Abstract
Neuronal migration and growth cone motility are essential aspects of the development and maturation of the nervous system. These cellular events result from dynamic changes in the organization and function of the cytoskeleton, in part due to the activity of cytoskeletal motor proteins such as myosins. Although specific myosins such as Myo2 (conventional or muscle myosin), Myo1, and Myo5 have been well characterized for roles in cell motility, the roles of the majority of unconventional (other than Myo2) myosins in cell motility events have not been investigated. To address this issue, we have undertaken an analysis of unconventional myosins in zebrafish, a premier model for studying cellular and growth cone motility in the vertebrate nervous system. We describe the characterization and expression patterns of several members of the unconventional myosin gene family. Based on available genomic sequence data, we identified 18 unconventional myosin- and 4 Myo2-related genes in the zebrafish genome in addition to previously characterized myosin (-1, -2, -3, -5, -6, -7) genes. Phylogenetic analyses indicate that these genes can be grouped into existing classifications for unconventional myosins from mouse and man. In situ hybridization analyses using EST probes for 18 of the 22 identified genes indicate that 11/18 genes are expressed in a restricted fashion in the zebrafish embryo. Specific myosins are expressed in particular neuronal or neuroepithelial cell types in the developing zebrafish nervous system, spanning the periods of neuronal differentiation and migration, and of growth cone guidance and motility.
Keywords: cytoskeleton, unconventional myosin, neuronal migration, axon guidance, growth cone motility, zebrafish, commissure, spinal cord, motor neuron, neural crest, somite, ear, eye, morphogenesis, in situ hybridization, phylogenetic tree, hindbrain, forebrain, midbrain, cranial muscles
1. INTRODUCTION
During embryogenesis, neuronal cell bodies and growth cones migrate extensively to establish precisely connected networks with target tissues. The functional properties of neural networks underlying physiology and behavior are critically dependent upon accurate positioning of neuronal cell bodies, and their axonal and dendritic processes. Not surprisingly, a number of brain disorders result from the loss of or aberrant migration of neurons and defective axon guidance (Copp and Harding, 1999; Gleeson and Walsh, 2000; Oster and Sretavan, 2003; ten Donkelaar et al., 2004). A large number of extracellular cues and their receptors have been intensively studied, and demonstrated to play essential roles in regulating the migration of neuronal growth cones and cell bodies (Chilton, 2006). Furthermore, the biochemical and molecular interactions between the signal transduction cascades initiated by these guidance cues and the cytoskeletal machinery that controls dynamic cellular and growth cone behaviors are being elucidated (Guan and Rao, 2003; Kalil and Dent, 2005). Nevertheless, the roles of specific components of the actin and microtubule cytoskeletons in generating particular responses to extracellular cues remain obscure. We are interested in characterizing the functions of the unconventional, non-muscle myosins in neuronal migration and axon guidance, and in elucidating whether these activities are regulated by guidance cues in the developing nervous system.
Unconventional non-muscle myosins are actin-binding motor proteins that lack the tail domain of conventional muscle myosin (myosin 2), and function in numerous processes including cell migration in lower eukaryotes, intracellular motility and trafficking, and sensory transduction (Libby and Steel, 2000; Tuxworth and Titus, 2000; Wu et al., 2000; Soldati, 2003; Hirokawa and Takemura, 2003). Furthermore, unconventional myosins have been localized to neuronal growth cones and regulate growth cone motility in vitro (Wang et al., 1996; Evans et al., 1997; Suter et al., 2000; Deifenbach et al., 2002; Sousa et al., 2006). Phylogenetic analysis of the catalytic head domain (containing the actin- and ATP-binding sites) of all available myosin heavy chain sequences shows that there are at least 18 families of myosins, including conventional (muscle) Myo2, several myosins found only in fungi or other lower eukaryotes (Myosins 4, 11, 12, 13, 14, and 17), and the plant-specific Myo8 (Hodge and Cope, 2000; Sellers, 2000; Berg et al., 2001; Reddy and Day, 2001; Volkmann et al., 2003). In addition to differences in the catalytic head domain, unconventional myosins belonging to various families differ in the presence and arrangement of specific domains and motifs that likely confer unique functions on each class of proteins (Fig. 1; Wu et al., 2000; De La Cruz and Ostap, 2004). Whereas only 4–6 unconventional myosin families have been identified in unicellular organisms and invertebrates like Caenorhabditis, comprehensive phylogenetic analyses have identified over 10 unconventional myosin families in Drosophila and mammals (Berg et al., 2001).
Figure 1.
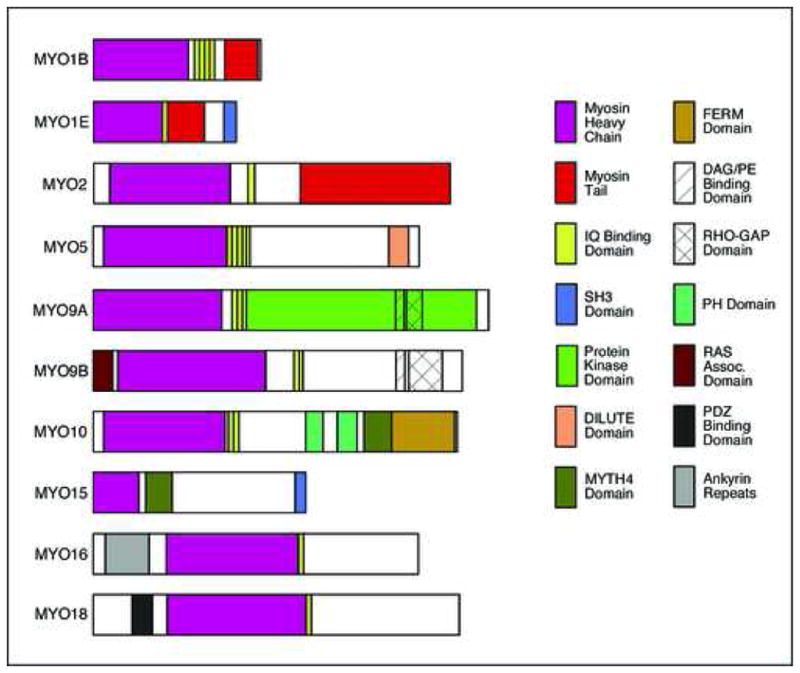
Domain organization of myosins examined in this report. Myosins are classified into several families based upon the occurrence and distribution of identified domains and sequence motifs in the heavy chain polypeptide. The nomenclature follows that for human and mouse proteins (Berg et al., 2001). This list contains only those myosins for which ESTs were obtained and expression analyzed in zebrafish embryos. The myosin head domain containing the actin- and ATP-binding sites (purple) is largely conserved, with differences identifying various classes. Other domains and motifs are found in only a subset of the proteins.
