Abstract
Since the discovery of microRNAs (miRNAs), the study of these small noncoding RNAs has steadily increased and more than 10,000 papers have already been published. The great interest in miRNAs reflects their central role in gene-expression regulation and the implication of miRNA-specific aberrant expression in the pathogenesis of cancer, cardiac, immune-related and other diseases. Another avenue of current research is the study of circulating miRNAs in serum, plasma, and other body fluids—miRNAs may act not only within cells, but also at other sites within the body. The presence of miRNAs in body fluids may represent a gold mine of noninvasive biomarkers in cancer. Since deregulated miRNA expression is an early event in tumorigenesis, measuring circulating miRNA levels may also be useful for early cancer detection, which can contribute greatly to the success of treatment. In this Review, we discuss the role of fluid-expressed miRNAs as reliable cancer biomarkers and treatment-response predictors as well as potential new patient selection criteria for clinical trials. In addition, we explore the concept that miRNAs could function as hormones.
Introduction
In 1993, Victor Ambros’1 and Gary Ruvkun’s2 groups discovered that the abundance of the protein LIN14 in Caenorhabditis elegans was regulated by a small RNA product encoded by the lin-4 gene. However, it was not until 2000, when another small RNA, let-7, was identified and found to be conserved in many species,3,4 that a new layer of complexity in the regulation of gene expression was unveiled. The discovery of the post-transcriptional silencing of target mRNAs5 by small RNAs was a revolutionary step in the understanding of genetic-information control. Further studies provided evidence that these microRNAs (miRNAs) are members of a large class of non-coding RNAs of approximately 22 nucleotides in length, which regulate most genes in the human genome.6 MiRNAs are strongly conserved between vertebrates, invertebrates, and plants,7 and are transcribed from individual genes sometimes clustered and located intergenic or in introns or exons of protein-coding genes.8
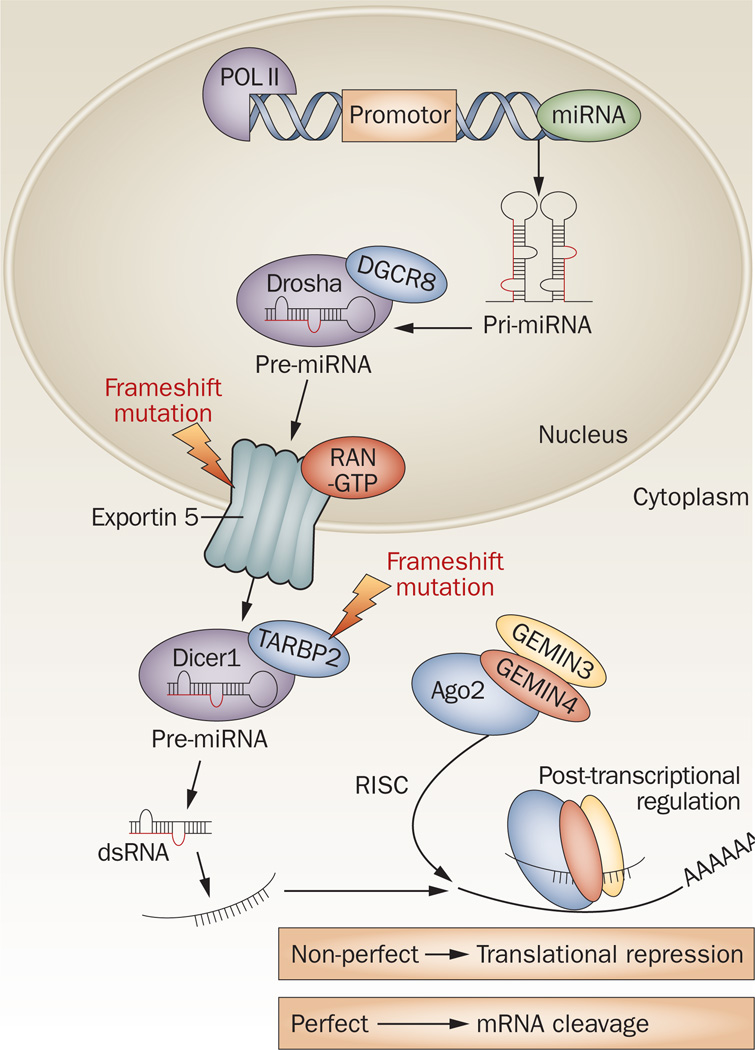
MiRNA biogenesis involves the maturation of miRNA precursors, assembly of the mature miRNA into micro-processor complexes and the regulation of gene expression of protein-coding genes by degrading or blocking translation of mRNA targets (Figure 1).9 First, miRNA is transcribed in the nucleus by RNA polymerase II as long and capped precursor primary miRNA (pri-miRNA).10,11 The next step is the production of a precursor miRNA (pre-miRNA) by the ribonuclease (RNase) III Drosha enzyme and the processing of the double-stranded DNA-binding protein DGCR8.12 Pre-miRNAs are actively exported to the cytoplasm by the nuclear export receptor exportin 5 and then processed by the RNase III endonuclease Dicer protein along with the double-stranded transactivation-responsive RNA-binding protein (TRBP), resulting in a small double-strand RNA structure of about 22 nucleotides.13,14 This miRNA duplex is unwound into the mature single-strand form and incorporated into the RNA-induced silencing complex (RISC), which guides the complex into the complementary 3' or 5'-untranslated region (UTR) of the target mRNA,15–18 open reading frames, and promoter regions.19 Variations in the expression of Dicer and Drosha mRNA and protein are extensive in human cancers,20 and frameshift functional mutations in the biogenesis machinery including XPO5 and TARBP2 occur in human tumors with microsatellite instability (Figure 1).21,22
Figure 1.
MiRNA biogenesis in the cell. MiRNAs are transcribed in the nucleus as pri-miRNA and then processed by Drosha into pre-miRNA. Pre-miRNA molecules are transported from the nucleus to the cytoplasm by exportin 5. Upon entering the cytoplasm, they are recognized by Dicer. Dicer modulates pre-miRNA and generates dsRNA, which are recognized by the RISC complex and converted into single-strand mature miRNA molecules. The RISC complex carries the mature miRNA molecule to complementary miRNA target sites within the mRNA molecule, where it affects gene expression by miRNA:mRNA sequence complementarity. A consequence of perfect complementarity between miRNA:mRNA molecules is mRNA cleavage and degradation. Imperfect alignment represses gene translation. Mutations in RNA processing are indicated with red lightning bolts. A frameshift mutation in exportin 5 caused premature codon termination and trapped pre-miRNAs in the nucleus.21 Other frameshift mutations in the RISC-loading complex subunit TARBP2 causes a loss of function of TARBP2, a secondary defect of Dicer activity and the loss of miRNA machinery regulation during tumorigenesis.22 Abbreviations: Ago2, Argonaute2; dsRNA, double-strand miRNA; miRNA, microRNA; POL II, RNA polymerase II; pre-miRNA, precursor miRNA; pri-miRNA, precursor primary miRNA; RISC, RNA-induced silencing complex.
Although negative regulation of gene expression occurs via mRNA cleavage or translational repression,23,24 studies have shown that miRNAs can upregulate the expression of their target genes25 and that a single gene can be targeted by multiple miRNAs.26 In mammals, gene regulation mediated by miRNAs is accomplished by imperfect base pairing together with protein translational repression of the target gene.27,28
MiRNAs are involved in virtually all biologic processes and, because a single miRNA can target hundreds of mRNAs, aberrant miRNA expression is involved in the initiation of many diseases, including cancer. In this Review, we focus on some paradigms of miRNA involvement in cancer, on the potential detection of tumor-specific miRNAs in body fluids and their applicability as diagnostic and prognostic markers in cancer. We also discuss the concept that miRNAs can act as hormones through their secretion in plasma and address the effects of their delivery to distant sites in the body.
