to note that the doxorubicin accumulation was unaltered by treatment of cells with PSC833 and FTC, confirming that ABCB1 and ABCG2 do not play a role in doxorubicin efflux in these cells. Likewise, cell toxicity assays demonstrated that doxorubicin sensitivity of HeLa, H460, and A549 cell lines is enhanced by pretreatment with MK571 or TBBz (Fig. 6C; Table 2). Moreover, similarly to doxorubicin accumulation assays, copretreatment with both MK571 and TBBz showed no additive effect, strongly suggesting that the effect of TBBz treatment results from inhibition of CK2α-mediated up-regulation of MRP1 function.
TABLE 2.
Cancer cell lines used, their doxorubicin sensitivity and its modulation by treatments with MK571 and TBBz
AUC was derived from fraction cell survival-doxorubicin concentration plot (not shown). C50 and IC90 values derived from four-parameter logistic nonlinear regression curve fit presented in Fig. 6C.
| Cell Line and Treatment | AUC | IC50 | 95% CI | IC90 | 95% CI | |
|---|---|---|---|---|---|---|
| nM | nM | nM | ||||
| HeLa | ||||||
| Control | 2083 | ∼1925 | (Very wide) | ∼ 1664 | (Very wide) | Ambiguous |
| 0 | 896 | 697 | 637.2–762.4 | 384.2 | 327.7–450.5 | |
| + TBBz | 1255 | 1057 | 903.6–1236 | 797.6 | 441.8–1440 | |
| + MK571 + TBBz | 699 | 613 | 521.1–721.1 | 281.4 | 203.2–389.7 | |
| H460 | ||||||
| Control | 1038 | 390.5 | 303.5–502.6 | 48.52 | 27.94–84.25 | |
| 0 | 575 | 116 | 105.1–127.9 | 53.38 | 43.35–65.72 | |
| + TBBz | 929 | 243.4 | 186.9–317.0 | 13.93 | 6.882–28.18 | |
| + MK571 + TBBz | 660 | 125.3 | 101.3–154.9 | 28.05 | 17.38–45.28 | |
| A549 | ||||||
| Control | 2117 | ∼ 1.634 × 106 | (Very wide) | ∼ 271.4 | (Very wide) | Ambiguous |
| 0 | 1557 | 280.8 | 112.4–701.6 | 4.36 | 0.2873–66.11 | |
| + TBBz | 1632 | ∼13,412 | (Very wide) | 0.87 | 0.0024–317.2 | Ambiguous |
| + MK571 + TBBz | 1466 | 98.81 | 42.72–228.5 | 1.59 | 0.0793–31.99 |
CI, confidence interval.
Discussion
Here we have examined the role of CK2α in regulating MRP1 function via phosphorylation of Thr249. CK2 kinase plays a major role in cell death/survival decisions; consequently, CK2α subunit knockout mice die in midembryogenesis, whereas CK2α knockdown in cell culture is associated with decreased cell survival (Di Maira et al., 2007; Seldin et al., 2008), and even modest reduction in its expression has a large impact on cancer cell homeostasis (Wang et al., 2001; Seeber et al., 2005; Duncan and Litchfield, 2008; Trembley et al., 2010). Thus, it was not surprising that 50% reduction in CK2α expression in our cells was reflected by increased doxorubicin sensitivity (Fig. 3, A and C; Table 1), significant changes in cells ability to efflux doxorubicin (Fig. 4), and reduced MRP1 transport ability (Fig. 3, E and F).