There has been no systematic characterization of unconventional myosins in zebrafish. Mutations generating defects in sensory neuron development and function have led to the cloning of members of the Myo6 and Myo7 families (Ernest et al., 2000; Kappler et al., 2004; Seiler et al., 2004; Coffin et al., 2007). Since the zebrafish embryo is an excellent model for studying cell migration and axon guidance (Kuwada, 1995; Hutson and Chien, 2002), the potential roles of unconventional myosins in these dynamic cellular processes can be readily investigated. Therefore, we have carried out an extensive characterization of the expression of unconventional myosin genes in the developing zebrafish nervous system. Our results demonstrate that specific myosins are expressed in particular neuronal or neuroepithelial cell types, spanning the periods of neuronal differentiation and migration, and of growth cone guidance and motility.
2. RESULTS AND DISCUSSION
2.1 Phylogenetic analysis of zebrafish unconventional myosins
Exhaustive search of the zebrafish genome assembly (Zv6, March 2006 and Zv7, July 2007) identified a total of 22 myosin genes representing classes 1, 2, 5, 9, 10, 15, 16 and 18. Zebrafish myosins 3, 6 and 7 were excluded from analyses because they have been described previously, whereas other myosin classes (4, 8, 11, 12, 13, 14, and 17) have been described only in fungi, other lower eukaryotes, or plants (Berg et al., 2001). In order to group the newly identified zebrafish (Danio rerio, Dr) myosins into appropriate classes, we constructed a phylogenetic tree with human (Homo sapiens, Hs) and mouse (Mus musculus, Mm) orthologs using amino acid sequences of the highly conserved core motor domains of various myosin classes, equivalent to residues 88–780 of chicken skeletal myosin II (Hodge and Cope, 2000; Berg et al., 2001). Alignment of core domains was performed in Megalign using Clustal-W method (Reddy and Day, 2001; see Methods for additional details). A rectangular tree for the aligned sequences rooted to Myo1 (Fig. 2) was generated by maximum parsimony method (Jiang and Ramachandran, 2004; see Methods). The tree identifies the presence of at least two genes each in the Myo1b, Myo1e, Myo5, Myo9a, Myo15 and Myo18 classes and three genes in the Myo10 class in the available zebrafish genome. Our study also identifies four conventional myo2 genes corresponding to different heavy chain genes as shown in the tree (Fig. 2). While most zebrafish myosins identified in this study group with one of the classes of human and mouse myosins with >95% bootstrap value, the core motor domains of zebrafish Myo9a1, Myo9a2 and Myo10b group with Myo16 and Myo18 with weak support (51% bootstrap value; Fig. 2). However, when the entire polypeptide sequences are used for the phylogenetic analysis, these genes group with their Myo9a and Myo10 orthologs (100% bootstrap value; cladogram not shown), respectively, due to the presence of class specific domains (Fig. 1).
Figure 2.
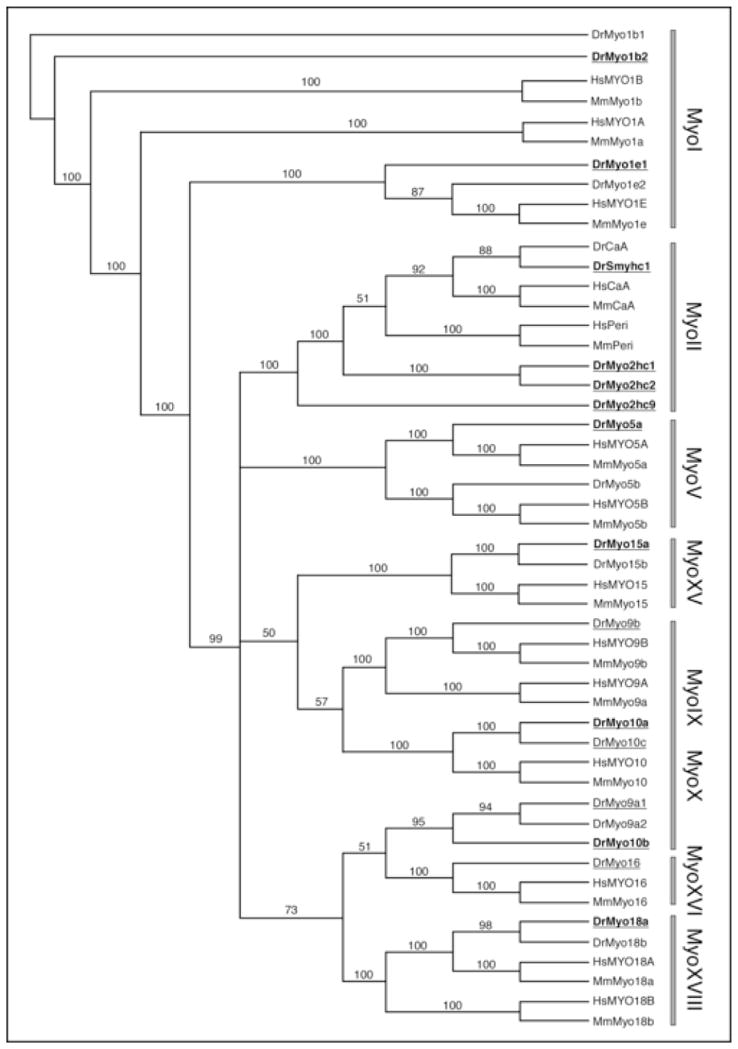
Phylogenetic relationship of unconventional myosin family members between zebrafish (Danio rerio), mouse (Mus musculus), and man (Homo sapiens). Rectangular tree rooted to Myo1, with bootstrap values for family members that were examined in this report (see Methods for details). Myosins for whom ESTs were characterized are identified by underlines. Those myosins among these that were expressed in zebrafish embryos (15–36 hpf) are identified by bold type (Table 1). Zebrafish myosins outside these two categories but included in this phylogenetic tree were identified solely from database searches. Abbreviations: CaA, cardiac myosin A; Peri, Perinatal skeletal myosin (Myhc8); Smyhc, Slow myosin heavy chain.
2.2 Expression analysis of zebrafish myosins
We carried out a detailed analysis of the expression patterns of various myosins for which ESTs were available in 2004 (Table 1). This includes all zebrafish genes described in Figs. 1 and 2, except myo1b1, myo1e2, myo5b, and myo15b, for which no ESTs were available, and myo3a (Thisse and Thisse, 2005), myo6 and myo7 whose expression patterns were previously described (Ernest et al., 2000; Seiler et al., 2004; Kappler et al., 2004). We mostly examined expression patterns in embryos fixed at 18, 24, 30, and 36 hpf. Expression at earlier stages was not systematically examined since we are primarily interested in potential roles for these molecules in neuronal migration and axon guidance, and older ages were examined as needed. The myosin genes exhibited restricted patterns of expression in various tissues. In the tissues examined, myo5a, myo10a, myo1b1 were expressed at all ages tested; myo1e1 was expressed only between 30–36 hpf; myo10b and myo18a were strongly expressed between 18–24 hpf, and weaker by 30 hpf; myo15a was expressed only between 12–18hpf; myo2hc1 was expressed strongly between 15–18 hpf, and weaker by 24–30 hpf; smyhc1, myo2hc2 and myo2hc9 were expressed in similar patterns between 18–36 hpf, with myo2hc2 and smyhc1 expressed at least until 72 and 120 hpf, respectively.