Paradigms of miRNAs in cancer
MiRNAs are altered in every type of cancer
Genome-wide miRNA-expression-profiling studies using high-throughput technologies have demonstrated that almost all cancer types present a specific profile of upregulated and downregulated miRNAs.29,30 Therefore, owing to the unique miRNA-expression profile for each tumor and the lack of complex transcriptional and translational modifications compared to mRNAs and proteins, the use of miRNAs as biomarkers for cancer has great potential. For example, by analyzing the chromosome region 13q14, a region that is deleted in more than half of all patients with B-cell chronic lymphocytic leukemia (B-CLL), it was demonstrated that miR-15a and miR-16a were either absent or downregulated in approximately 68% of patients with B-CLL.31 One of the first studies of miRNA in solid tumors identified 28 miRNAs differentially expressed between colonic adenocarcinoma and normal mucosa and also found that levels of miR-143 and miR-145 were significantly lower in tumors than in normal tissue.32 Similarly, other studies found specific signatures of miRNA expression in breast carcinoma,33 primary glioblastoma,34 hepatocellular carcinoma,35 papillary thyroid carcinoma,36 and lung cancer.37 In a large profiling analysis of 540 samples of six solid tumors (lung, breast, stomach, prostate, colon, and pancreas), a group of 43 miRNAs was found to be deregulated compared to matched normal tissues.38 MiRNA-expression signatures also correlate with tumor classification and have proven useful in determining the primary site of cancers of unknown origin.29 Metastatic cancers of unknown primary origin were classified with >90% accuracy based on an expression profile of 48 miRNAs.39 Furthermore, the global-expression pattern of miRNAs distinguished between long-term and short-term survivors of pancreatic cancer and also differentiated ductal adenocarcinomas of the pancreas from normal pancreas and chronic pancreatitis with 95% accuracy.40 Following the discovery of different expression levels of miRNAs between normal and cancerous tissues, the next step has been to attempt to ascertain the impact of these small RNAs on tumorigenesis.
Angels and devils
Growing evidence has demonstrated that miRNAs can act as either oncogenes or tumor-suppressor genes. For instance, miR-21 is upregulated in various solid tumors as well as hematologic malignancies, and this potentially oncogenic miRNA regulates important tumor-suppressor genes such as PTEN41 and PDCD4.42 Another well-characterized oncogenic miRNA is miR-10b that promotes metastasis by suppressing HOXD10, which is the negative regulator of a gene associated with tumor cell proliferation and metastasis.43 In addition to single miRNAs, clusters of miRNAs such as the miR-17-92 cluster also promote proliferation, increase angiogenesis, and sustain cancer cell survival via post-transcriptional repression of target mRNAs.44
There are also miRNAs that regulate oncogenes and thus act as tumor suppressors. For example, members of the tumor-suppressor miRNA let-7 family are down-regulated in many malignancies and inhibit cancer growth by targeting various oncogenes and key regulators of mitogenic pathways, such as RAS and HMGA2.45 Other well-established tumor suppressors are the effectors of TP53 activation in the miR-34 family.46
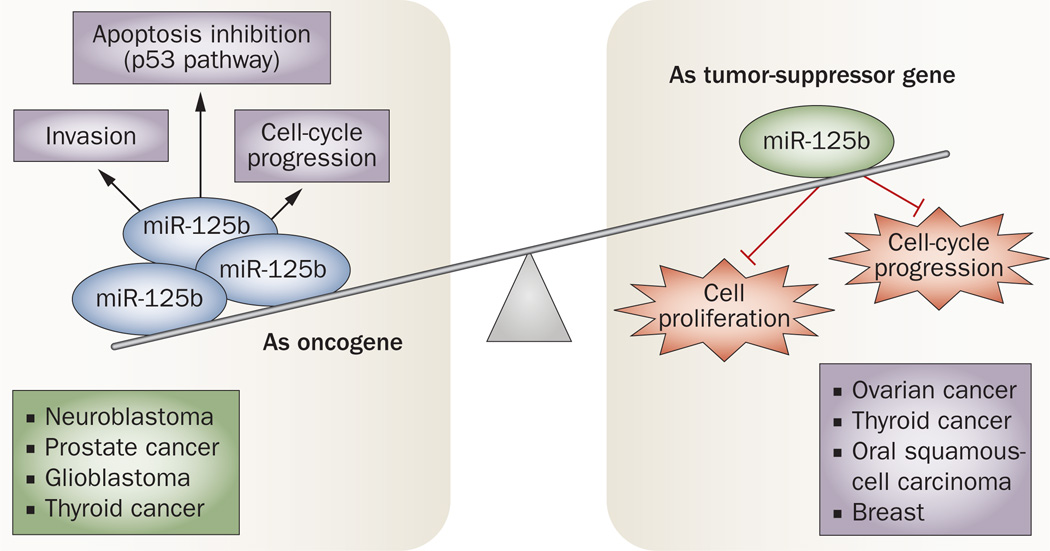
Nonetheless, the initial categorization of miRNAs as oncogenes or tumor-suppressor genes based on their levels of expression in tumors versus normal tissues has proven to be inaccurate, as experiments have shown that many function dually as both oncogenes and tumor-suppressor genes depending on the cancer type and cellular context. An example is miR-125b, which was reported to function as either an oncogene or tumor-suppressor gene in different cancer types or cell lines (Figure 2). In ovarian, thyroid, and oral squamous-cell carcinomas, miR-125b is downregulated and has been shown to inhibit cell proliferation and cell-cycle progression.47,48 On the other hand, miR-125b inhibited apoptosis in neuroblastoma cells in a p53-dependent manner49 and promoted cell proliferation and invasion in prostate cancer cells.50 A similar duality of function in distinct types of cancers has also been found for miR-181a, miR-181c, and miR-220.51 In line with these findings, it seems that the role of a given miRNA is dependent on cancer type and target specificity and, therefore, it is important to elucidate the role of aberrantly expressed miRNAs in each type of cancer.