Cell survival when exposed to a known MRP1 substrate, doxorubicin, was assessed by MTT assay; IC50 and IC90 values were extrapolated from this plot (Fig. 3, A and B; Table 1). Although the statistics listed in Table 1 demonstrate significant reduction in doxorubicin sensitivity in MRP1-expressing cells upon CK2α knockdown, survival curves for MRP1 and MRP1 mutant lines show little difference at lower doxorubicin concentrations (including IC50), but they start to differentiate toward higher concentrations, as depicted by the IC90 values. To better understand this phenomenon we plotted the data directly as fraction of cell survival versus concentration (Fig. 3, C and D) and calculated the area under the resulting fit curves (AUC; Table 1), as described by others (Moon et al., 1981). Figure 3D clearly shows that the survival curve for MRP1-T249E mutant reaches a plateau much earlier than for either MRP1 or MRP1-T249A, represented by a larger AUC value. The earlier plateau for MRP1-T249E indicates reduction in lethality beyond 100 to 200 nM and suggests the presence of cell subpopulations biochemically or kinetically resistant to doxorubicin (Moon et al., 1981). Taken together, we postulate that there are differences in doxorubicin sensitivity among MRP1, MRP1-T249A, and MRP1-T249E cell lines, although they are difficult to demonstrate with this assay for several reasons (Chambers et al., 1984; Cole, 1986; Campling et al., 1988; Campling et al., 1991). Moreover, toxicity of doxorubicin does not depend simply on total amount of drug within the cell but also on the proliferative state of the cell (Chambers et al., 1984). It is noteworthy that cell density is also known to affect localization of MRP1 protein. Roelofsen et al. (1997) showed that MRP1 localized to the lateral membrane of adjacent cells but was not detectable in membranes of separate cells MRP1, also seen in Fig. 2B. In our doxorubicin accumulation assays, cells are assayed at 80 to 90% confluence, whereas in MTT assays, cells are plated to be in exponential phase and reach ≤70% confluence on a third day, before addition of MTT. If indeed MRP1 localized to a different compartment of the cell in nonadjacent cells, it would not efflux drug from the cell, and depending on the compartment, it might actually contribute to drug sequestration inside, resulting in altered apparent drug cytotoxicity. In this light, the most reasonable explanation for the apparent discrepancies between the results of cytotoxicity, doxorubicin accumulation, and transport assay is that each assay measures different parameters under different conditions. The transport assay measures apparent saturable kinetics of efflux in vitro, whereas the doxorubicin accumulation experiment measures MRP1-dependent efflux from the cell and is tied directly to the ability of the drug to enter the cell. At the conditions used in the doxorubicin accumulation assay, it is unlikely that our system was saturated, and therefore the assay is likely to be much more sensitive to loss of function rather than gain of function. In addition, because the results of doxorubicin accumulation and cytotoxicity assays are sensitive to cell-seeding density and MRP1 cellular localization, even if performed at very similar conditions, these factors would not allow for direct correlation. With respect to our cytotoxicity experiments, those are carried out over 72 h and therefore reflect not only drug induced cytotoxicity but also inhibition of cell growth. For these reasons, we feel that the doxorubicin accumulation and transport assays are more representative of MRP1 function in vivo. Analysis of MRP1-dependent in vitro transport (Fig. 3, E and F) and MRP1-dependent doxorubicin intracellular accumulation in tissue culture (Fig. 4) strongly suggests that MRP1 function is regulated by CK2α in a Thr249-dependent manner. The regulation of MRP1 via CK2α seems to be general, not substrate-specific, because both CK2α knockdown and Thr249Ala mutation similarly alter MRP1 function toward different substrates: LTC4, E217βG, and doxorubicin (Figs. 3 and 4). It is noteworthy that transport analysis suggests that in MCF7 cells, MRP1 is phosphorylated at Thr249 at approximately 50% of capacity, because T249E mutation increases transport by 2-fold and T249A mutation decreases it by 50%. However, tissue culture experiments show similar doxorubicin accumulation for MRP1 and MRP1-T249E, whereas the MRP1-T249A and MRP1 CK2α(−) cells accumulate 2-fold more doxorubicin compared with MRP1 cells, suggesting that a large fraction of MRP1 cellular pool is phosphorylated (50–100%).
We provide strong evidence that CK2α regulates MRP1 function via a Thr249-dependent mechanism involving direct phosphorylation of Thr249 by CK2α. Support for our model comes from coIPs showing that MRP1 and CK2α physically interact (Fig. 5A), CK2α kinase assays showing that CK2 consensus site of MRP1 is phosphorylated in a Thr249-dependent manner (Fig. 5B), and immunoprecipitations with phosphospecific antibody demonstrating absent phosphorylation of Thr249 in MRP1-T249A and reduced in MRP1 CK2α(−) cells (Fig. 5E).