Table 1.
Zebrafish Myosin Genes Characterized in this Report
| Gene Ensembl ID | Chromosome | ESTs (source) *, % | Length of ORF | % Identity to | Gene Expressed | |
|---|---|---|---|---|---|---|
| Human | Mouse | |||||
|
myo1b2 ENSDARG00000024694 |
9 | MPMGp609P206Q8 (B) MPMGp609G1826Q8 (B) A1G (A) |
Partial (90%) | 81.4 | 81.5 | Yes |
|
myo1e1 ENSDARG00000036179 |
7 | MPMGp609L0725Q8 (B) MPMGp609L0281Q8 (B) MPMGp637H137Q2 (B) IMAGp998P1112400Q3 (B) IMAGp998G1514300Q3 (B) |
Full-length | 76.3 | 76.3 | Yes |
|
| ||||||
|
smyhc1 ENSDARG00000071430 |
24 | MPMGp609B1310Q8 (B) | Full-length | 84.8 | 84.6 | Yes |
|
myo2hc1 (myhz1) ENSDARG00000067990 |
5 | MPMGp609P1624Q8 (B) | Full-length | 80.8 | 82.1 | Yes |
|
myo2hc2 (myhz2) ENSDARG00000012944 |
5 | MPMGp609M1825Q8 (B) | Full-length | 80.7 | 82.1 | Yes |
|
myo2hc9 (myhz9) ENSDARG00000001014 |
3 | IMAGp998D0714289Q3 (B) | Full-length | 84.3 | 81.9 | Yes |
|
| ||||||
|
myo5a ENSDARG00000061635 |
18 | 129-E08-02 (S) | Full-length | 79.9 | 79.9 | Yes |
|
| ||||||
|
myo6a ENSDARG00000044016 |
20 | IMAGp998H1712050Q3 (B) | Full-length | 80.9 | 80.6 | No |
|
| ||||||
|
myo9a1 ENSDARG00000019585 |
7 | 46-E08-02 (S) | Partial (90%) | 56.7 | 56.3 | No |
|
myo9a2 ENSDARG00000063639 |
25 | IMAGp998N0912391Q3 (B) | Partial (80%) | 54.8 | 54.3 | No |
|
myo9b ENSDARG00000062159 |
10 | MPMGp609G0210Q8 (B) | Full-length | 57.3 | 58.1 | No |
|
| ||||||
|
myo10a ENSDARG00000062580 |
6 | IMAGp998M2314313Q (B) | Partial (98%) | 61.6 | 62.1 | Yes |
|
myo10b ENSDARG00000017004 |
2 | MPMGp609N1624Q8 (B) MPMGp609C1781Q8 (B) 88-A02-02 (S) |
Partial (84%) | 68.3 | 67.7 | Yes |
|
myo10c ENSDARG00000063138 |
9 | 112-H11-02 (S) | Full-length | 62.1 | 62.6 | No |
|
| ||||||
|
myo15a ENSDARG00000059709 |
3 | 62-G10-02 (S) | Full-length | 62.2 | 61.2 | Yes |
|
| ||||||
|
myo16 ENSDARG00000004512 |
9 | 86-H05-02 (S) 100-C07-02 (S) |
Full-length | 59.8 | 57.8 | No |
|
| ||||||
|
myo18a ENSDARG00000061862 |
15 | DKFZp717N112Q2 (B) | Full-length | 78 | 76.8 | Yes |
|
myo18b ENSDARG00000061225 |
10 | 54-D04-02 (S) 98-C07-02 (S) |
Partial (87%) | 73.7 | 73.3 | No |
, Source, B: RZPD, Berlin; S: Dr. Jinrong Peng, IMCB, Singapore; A: Research Genetics (Invitrogen), Alabama.
For genes with multiple ESTs, all ESTs were tested, and generated similar in situ results.
2.2.1 Expression in Hindbrain segments
Two myosins are transiently expressed in a segmented manner in specific rhombomeres. At 18 hpf, myo10b is strongly expressed in r5, and more weakly in r2, r3 and r4, and at the midbrain-hindbrain boundary (Fig. 3A). At 12 hpf, myo15a is expressed in r3, r5, and at the midbrain-hindbrain boundary (Fig. 3B), with weaker expression at 18 hpf (data not shown). Thus, these genes are expressed in cells exhibiting sorting behaviors to set up well-defined rhombomere boundaries (Cooke et al., 2005). From 30–36 hpf, myo1e1 is expressed in a small patch of mesendodermal cells located at the somite 2–3 boundary (Fig. 3C), either the developing pancreas (Argenton et al., 1999; Stafford and Prince, 2002) or the pronephros (Drummond et al., 1998).
Figure 3.
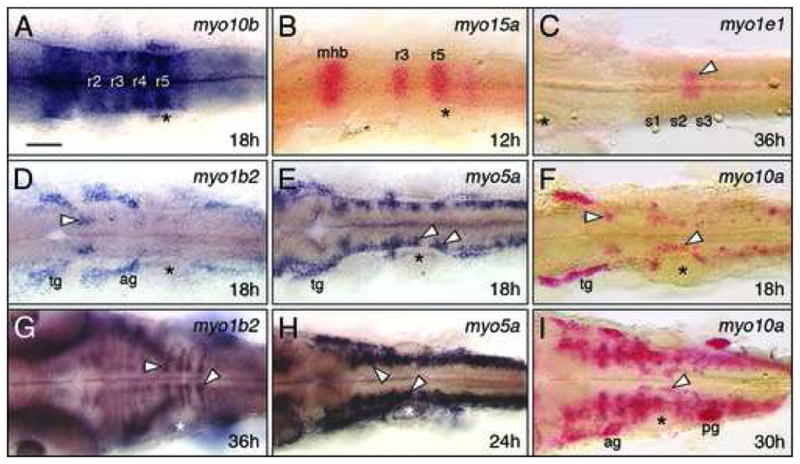
Myosins exhibiting restricted expression patterns in the hindbrain. All panels show dorsal views of the hindbrain with anterior to the left. Asterisks mark the otic vesicle in each panel. (A, B) myo10b is expressed at varying levels in rhombomeres 2–5 (A), and myo15a is expressed in r3 and r5 (B), with both expressed at the midbrain-hindbrain boundary (mhb). (C) myo1e1 is weakly but specifically expressed in a mesendodermal domain spanning the 2nd and 3rd somites (arrowhead). (D, G) At 18 hpf, myo1b2 is expressed in sensory ganglia (tg, ag) and a cluster of cells in r2 (arrowhead, D). By 36 hpf (G), there is prominent expression in cells (arrowheads) at the boundaries of r5 and r6. (E, H) At 18 hpf, myo5a is expressed in sensory ganglia, and in differentiating neurons (arrowheads, E) within every rhombomere. By 24 hpf (H), expressing cells form a continuous column along the neural tube (arrowheads). (F, I) At 18 hpf, myo10a is expressed in the sensory ganglia (tg), and in the branchiomotor neurons in r2 and r5 (arrowheads, F). By 30 hpf (I), expression persists in the sensory ganglia and migrating motor neurons (arrowhead), and has expanded to large clusters of cells in the anterior hindbrain. tg, trigeminal ganglion, ag/pg, anterior and posterior lateral line ganglion. Scale bar, 75 μm.