Figure 2.
A miRNA can function dually as both an oncogene and tumor-suppressor gene depending on the cancer type and cellular context. A duality of function in distinct types of cancer has been found for many miRNAs. An example is miR-125b, which has opposite roles (oncogene and tumor suppressor) in different cancer types or cell lines. As a tumor suppressor, miR-125b is downregulated in ovarian, thyroid, breast, and oral squamous-cell carcinomas, which promotes cell proliferation and cell-cycle progression.47,48 On the other hand, miR-125b is an oncogene in cancers such as prostate, thyroid, glioblastoma, and neuroblastoma. In neuroblastoma cells, miR-125b inhibits apoptosis in a p53-dependent manner,49 and promotes cell proliferation and invasion in prostate cancer cells.50
MiRNAs and cancer predisposition
Unlike the aberrant miRNA expression in somatic cells that can promote tumorigenesis, altered expression of miRNA in germline cells may predispose to cancer development. An explanation for this well-characterized difference in miRNA expression in cancer compared with normal cells is that these small RNAs are frequently located in cancer-associated genomic regions (CAGRs) and are often subject to rearrangements, breakpoints, loss of heterozygosity, and deletions.52 A paradigm for this model is B-CLL, in which miR-15a and miR-16-1 are located in the most frequently deleted genomic region, are downregulated in the majority of cases, and harbor germline mutations in familial cases of CLL and breast cancer.31 Another example is the miR-17-92 cluster, which is located in intron 3 of the C13orf25 gene on 13q31.3, a chromosomal region amplified in many types of cancer.53 Moreover, specific miRNA-expression signatures have been associated with specific translocations in hematopoietic malignancies.54 For example, the fusion gene AML1–ETO produced by the t(8;21) translocation promotes heterochromatic silencing of pre-miR-223 in patients with leukemia.55
Along with the CAGRs harboring miRNA genes and epigenetic changes, several studies have indicated that single nucleotide polymorphisms (SNPs) in both miRNA genes and miRNA-target genes also increase the predisposition to specific types of cancers. Although SNPs are rare in miRNA genes, they can affect miRNA function in pri-miRNA transcription, pri-miRNA and pre-miRNA processing, and miRNA binding sites.56 For example, a SNP found in the binding site of let-7 in the 3'-UTR of the KRAS gene increased the risk of lung cancer in moderate smokers.57 The presence of the SNP rs531564 in pri-miR-124-1 was associated with increased bladder and esophageal cancer risk.58,59 In breast cancer, SNPs in miRNA genes have been associated with both increased and decreased risk of cancer, such as SNP rs11614913 in pre-miR-196a-2,60 and SNP rs895819 in pre-miR-27a,61 respectively. SNPs affecting the function of a miRNA can also increase the risk of cancer. The SNP rs2910164 located in the 3' strand of miR-146a promotes mispairing in the hairpin of the precursor, altering its expression and leading to an increased risk of papillary thyroid carcinoma.62 Since aberrant expression and sequence variations in miRNAs are related to cancer risk, it is thought that these noncoding RNAs may function as useful biomarkers for cancer predisposition.