The basis for our study came from discovery of a CK2 phosphorylation site in Ycf1p and previous reports of homologous site in murine MRP6 phosphorylated in vivo (Villén et al., 2007; Paumi et al., 2008; Pickin et al., 2010). As for other potential CK2 consensus sites on MRP1, prediction engines (e.g., GPS, KinasePhos) most consistently list, among the multitude of possibilities, Ser1268 (Xue et al., 2008). Ser1268 is localized toward the C terminus of the protein, between membrane-spanning domain 2 and nucleotide-binding domain 2, but thus far, high-throughput studies have not confirmed that this residue is phosphorylated. As discussed in our recent review (Stolarczyk et al., 2010), the majority of protein kinase A and C phosphorylation sites identified so far lie within the regulatory domain (R), located on the cytosolic linker connecting the two “halves” of the full-length ABC transporters. The case for MRP1 is expected to be similar, because there are multiple potential protein kinase A and C consensus sites within the R-like linker region on MRP1, and high-throughput detection studies found 11 phosphorylated residues in this region. However, identity of the kinase and the biological effect of the phosphorylation remain to be elucidated.
Immunoprecipitation analysis shows that in MRP1 CK2α(−) cells, phosphorylation is reduced but not absent, consistent with reduced but not eliminated CK2α protein expression, confirming that Thr249 phosphorylation is CK2α-dependent. This observation is in agreement with our finding that vesicles derived from MRP1 CK2α cells have an intermediate ability to transport LTC4 and E217βG compared with MRP1 and MRP1-T249A. Our model for CK2α phosphorylation of Thr249 is further supported by results from doxorubicin accumulation assays in the presence of DMAT, a specific CK2 inhibitor. Pretreatment of MRP1 cells with DMAT increased doxorubicin cellular accumulation by 2-fold, whereas no change was observed for pretreatments of MRP1 CK2α(−), MRP1-T249A, and MRP1-T249E cells, indicating that Thr249 is the primary site of CK2α-mediated phosphorylation of MRP1. However, without further investigation, we cannot eliminate the possibility that other CK2 sites (perhaps dependent or secondary to Thr249) exist, that other kinases also phosphorylate Thr249, or that other condition-specific phosphorylation events occur.
It remains to be determined whether other pathways of MRP1 regulation involve Thr249 and intersect or compete with CK2α. One interesting possibility is that CK2α may interact with MRP1 as a member of a bigger protein complex. CK2α has been shown to be recruited to the plasma membrane by a pleckstrin homology domain protein, CK2-interacting protein-1 (Olsten et al., 2004). It is reasonable to believe that CK2-interacting protein-1 is required to recruit CK2α to the plasma membrane to interact with MRP1. We examined this possibility in our coIPs; however, the results were inconclusive and we therefore cannot rule out this possibility.
We believe that this novel observation linking two important players in cancer, the oncogenic kinase and the ABC transporter conferring resistance to therapeutic agents, opens a promising opportunity for improvement in cancer therapies. To this end, we have investigated the potential use of CK2 inhibitors as a mechanism of reducing MRP1-mediated drug resistance in a small panel of cancer cell lines that naturally express high levels of MRP1 (Fig. 6A). Here we show that MRP1-dependent doxorubicin efflux from HeLa, H460, and A549 cells can be reversed not only by direct MRP1 inhibition but also through inhibition of CK2 kinase by TBBz. Each of these agents separately increases drug accumulation in tested cell lines and enhances doxorubicin sensitivity. However, as demonstrated by cotreatment with both MK571 and TBBz, this effect is not additive, strongly suggesting that TBBz inhibits CK2-mediated up-regulation of MRP1 function. In conclusion, we believe that CK2 kinase inhibitor coadministration with currently used chemotherapies would improve overall treatment efficacy and decrease side effects of chemotherapy in part by reduction in MRP1-dependent drug resistance.