2.2.2 Expression in Hindbrain neurons
Three myosins are expressed in specific neuronal populations in the hindbrain. At 18 hpf, myo1b2 is expressed in a subset of the cranial sensory ganglia, as well as in a small group of unidentified neurons in r2 (Fig. 3D). Analysis of myo1b2 expression in the hedgehog pathway mutant detour (gli1−/−; Vanderlaan et al., 2005) indicates that these cells are not cranial motor neurons (data not shown). At 36 hpf, myo1b2 is expressed by a small number of cells within each rhombomere, with strong expression in cells flanking the boundaries of rhombomeres 5 and 6 (Fig. 3G). At 18 hpf, myo5a is expressed in a subset of the cranial ganglia, and in differentiating neurons in every rhombomere (Fig. 3E), similar to expression of the neurogenesis marker huC (Bingham et al., 2003). By 24 hpf, myo5a expression expands into a continuous column, as additional neurons differentiate in these regions (Fig. 3H). At 18 hpf, myo10a is expressed in the trigeminal ganglion, and is also expressed at high levels in the lateral line ganglia by 30 hpf (Figs. 3F, I). In addition, myo10a is expressed weakly by trigeminal and facial branchiomotor neurons from 18–30 hpf, and by other differentiating neurons by 30 hpf (Fig. 3I). The expression of these myosins in cranial ganglia and in differentiating neurons coincides with axonogenesis or soma migration in these neuronal populations (Metcalfe et al., 1990; Higashijima et al., 2000).
2.2.3 Expression in the Forebrain and midbrain
Two myosins, myo5a and myo10a, are expressed in specific populations of forebrain and midbrain neurons (Fig. 4). In the forebrain, myo5a- and myo10a-expressing cells from 18–30 hpf (Fig. 4A–C; data not shown) appear to correspond to the telencephalic and diencephalic neurons that generate the anterior and post-optic commissures, respectively (Chitnis et al., 1990; Wilson et al., 1991). The myo5a- and myo10a-expressing cells in the midbrain are likely the nucleus of the medial longitudinal fasicle (nucMLF; Chitnis et al., 1990; Fig. 4A–C), and this was confirmed by anti-tubulin antibody labeling of the MLF in embryos processed for myo10a in situs (Fig. 4D). Myosin gene expression in telencephalic and nucMLF neurons from 18–30 hpf coincides with the period of growth cone motility and axon guidance in these neurons. Myo2hc1 is strongly expressed at the midbrain-hindbrain boundary at 18 hpf, and becomes restricted to a narrow domain in the caudal optic tectum by 32 hpf (Fig. 4E, F).
Figure 4.
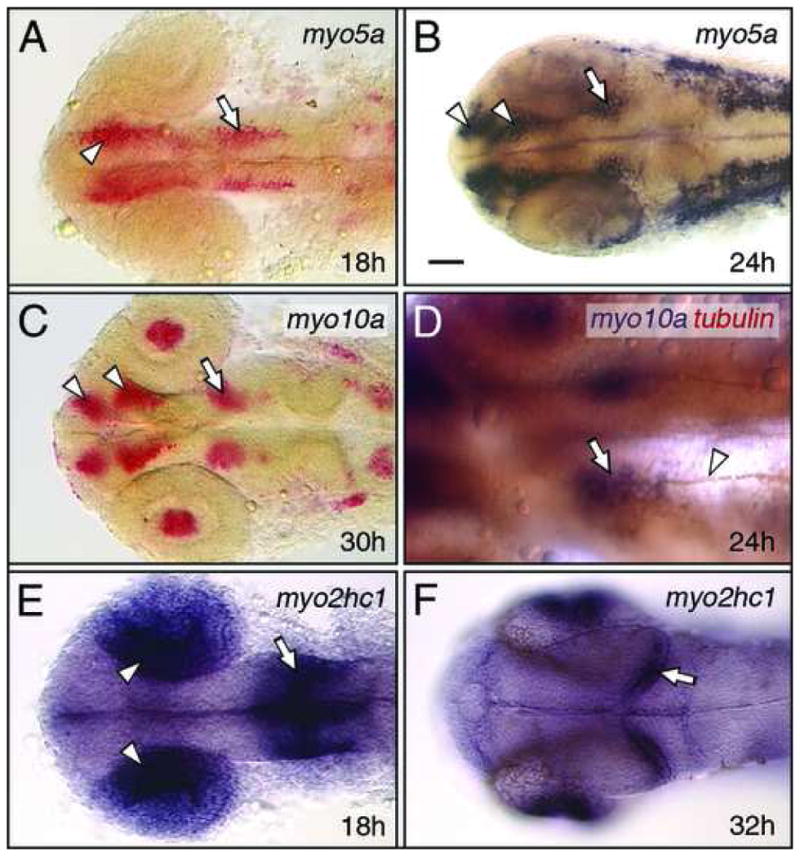
Myosin expression in the forebrain and midbrain. All panels show dorsal views of the head with anterior to the left. (A, B) At 18 hpf, myo5a is expressed in nascent neurons (arrowhead, A) in the forebrain, and in the nucMLF neurons (arrow) in the midbrain. By 24 hpf (B), forebrain expression has refined into distinguishable clusters representing the neurons of the anterior and post-optic commissures (arrowheads, B), and is maintained in the nucMLF neurons (arrow). (C) At 30 hpf, myo10a is expressed in the neurons of the nucMLF (arrow), and of the anterior and post-optic commissures (arrowheads), and in the lens (see Fig. 6). (D) Double-labeling shows that tubulin antibody-labeled axons (arrowhead) of the MLF arise from the cluster of myo10a-expressing neurons (arrow), corresponding to the nucMLF. (E, F) At 18 hpf, myo2hc1 is expressed in the medial layer of the optic cup (arrowheads), and at the mid-hindbrain boundary (arrow). By 32 hpf (F), expression is restricted to the retinal ganglion layer (see Fig. 6), and to the caudal edge of the developing optic tectum (arrow). Scale bar, 25 μm (D), 50 μm (A–F).