MiRNAs as biomarkers
Based on the tissue-specific deregulation of miRNA expression in cancer, multiple studies have explored the potential usefulness of miRNA-expression profiles as biomarkers of cancer diagnosis, prognosis, and response to treatment. In a study of 143 lung cancer samples from patients who underwent potentially curative resection, patients could be classified into two major groups according to let-7 expression, with reduced let-7 expression associated with significantly shorter survival after resection.63 High miR-21 expression levels were associated with poor survival and therapeutic outcome in 84 patients with colon adenocarcinoma.64 Expression levels of miR-15b, miR-34c, and miR-361 may predict a low risk of tumor recurrence following curative resection, with an overall accuracy of 90% in hepatocellular carcinoma.65 In addition, since the loss of specific miRNAs provides a selective advantage for cells destined for metastatic colonization,66 these small RNAs are valuable biomarkers of cancer progression and metastasis. The loss of miR-335 and miR-126 expression in the majority of primary breast tumors in patients who relapse is associated with poor distal-metastasis-free survival.66 In liver cancer, the loss of miR-122 expression in tumor cells segregates with specific gene-expression profiles linked to cancer progression and gain of metastatic properties.67 Although these findings suggest the possibility of using small RNAs as biomarkers for noninvasive diagnostic screening and early cancer detection, these findings do not yet eliminate the necessity of using invasive cancer screening techniques.
MiRNA levels in body fluids
Tumor-specific miRNAs were first discovered in the serum of patients with diffuse large B-cell lymphoma; high levels of miR-21 correlated with improved relapse-free survival.68 In an elegant experiment in a xenograft mouse prostate cancer model, the presence of circulating tumor-derived miR-629 and miR-660 was confirmed in blood with 100% sensitivity and specificity.69 In addition to showing that both serum and plasma samples are adequate for measuring specific miRNA levels, the investigators reported that by measuring serum levels of miR-141, they were able to distinguish patients with prostate cancer from healthy subjects. Since then, over 100 studies have assessed the potential use of serum or plasma miRNAs as biomarkers in different types of cancer (Table 1). Confirming these data, another study found that miR-141 was overexpressed in sera from patients with prostate cancer compared to normal tissue samples.70 Moreover, 15 serum miRNAs were over-expressed in patients with prostate cancer, including miR-16, miR-92a and miR-92b.70 In a comprehensive study, miRNA-expression profiles were identified in the sera of patients with lung or colorectal cancer, or diabetes by extracting miRNA from the serum.71 For example, 63 new miRNAs that were absent in normal controls were detected in the sera of patients with non-small-cell lung cancer (NSCLC). Although a unique expression profile of serum miRNAs was identified for each cancer type, an overlap was found in the profiles of specimens from all diseases analyzed in the study, including diabetes. In addition, this study also showed that miRNA-expression profiles differed between the serum and blood cells of lung cancer patients, while similar miRNA-expression profiles were seen in the serum and blood cells of healthy controls. These findings suggest that tumor-specific miRNAs in serum are derived not only from circulating blood cells but also cancer cells.
Table 1.