Supplementary Material
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This project was supported in part by the National Institutes of Health National Center for Research Resources [Grant 5P20-RR020171-09]; and the National Institutes of Health National Institute of General Medical Sciences [Grant 8P20-GM103486-09].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- ABC
- ATP-binding cassette
- MRP
- multidrug resistance-associated protein
- NTE
- N-terminal extension
- LTC4
- leukotriene C4
- E217βG
- estradiol-17β-d-glucuronide
- CK2
- casein kinase 2
- Ycf1p
- yeast cadmium factor 1
- Cka1p
- yeast casein kinase CK2α
- CHX
- cycloheximide
- DMAT
- 2-dimethylamino-4,5,6,7-tetrabromobenzimidazole
- TBBz
- 4,5,6,7-tetrabromo-benzimidazole
- MK571
- (E)-3-[[[3-[2-(7-chloro-2-quinolinyl)ethenyl]phenyl]-[[3-dimethylamino)-3-oxopropyl]thio]methyl]thio]-propanoic acid
- PSC883
- valspodar
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- G418
- Geneticin
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- RIPA
- radioimmunoprecipitation assay
- PAGE
- polyacrylamide gel electrophoresis
- AUC
- area under the curve
- TS
- Tris-sucrose
- shRNA
- short hairpin RNA
- FTC
- fumitremorgin C
- coIP
- coimmunoprecipitation
- ANOVA
- analysis of variance.
Authorship Contributions
Participated in research design: Stolarczyk and Paumi.
Conducted experiments: Stolarczyk, Reiling, Pickin, and Paumi.
Contributed new reagents or analytic tools: Coppage, Knecht, and Paumi.
Performed data analysis: Stolarczyk and Paumi.
Wrote or contributed to the writing of the manuscript: Stolarczyk and Paumi.
References
- Ausubel FM, Brent R, Kingston RE, Moore FF, Seidman JG, Smith JA, Struhl K. (1987) Current Protocols in Molecular Biology, Greene Publishing Associates/Wiley Interscience, New York [Google Scholar]
- Bakos E, Evers R, Calenda G, Tusnády GE, Szakács G, Váradi A, Sarkadi B. (2000) Characterization of the amino-terminal regions in the human multidrug resistance protein (MRP1). J Cell Sci 113:4451–4461 [DOI] [PubMed] [Google Scholar]
- Burnett G, Kennedy EP. (1954) The enzymatic phosphorylation of proteins. J Biol Chem 211:969–980 [PubMed] [Google Scholar]
- Campling BG, Pym J, Baker HM, Cole SP, Lam YM. (1991) Chemosensitivity testing of small cell lung cancer using the MTT assay. Br J Cancer 63:75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campling BG, Pym J, Galbraith PR, Cole SP. (1988) Use of the MTT assay for rapid determination of chemosensitivity of human leukemic blast cells. Leuk Res 12:823–831 [DOI] [PubMed] [Google Scholar]
- Chambers SH, Bleehen NM, Watson JV. (1984) Effect of cell density on intracellular adriamycin concentration and cytotoxicity in exponential and plateau phase EMT6 cells. Br J Cancer 49:301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TC, Pohl J, Glass DB, Kuo JF. (1994) Phosphorylation by protein kinase C and cyclic AMP-dependent protein kinase of synthetic peptides derived from the linker region of human P-glycoprotein. Biochem J 299:309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappe V, Hinkson DA, Zhu T, Chang XB, Riordan JR, Hanrahan JW. (2003) Phosphorylation of protein kinase C sites in NBD1 and the R domain control CFTR channel activation by PKA. J Physiol 548:39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZS, Tiwari AK. (2011) Multidrug resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic diseases. FEBS J 278:3226–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SP. (1986) Rapid chemosensitivity testing of human lung tumor cells using the MTT assay. Cancer Chemother Pharmacol 17:259–263 [DOI] [PubMed] [Google Scholar]
- Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. (1992) Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 258:1650–1654 [DOI] [PubMed] [Google Scholar]
- Di Maira G, Brustolon F, Bertacchini J, Tosoni K, Marmiroli S, Pinna LA, Ruzzene M. Pharmacological inhibition of protein kinase CK2 reverts the multidrug resistance phenotype of a CEM cell line characterized by high CK2 level. Oncogene 26(48):6915–6926, 2007 [DOI] [PubMed] [Google Scholar]
- Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. (1998) A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA 95:15665–15670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Litchfield DW. (2008) Too much of a good thing: the role of protein kinase CK2 in tumorigenesis and prospects for therapeutic inhibition of CK2. Biochim Biophys Acta 1784:33–47 [DOI] [PubMed] [Google Scholar]
- Fernández SB, Holló Z, Kern A, Bakos E, Fischer PA, Borst P, Evers R. (2002) Role of the N-terminal transmembrane region of the multidrug resistance protein MRP2 in routing to the apical membrane in MDCKII cells. J Biol Chem 277:31048–31055 [DOI] [PubMed] [Google Scholar]
- Grant CE, Valdimarsson G, Hipfner DR, Almquist KC, Cole SP, Deeley RG. (1994) Overexpression of multidrug resistance-associated protein (MRP) increases resistance to natural product drugs. Cancer Res 54:357–361 [PubMed] [Google Scholar]
- Hunter T, Sefton BM. (1991) Protein Phosphorylation, Academic Press, San Diego [Google Scholar]
- Juliano RL, Ling V. (1976) A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta 455:152–162 [DOI] [PubMed] [Google Scholar]
- Keppler D, Jedlitschky G, Leier I. (1998) Transport function and substrate specificity of multidrug resistance protein. Methods Enzymol 292:607–616 [DOI] [PubMed] [Google Scholar]
- Keppler D, Leier I, Jedlitschky G. (1997) Transport of glutathione conjugates and glucuronides by the multidrug resistance proteins MRP1 and MRP2. Biol Chem 378:787–791 [PubMed] [Google Scholar]
- Kramerov AA, Saghizadeh M, Caballero S, Shaw LC, Li Calzi S, Bretner M, Montenarh M, Pinna LA, Grant MB, Ljubimov AV. (2008) Inhibition of protein kinase CK2 suppresses angiogenesis and hematopoietic stem cell recruitment to retinal neovascularization sites. Mol Cell Biochem 316:177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautier D, Canitrot Y, Deeley RG, Cole SP. (1996) Multidrug resistance mediated by the multidrug resistance protein (MRP) gene. Biochemical pharmacology 52:967–977 [DOI] [PubMed] [Google Scholar]
- Litchfield DW. (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J 369:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loe DW, Almquist KC, Cole SP, Deeley RG. (1996) ATP-dependent 17 beta-estradiol 17-(beta-d-glucuronide) transport by multidrug resistance protein (MRP). Inhibition by cholestatic steroids. J Biol Chem 271:9683–9689 [DOI] [PubMed] [Google Scholar]
- Mason DL, Michaelis S. (2002) Requirement of the N-terminal extension for vacuolar trafficking and transport activity of yeast Ycf1p, an ATP-binding cassette transporter. Molecular biology of the cell 13:4443–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J 17:349–368 [DOI] [PubMed] [Google Scholar]
- Mishra S, Pertz V, Zhang B, Kaur P, Shimada H, Groffen J, Kazimierczuk Z, Pinna LA, Heisterkamp N. (2007) Treatment of P190 Bcr/Abl lymphoblastic leukemia cells with inhibitors of the serine/threonine kinase CK2. Leukemia 21:178–180 [DOI] [PubMed] [Google Scholar]
- Moon TE. (1980) Quantitative and statistical analysis of the association between in vitro and in vivo studies. Prog Clin Biol Res 48:209–221 [PubMed] [Google Scholar]
- Moon TE, Salmon SE, White CS, Chen HS, Meyskens FL, Durie BG, Alberts DS. (1981) Quantitative association between the in vitro human tumor stem cell assay and clinical response to cancer chemotherapy. Cancer Chemother Pharmacol 6:211–218 [DOI] [PubMed] [Google Scholar]
- Munoz M, Henderson M, Haber M, Norris M. (2007) Role of the MRP1/ABCC1 multidrug transporter protein in cancer. IUBMB Life 59:752–757 [DOI] [PubMed] [Google Scholar]
- Nooter K, Westerman AM, Flens MJ, Zaman GJ, Scheper RJ, van Wingerden KE, Burger H, Oostrum R, Boersma T, Sonneveld P. (1995) Expression of the multidrug resistance-associated protein (MRP) gene in human cancers. Clin Cancer Res 1:1301–1310 [PubMed] [Google Scholar]
- Olsten ME, Canton DA, Zhang C, Walton PA, Litchfield DW. (2004) The Pleckstrin homology domain of CK2 interacting protein-1 is required for interactions and recruitment of protein kinase CK2 to the plasma membrane. J Biol Chem 279:42114–42127 [DOI] [PubMed] [Google Scholar]