2.2.4 Expression in the Otic vesicle
In addition to myo6b and myo7a, whose expression patterns have been previously described (Ernest et al., 2000; Seiler et al., 2004; Kappler et al., 2004), myo10b and myo2hc1 are also expressed in the otic vesicle. However, their expression is not restricted to the sensory neurons of the epithelium as for myo6b and myo7a. At 18 hpf, myo2hc1 transcripts are localized to the apical regions of cells throughout the otic epithelium (Fig. 5B, D), whereas myo10b transcripts are localized to apical regions of cells only in the rostroventral epithelium (Fig. 5A, C; see also Fig. 3A). These genes are expressed in the otic vesicle from 15–24 hpf, during morphogenesis of the vesicle (Kimmel et al., 1995).
Figure 5.
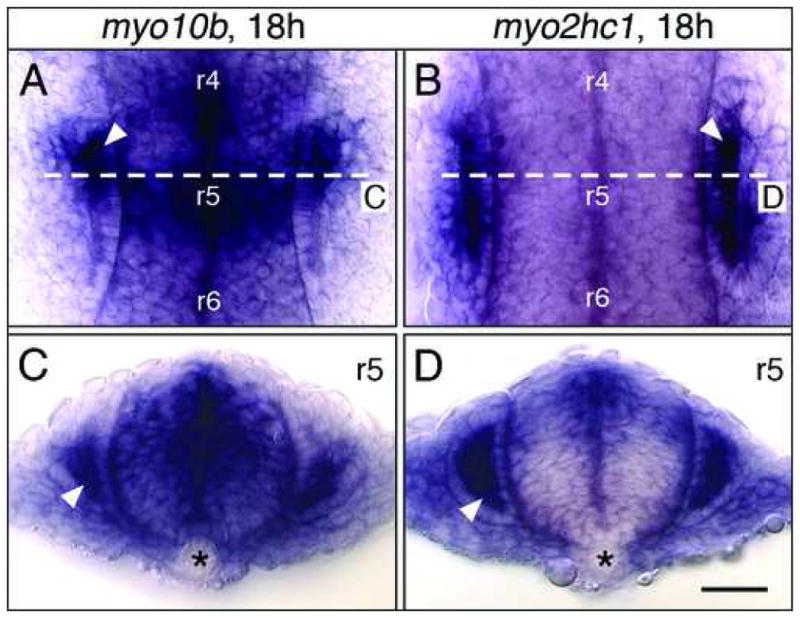
Myosin expression in the developing ear. Upper panels show dorsal views, with anterior at top. Lower panels show cross-sections (dorsal up) at the levels indicated in the upper panels. (A, C) myo10b is expressed in epithelial cells (arrowhead) lining the anterior third (A), and the ventral half (C) of the otic vesicle. (B, D) myo2hc1 is expressed in the epithelial cells of the otic vesicle (arrowheads), adjacent to the hindbrain. Asterisks (C, D) indicate the notochord. Scale bar, 30 μm.
2.2.5 Expression in the Eye
Three myosins are expressed in the eye, two in the developing lens (myo1b2, myo10a), and one in the retina (myo2hc1). Interestingly, myo1b2 is expressed by cells only in the medial half of the developing lens (Fig. 6A, B), whereas myo10a is expressed only in the dorsal half of the developing lens (Fig. 6C; see also Fig. 4C). These localized patterns may reflect the progression of differentiation of lens fibers, or the inability of transcripts to distribute uniformly within differentiating fibers. At 18 hpf, myo2hc1 is strongly expressed in the medial layer of the optic cup (Fig. 4E) during the period of eye morphogenesis (Li et al., 2000), and becomes restricted to the ganglion cell layer by 32 hpf (Figs. 4F and 6D), when retinal axons are extending into the brain (Hutson and Chien, 2002).
Figure 6.
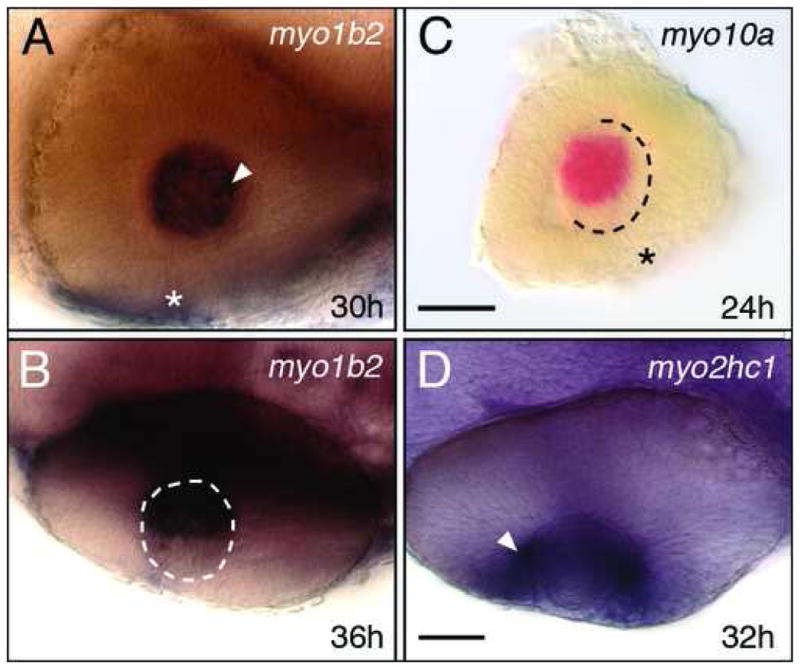
Myosin expression in the eye. (A, B) myo1b2 is expressed in the differentiating lens fibers (arrowhead, A) located in the medial half of the lens (B, dorsal view). (C) myo10a is expressed in a subset of lens cells at 24 hpf, and at 30 hpf (see Fig. 4C). (D) myo2hc1 is expressed in retinal ganglion cells (arrowhead) adjacent to the lens. Asterisks (A, C) indicate the choroid fissure. Scale bar (A, B, D), 40 μm. Scale bar (C), 30 μm.