A compendium of circulating miRNAs with potential as biomarkers for cancer
| miRNAs | Cancer type | Body fluid source |
Healthy subjects (n) |
Patients (n) |
Clinical correlations |
|---|---|---|---|---|---|
| miRs-21, 155 and 210 | Diffuse large B-cell lymphoma | Serum | 43 | 60 | High miR-21 expression was associated with relapse-free survival68 |
| miR-141 | Prostate | Serum | 25 | 25 | Serum levels of miR-141 distinguished patients from healthy subjects69 |
| miRs-141, 16, 92a, 92b, 103, 107, 197, 34b, 328, 485-3p, 486-5p, 574-3p, 636, 640, 766, and 885-5p | Prostate | Serum | 15 | 6 | Serum levels were significantly higher in patients compared to controls70 |
| miRs-486, 30d, 1 and 499 | Lung | Serum | – | 243 | Serum levels were differentially expressed between patients with longer and shorter survival. The four-miRNA signature was an independent predictor of overall survival72 |
| miRs-21, 92, 93, 126 and 29a | Ovarian | Serum | 11 | 19 | miRs-21, 92 and 93 were overexpressed in patients with normal preoperative cancer antigen 12573 |
| miRs-17-3p and 92 | Colorectal | Plasma | 50 | 90 | Plasma levels decrease after surgery; differentiated colorectal from gastric cancer and normal individuals74 |
| miRs-92a and 29a | Colorectal | Plasma | 59 | 157 | Plasma levels significantly higher in patients with advanced-stage cancer than healthy controls75 |
| miRs-17-5p, 21, 106a and 106b | Gastric | Plasma | 69 | 30 | Plasma miRNA levels reflected the tumor miRNAs in most cases; miRNAs were significantly reduced in post-operative samples76 |
| miR-195 and let7-a | Breast | Serum | 44 | 83 | Serum levels were decreased after tumor resection and correlated with nodal and estrogen-receptor status77 |
| miRs-21, 210, 155, and 196a | Pancreas | Plasma | 36 | 49 | Plasma levels discriminate patients from healthy controls78 |
| miR-210 | Pancreas | Plasma | 25 | 22 | Plasma levels were significantly elevated in two independent patient cohorts79 |
| miR-500 | Liver | Serum | 40 | 40 | Increased levels found in patients with hepatocellular carcinoma; miR-500 serum levels returned to normal after surgical treatment80 |
| miR-206 | Rhabdomyo-sarcoma | Serum | 17 | 8 | Serum levels of the muscle-specific miRNA miR-206 were significantly higher in patients with rhabdomyosarcoma tumors than in patients with other types of tumors or in the control group81 |
| miR-184 | Tongue | Serum | 20 | 20 | Serum levels were significantly reduced after surgical removal of the primary tumors82 |
| miR-92a | Acute leukemia | Plasma | 20 | 20 | Decreased levels in plasma samples of acute leukemia patients84 |
| miRs-125a and 200a | Oral squamous-cell | Saliva | 50 | 50 | Lower levels in the saliva of patients than control subjects86 |
| miR-31 | Oral squamous-cell | Plasma Saliva |
21 8 |
43 9 |
Increased levels in patients compared with controls; level in most patients declined after surgery87 |
| miRs-126, 152 and 182 | Bladder | Urine | 9 | 47 | Increased levels in patients compared with controls88 |
| miR-141 | Colorectal | Plasma | – | 102 | High levels were associated with poor prognosis123 |
Another study in NSCLC demonstrated that a set of 11 serum miRNAs were differentially expressed between patients with longer or shorter survival, and among this set, four (miR-486, miR-30d, miR-1, and miR-499) were associated with decreased overall survival of patients.72 In a study of epithelial ovarian cancer, eight serum miRNAs, among them miR-21, miR-92, miR-93, miR-126, and miR-29a, were significantly overexpressed in a set of 19 samples collected before therapy compared with 11 healthy controls.73 This study also provided evidence that miRNAs may be used as biomarkers for early detection by showing that miR-21, miR-92, and miR-93 were overexpressed in three patients with normal levels of preoperative cancer antigen 125, a biomarker used for detecting the recurrence of ovarian cancer. In colorectal cancer, a study demonstrated that a set of miRNAs, including miR-17-3p and miR-92, were simultaneously upregulated in plasma and tissue samples.74 By analyzing an independent group of plasma samples, the researchers also demonstrated that miR-92 was differentially expressed in colorectal cancer compared with gastric cancer, inflammatory bowel disease, and tissues from normal controls and may be a potential molecular marker for detecting colorectal cancer in plasma samples. Interestingly, another study also found that the levels of miR-92a (and of miR-29a) were significantly higher in plasma samples from patients with advanced-stage colorectal cancer than in those from healthy controls.75