- Pagano MA, Marin O, Cozza G, Sarno S, Meggio F, Treharne KJ, Mehta A, Pinna LA. (2010) Cystic fibrosis transmembrane regulator fragments with the Phe508 deletion exert a dual allosteric control over the master kinase CK2. Biochem J 426:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano MA, Meggio F, Ruzzene M, Andrzejewska M, Kazimierczuk Z, Pinna LA. (2004) 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: a novel powerful and selective inhibitor of protein kinase CK2. Biochem Biophys Res Commun 321:1040–1044 [DOI] [PubMed] [Google Scholar]
- Paumi CM, Chuk M, Chevelev I, Stagljar I, Michaelis S. (2008) Negative regulation of the yeast ABC transporter Ycf1p by phosphorylation within its N-terminal extension. J Biol Chem 283:27079–27088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi CM, Chuk M, Snider J, Stagljar I, Michaelis S. (2009) ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol Mol Biol Rev 73:577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi CM, Ledford BG, Smitherman PK, Townsend AJ, Morrow CS. (2001) Role of multidrug resistance protein 1 (MRP1) and glutathione S-transferase A1–1 in alkylating agent resistance. Kinetics of glutathione conjugate formation and efflux govern differential cellular sensitivity to chlorambucil versus melphalan toxicity. J Biol Chem 276:7952–7956 [DOI] [PubMed] [Google Scholar]
- Paumi CM, Wright M, Townsend AJ, Morrow CS. (2003) Multidrug resistance protein (MRP) 1 and MRP3 attenuate cytotoxic and transactivating effects of the cyclopentenone prostaglandin, 15-deoxy-Delta(12,14)prostaglandin J2 in MCF7 breast cancer cells. Biochemistry 42:5429–5437 [DOI] [PubMed] [Google Scholar]
- Pickin KA, Ezenwajiaku N, Overcash H, Sethi M, Knecht MR, Paumi CM. (2010) Suppression of Ycf1p function by Cka1p-dependent phosphorylation is attenuated in response to salt stress. FEMS Yeast Res 10:839–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofsen H, Vos TA, Schippers IJ, Kuipers F, Koning H, Moshage H, Jansen PL, Müller M. (1997) Increased levels of the multidrug resistance protein in lateral membranes of proliferating hepatocyte-derived cells. Gastroenterology 112:511–521 [DOI] [PubMed] [Google Scholar]
- Ruzzene M, Pinna LA. (2010) Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta 1804:499–504 [DOI] [PubMed] [Google Scholar]
- Seeber S, Issinger OG, Holm T, Kristensen LP, Guerra B. (2005) Validation of protein kinase CK2 as oncological target. Apoptosis 10:875–885 [DOI] [PubMed] [Google Scholar]
- Seldin DC, Lou DY, Toselli P, Landesman-Bollag E, Dominguez I. (2008) Gene targeting of CK2 catalytic subunits. Mol Cell Biochem 316:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolarczyk EI, Reiling CJ, Paumi CM. (2011) Regulation of ABC transporter function via phosphorylation by protein kinases. Curr Pharm Biotechnol 12:621–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trembley JH, Chen Z, Unger G, Slaton J, Kren BT, Van Waes C, Ahmed K. (2010) Emergence of protein kinase CK2 as a key target in cancer therapy. Biofactors 36:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villén J, Beausoleil SA, Gerber SA, Gygi SP. (2007) Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci USA 104:1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Davis A, Yu S, Ahmed K. (2001) Response of cancer cells to molecular interruption of the CK2 signal. Mol Cell Biochem 227:167–174 [PubMed] [Google Scholar]
- Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. (2008) GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 7:1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Kinsella TJ. (2005) CK2 inhibits apoptosis and changes its cellular localization following ionizing radiation. Cancer Res 65:4362–4367 [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu Y, Dong Z, Xu J, Peng H, Liu Z, Zhang JT. (2007) Regulation of function by dimerization through the amino-terminal membrane-spanning domain of human ABCC1/MRP1. J Biol Chem 282:8821–8830 [DOI] [PubMed] [Google Scholar]
- Yang Y, Mo W, Zhang JT. (2010) Role of transmembrane segment 5 and extracellular loop 3 in the homodimerization of human ABCC1. Biochemistry 49:10854–10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.