2.2.5 Expression in the Spinal cord and Trunk
As expected, myo2hc2 is expressed strongly in all muscles (Figs. 7A and 8), and our probe labels expressing tissue more effectively than previously described (Peng et al., 2002). Interestingly, only one other myosin among 18 tested was expressed in the presumptive musculature. At 18 hpf, myo18a is expressed throughout the caudal (newborn) somites, and continues to be expressed strongly in cells at the boundaries between the rostral (older) somites (Fig. 7B). At 18 hpf, myo1b2 is expressed in the superficial cells of the somites (Fig. 7C, D), and weakly expressed in a few cells in the spinal cord, likely neurons. At 18 hpf, myo5a is expressed by several presumptive neurons throughout the spinal cord, except in the most dorsal and ventral regions (Fig. 7E, F). By 30 hpf, myo5a-expressing cells are located mostly in the dorsal half of the neural tube, with several putative migrating neural crest cells in characteristic locations beside the somites (Fig. 7G, H). At 18 hpf, myo10a is expressed by cells in the dorsal spinal cord in putative neural crest cells (Fig. 7I, J). These results indicate that specific myosins are expressed in neurons and neural crest cells during the period of cell migration and axon guidance.
Figure 7.
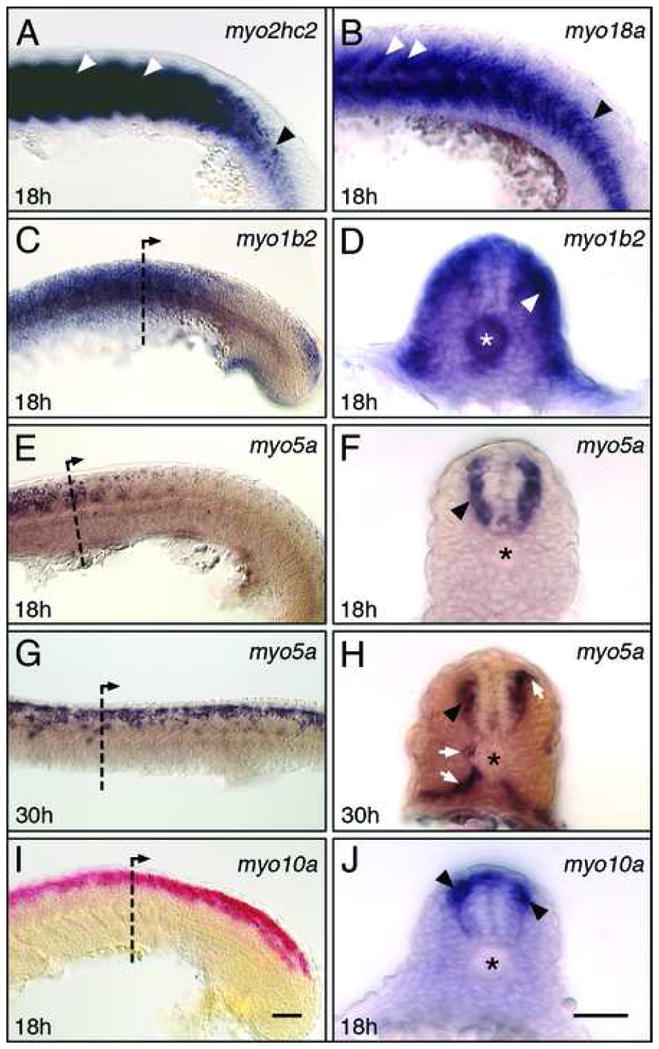
Myosin expression in the trunk and spinal cord. Panels A–C, E, G, I show side views with anterior to left. Panels D, F, H, J show cross-sections at levels indicated in C, E, G, I, respectively. (A) myo2hc2 is strongly expressed in the differentiating muscles of the somites (white arrowheads), but only weakly in the newly formed somites (black arrowhead). (B) myo18a is expressed in the mesenchyme of newly-formed and differentiating somites (black arrowhead), and strongly in the cells at the boundaries of older somites (white arrowheads). (C, D) myo1b2 is expressed broadly in the trunk, excluding the presomitic mesoderm (C). Expressing cells are located superficially within the somitic mesoderm (arrowhead, D), and in the notochord (asterisk). (E, F) At 18 hpf, myo5a expression is restricted to the spinal cord at anterior trunk levels (E), and represents differentiating neurons (arrowhead, F). (G, H) At 30 hpf, myo5a is expressed by scattered cells located in the dorsal half of the spinal cord (G). A cross-section in the anterior trunk (H) shows that expressing cells include neurons (arrowhead) and migrating neural crest cells (arrows). (I, J) myo10a is expressed in a continuous column of cells in the dorsal half of the spinal cord (I), including neurons (arrowheads, J). Asterisks in D, F, H, J indicate the notochord. Scale bar (I), 50 μm (A–C, E, G, I). Scale bar (J), 50 μm (D, F, H, J).
Figure 8.
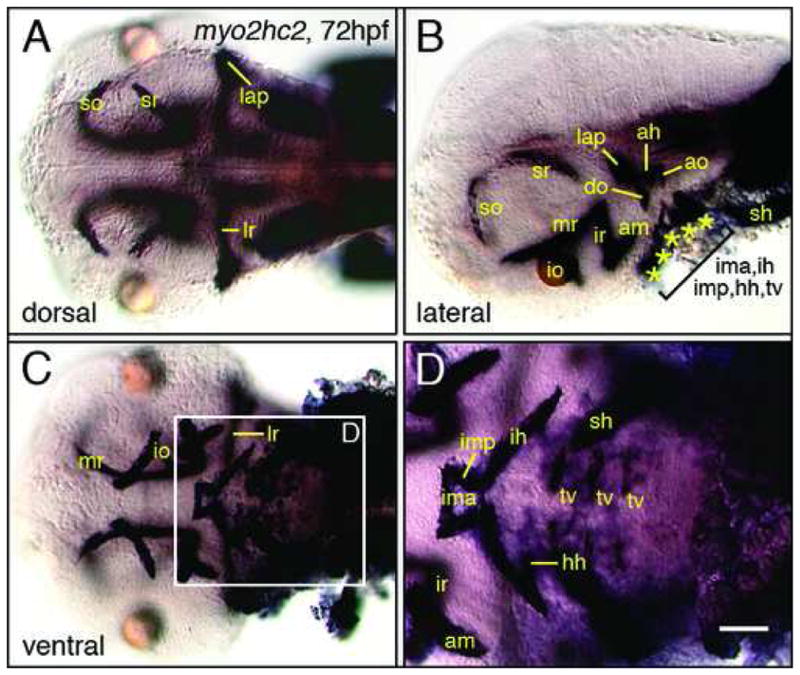
Myosin2 expression in head muscles. (A) In a dorsal view, some eye (so, sr, lr) and jaw (lap) muscles can be identified. (B) In a side view, several eye (so, sr, mr, io, ir, am) and jaw (ah, ao, do, lap) muscles can be distinguished. (C, D) In a ventral view, several eye (am, io, ir, lr, mr), jaw (ima, imp, ih, hh) and gill (tv, sh) muscles can be identified. Scale bar, 100 μm (A–C), 50 μm (D).
2.2.6 Expression of myo2hc2 in cranial muscles
Previous work (Peng et al., 2002) noted weak myo2hc2 (myhz2) expression in two cranial muscles. We found that myo2hc2 is expressed robustly in all cranial muscles (Fig. 8). By comparing the expression pattern to that of anti-myosin antibody labeling of cranial muscles (Schilling et al., 1997), we identified most of the muscles in the zebrafish head at 72 hpf. Our myo2hc2 probe represents an excellent marker for studies of cranial muscle biology.
3. EXPERIMENTAL PROCEDURES
3.1 Animals
Maintenance of zebrafish stocks, and collection and development of embryos in E3 embryo medium were carried out as described previously (Westerfield, 1995; Chandrasekhar et al., 1997; Bingham et al., 2002). Throughout the text, the developmental age of the embryos corresponds to the hours elapsed since fertilization (hours post fertilization, hpf, at 28.5°C).
3.2 Cloning and phylogenetic analysis of zebrafish unconventional myosins
We used the nucleotide and amino acid sequences for the various unconventional myosins from human and mouse databases (Hodge and Cope, 2000, and http://www.mrc-lmb.cam.ac.uk/myosin/; Berg et al., 2001) to search the zebrafish EST databases at Washington University and RZPDfor orthologs. Using this information, we obtained EST clones for the various myosins from RZPD (Berlin, Germany), Open Biosystems (Huntsville, AL) and Dr. Jinrong Peng (IMCB, Singapore) (Table 1). The EST clones were verified by sequencing prior to making in situ probes. The EST sequences were used to BLAST the latest assembly of the zebrafish genomic sequence (Zv7, July 2007) using BLASTN to retrieve the most complete annotated cDNA sequences for the various myosin genes. The newly identified zebrafish and previously described human and mouse myosins were BLASTed against chicken skeletal myosin 2 to delineate the core domain (composed of the actin- and ATP-binding sites) corresponding to amino acids 88–780 of the chicken protein. The core domains of zebrafish, human and mouse myosins were aligned using Clustal-W method in Megalign program of LASERGENE (DNASTAR Inc, Madison, WI; Reddy and Day, 2001). The aligned core domains were further analyzed using Mac PAUP 4.0 b10 (http://paup.csit.fsu.edu/; Jiang and Ramachandran, 2004) to generate a phylogenetic tree (Fig. 2). The heuristic search option was used in maximum parsimony method to create a tree with strict consensus. Bootstrap analyses were replicated 1000 times to obtain a robust support value at each node which indicates the percent times that the grouping is supported out of 1000 trials (Baker and Titus, 1997; Hodge and Cope, 2000; Sellers, 2000). Bootstrap values of greater than 70 indicate a strongly supported node with 95% probability (Hills and Bull, 1993).
3.3 Histological Procedures
The EST clones were verified by sequencing prior to probe synthesis. Synthesis of the digoxygenin-labeled probes, and whole-mount in situ hybridization was carried out as described previously (Bingham et al., 2003). Given the large number of clones and time points, embryos of different ages were pooled for the hybridization step following protease treatment. Whole-mount immunohistochemistry (tubulin and GFP antibodies) was performed as described previously (Vanderlaan et al., 2005). For combined in situ and antibody labeling, embryos were first processed for in situs before antibody labeling. Embryos were deyolked, mounted in 70% glycerol, and examined with an Olympus BX60 microscope. At least ten embryos were examined for each gene and time point. Images were acquired using an Optronics camera and Scion Imaging Software on a MacOS computer equipped with an LG3 frame grabber. Images were processed in Adobe Photoshop to adjust brightness and contrast only.
Acknowledgments
We thank Jinrong Peng (IMCB, Singapore) for providing several myosin ESTs used in our analysis, Tom Schilling (UC, Irvine) for help with muscle identification, and Chris Pires and Robert Snyder (University of Missouri) for help with the phylogenetic analysis. We thank Gary Vanderlaan for help in the early stages of the project, and Moe Baccam, Amy Foerstel and other members of our lab for excellent fish care. This work was supported by an Interdisciplinary Neuroscience Program Fellowship from the University of Missouri Graduate School to VS, and grants from the University of Missouri System (RB07-03) and NIH (NS40449) to AC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Argenton F, Zecchin E, Bortolussi M. Early appearance of pancreatic hormone-expressing cells in the zebrafish embryo. Mech Dev. 1999;87:217–221. doi: 10.1016/s0925-4773(99)00151-3. [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham S, Chaudhari S, Vanderlaan G, Itoh M, Chitnis A, Chandrasekhar A. Neurogenic phenotype of mind bomb mutants leads to severe patterning defects in the zebrafish hindbrain. Dev Dyn. 2003;228:451–463. doi: 10.1002/dvdy.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham S, Higashijima S, Okamoto H, Chandrasekhar A. The Zebrafish trilobite gene is essential for tangential migration of branchiomotor neurons. Dev Biol. 2002;242:149–160. doi: 10.1006/dbio.2001.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar A, Moens CB, Warren JT, Jr, Kimmel CB, Kuwada JY. Development of branchiomotor neurons in zebrafish. Development. 1997;124:2633–2644. doi: 10.1242/dev.124.13.2633. [DOI] [PubMed] [Google Scholar]
- Chilton JK. Molecular mechanisms of axon guidance. Dev Biol. 2006;292:13–24. doi: 10.1016/j.ydbio.2005.12.048. [DOI] [PubMed] [Google Scholar]
- Chitnis AB, Kuwada JY. Axonogenesis in the brain of zebrafish embryos. J Neurosci. 1990;10:1892–1905. doi: 10.1523/JNEUROSCI.10-06-01892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin AB, Dabdoub A, Kelley MW, Popper AN. Myosin VI and VIIa distribution among inner ear epithelia in diverse fishes. Hear Res. 2007;224:15–26. doi: 10.1016/j.heares.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke JE, Kemp HA, Moens CB. EphA4 is required for cell adhesion and rhombomere-boundary formation in the zebrafish. Curr Biol. 2005;15:536–542. doi: 10.1016/j.cub.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Harding BN. Neuronal migration disorders in humans and in mouse models--an overview. Epilepsy Res. 1999;36:133–141. doi: 10.1016/s0920-1211(99)00047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Diefenbach TJ, Latham VM, Yimlamai D, Liu CA, Herman IM, Jay DG. Myosin 1c and myosin IIB serve opposing roles in lamellipodial dynamics of the neuronal growth cone. J Cell Biol. 2002;158:1207–1217. doi: 10.1083/jcb.200202028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond IA, Majumdar A, Hentschel H, Elger M, Solnica-Krezel L, Schier AF, Neuhauss SC, Stemple DL, Zwartkruis F, Rangini Z, Driever W, Fishman MC. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- Ernest S, Rauch GJ, Haffter P, Geisler R, Petit C, Nicolson T. Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum Mol Genet. 2000;9:2189–2196. doi: 10.1093/hmg/9.14.2189. [DOI] [PubMed] [Google Scholar]
- Evans LL, Hammer J, Bridgman PC. Subcellular localization of myosin V in nerve growth cones and outgrowth from dilute-lethal neurons. J Cell Sci. 1997;110:439–449. doi: 10.1242/jcs.110.4.439. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Walsh CA. Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci. 2000;23:352–359. doi: 10.1016/s0166-2236(00)01607-6. [DOI] [PubMed] [Google Scholar]
- Guan KL, Rao Y. Signalling mechanisms mediating neuronal responses to guidance cues. Nat Rev Neurosci. 2003;4:941–956. doi: 10.1038/nrn1254. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Biochemical and molecular characterization of diseases linked to motor proteins. Trends Biochem Sci. 2003;28:558–565. doi: 10.1016/j.tibs.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Hills DM, Bull JJ. An empirical-test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Hodge T, Cope MJ. A myosin family tree. J Cell Sci. 2000;113:3353–3354. doi: 10.1242/jcs.113.19.3353. [DOI] [PubMed] [Google Scholar]
- Hutson LD, Chien CB. Wiring the zebrafish: axon guidance and synaptogenesis. Curr Opin Neurobiol. 2002;12:87–92. doi: 10.1016/s0959-4388(02)00294-5. [DOI] [PubMed] [Google Scholar]
- Jiang S, Ramachandran S. Identification and molecular characterization of myosin gene family in Oryza sativa genome. Plant Cell Physiol. 2004;45:590–599. doi: 10.1093/pcp/pch061. [DOI] [PubMed] [Google Scholar]
- Kalil K, Dent EW. Touch and go: guidance cues signal to the growth cone cytoskeleton. Curr Opin Neurobiol. 2005;15:521–526. doi: 10.1016/j.conb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Kappler JA, Starr CJ, Chan DK, Kollmar R, Hudspeth AJ. A nonsense mutation in the gene encoding a zebrafish myosin VI isoform causes defects in hair-cell mechanotransduction. Proc Natl Acad Sci USA. 2004;101:13056–13061. doi: 10.1073/pnas.0405224101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kuwada JY. Development of the zebrafish nervous system: genetic analysis and manipulation. Curr Opin Neurobiol. 1995;5:50–54. doi: 10.1016/0959-4388(95)80086-7. [DOI] [PubMed] [Google Scholar]
- Li Z, Joseph NM, Easter SS., Jr The morphogenesis of the zebrafish eye, including a fate map of the optic vesicle. Dev Dyn. 2000;218:175–188. doi: 10.1002/(SICI)1097-0177(200005)218:1<175::AID-DVDY15>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Libby RT, Steel KP. The roles of unconventional myosins in hearing and deafness. Essays Biochem. 2000;35:159–174. doi: 10.1042/bse0350159. [DOI] [PubMed] [Google Scholar]
- Metcalfe WK, Myers PZ, Trevarrow B, Bass MB, Kimmel CB. Primary neurons that express the L2/HNK-1 carbohydrate during early development in the zebrafish. Development. 1990;110:491–504. doi: 10.1242/dev.110.2.491. [DOI] [PubMed] [Google Scholar]
- Oster SF, Sretavan DW. Connecting the eye to the brain: the molecular basis of ganglion cell axon guidance. Br J Ophthalmol. 2003;87:639–645. doi: 10.1136/bjo.87.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng MY, Wen HJ, Shih LJ, Kuo CM, Hwang SP. Myosin heavy chain expression in cranial, pectoral fin, and tail muscle regions of zebrafish embryos. Mol Reprod Dev. 2002;63:422–429. doi: 10.1002/mrd.10201. [DOI] [PubMed] [Google Scholar]
- Reddy AS, Day IS. Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2001;2:RESEARCH0024. doi: 10.1186/gb-2001-2-7-research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- Seiler C, Ben-David O, Sidi S, Hendrich O, Rusch A, Burnside B, Avraham KB, Nicolson T. Myosin VI is required for structural integrity of the apical surface of sensory hair cells in zebrafish. Dev Biol. 2004;272:328–338. doi: 10.1016/j.ydbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- Soldati T. Unconventional myosins, actin dynamics and endocytosis: a menage a trois? Traffic. 2003;4:358–366. doi: 10.1034/j.1600-0854.2003.t01-1-00095.x. [DOI] [PubMed] [Google Scholar]
- Sousa AD, Berg JS, Robertson BW, Meeker RB, Cheney RE. Myo10 in brain: developmental regulation, identification of a headless isoform and dynamics in neurons. J Cell Sci. 2006;119:184–194. doi: 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Suter DM, Espindola FS, Lin CH, Forscher P, Mooseker MS. Localization of unconventional myosins V and VI in neuronal growth cones. J Neurobiol. 2000;42:370–382. [PubMed] [Google Scholar]
- ten Donkelaar HJ, Lammens M, Wesseling P, Hori A, Keyser A, Rotteveel J. Development and malformations of the human pyramidal tract. J Neurol. 2004;251:1429–1442. doi: 10.1007/s00415-004-0653-3. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 ( http://zfin.org)
- Tuxworth RI, Titus MA. Unconventional myosins: anchors in the membrane traffic relay. Traffic. 2000;1:11–18. doi: 10.1034/j.1600-0854.2000.010103.x. [DOI] [PubMed] [Google Scholar]
- Vanderlaan G, Tyurina OV, Karlstrom RO, Chandrasekhar A. Gli function is essential for motor neuron induction in zebrafish. Dev Biol. 2005;282:550–570. doi: 10.1016/j.ydbio.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann D, Mori T, Tirlapur UK, Konig K, Fujiwara T, Kendrick-Jones J, Baluska F. Unconventional myosins of the plant-specific class VIII: endocytosis, cytokinesis, plasmodesmata/pit-fields, and cell-to-cell coupling. Cell Biol Int. 2003;27:289–291. doi: 10.1016/s1065-6995(02)00330-x. [DOI] [PubMed] [Google Scholar]
- Wang FS, Wolenski JS, Cheney RE, Mooseker MS, Jay DG. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. University of Oregon; Eugene, OR: 1995. [Google Scholar]
- Wilson SW, Ross LS, Parrett T, Easter SJ. The development of a simple scaffold of axon tracts in the brain of the embryonic zebrafish, Brachydanio rerio. Development. 1990;108:121–145. doi: 10.1242/dev.108.1.121. [DOI] [PubMed] [Google Scholar]
- Wu X, Jung G, Hammer JA., 3rd Functions of unconventional myosins. Curr Opin Cell Biol. 2000;12:42–51. doi: 10.1016/s0955-0674(99)00055-1. [DOI] [PubMed] [Google Scholar]