In a report on gastric cancer, serum levels of upregulated miRNAs such as miR-21 and miR-106b were significantly higher in patients with gastric cancer than in controls before resection, and were reduced after resection.76 A prospective study reported that tumor-specific miRNAs such as miR-195 were detected and significantly altered in the circulation by using prospectively collected samples from 127 women, including 83 patients with breast cancer and 44 healthy age-matched controls.77 Furthermore, serum levels of miR-195 and let-7a were decreased after tumor resection and specific circulating miRNAs were correlated with nodal and estrogen receptor status. The combined expression analyses of miR-21, miR-210, miR-155, and miR-196a in plasma can discriminate pancreatic adenocarcinoma patients from controls.78 The plasma levels of the hypoxia-related miRNA miR-210 were also altered in patients with pancreatic cancer compared to healthy controls from two independent cohorts.79 High levels of miR-500 were found in the serum of patients with hepatocellular carcinoma, and after tumor resection these levels returned to normal in three out of 40 patients.80 Serum levels of the muscle-specific miRNA miR-206 were significantly higher in patients with rhabdomyosarcoma than in patients with other types of tumors or those in the control group.81 Nonetheless, more studies are necessary to identify new miRNAs as biomarkers in rhabdomyosarcoma because other rare myogenic tumors such as leoimyosarcoma and rhabdomyoma also overexpress miR-206. In patients with squamous-cell carcinoma of the tongue, plasma levels of miR-184 were significantly higher than those in healthy individuals and miR-184 levels were significantly reduced after surgical removal of the primary tumors.82
Recently, a high-throughput study generated miRNA signatures from plasma samples collected 12–28 months before and at the time of lung cancer detection.83 In this study, 21 miRNAs were identified as risk, diagnosis, and prognosis predictors and are potentially useful in the monitoring of high-risk disease-free smokers. Furthermore, this study is one of the first to demonstrate that specific pre-disease signatures of miRNA expression in plasma samples can predict the development of lung cancer before diagnosis by conventional techniques and in a noninvasive manner.
MiRNAs are likely to be useful as noninvasive biomarkers not only in solid tumors, but also in hematologic malignancies. For instance, in a study in acute leukemias, there was a decrease of miR-92a in the plasma samples of all patients compared with controls.84 A specific profile of plasma miRNAs was also found in CLL compared with multiple myeloma, hairy-cell leukemia and healthy-control samples.85 The results of this study indicated that circulating miRNAs correlated with the prognosis marker ZAP-70 status and might be used to detect and stratify individuals with CLL.
Although the majority of studies assessed circulating miRNAs in serum and plasma, recent studies have confirmed the potential use of tumor-specific miRNAs as diagnostic markers for cancer in other body fluids. For instance, a study analyzed a panel of four miRNAs in the saliva of patients with oral squamous-cell carcinoma compared with matched healthy controls; miR-125a and miR-200a were present in significantly lower levels in the saliva of the cancer patients.86 In another study, miRNA expression was analyzed in the saliva collected in the week before surgery and 6 weeks after surgery from nine patients with oral squamous-cell carcinoma and compared with eight normal individuals.87 Cancer patients had significantly higher salivary levels of miR-31 than the controls, and eight of nine patients had a decrease in salivary miR-31 levels after tumor resection. These results were also found for miR-31 plasma levels. Increased levels of miR-126, miR-152, and miR-182 were found in urine samples from patients with bladder cancer, and the ratios of miR-126 to miR-152 and miR-182 to miR-152 indicated the presence of bladder cancer with a specificity of 82% and a sensitivity of 72%.88
MiRNAs have also been detected in other body fluids, such as tears, breast milk, bronchial lavage, colostrum, and seminal, amniotic, pleural, peritoneal, and cerebro-spinal fluids.89 Specific compositions and concentrations were found for each body fluid analyzed. These findings might be useful if a correlation between specific miRNA levels in body fluids and various disease states is proven.
Body-fluid miRNA stability and activity
The diagnostic and prognostic potential of miRNAs as cancer biomarkers relies mainly on their high stability and resistance to storage handling. It has been consistently shown that serum miRNAs remain stable after being subjected to severe conditions that would normally degrade most RNAs, such as boiling, very low or high pH levels, extended storage, and 10 freeze–thaw cycles.71 In addition, recent studies have demonstrated that miRNAs are preserved in archived 10-year-old human serum samples,90 and in unrefrigerated dried serum blots —which may be a more convenient and safer way to save, transport, and store serum and other body fluids, such as saliva and urine, for miRNA assays.91 This stability can be partially explained by the discovery of lipoprotein complexes, including small membrane vesicles of endocytic origin called exosomes or microvesicles (30–100 nm), containing miRNAs,92 mRNAs,93,94 and proteins.94 Exosomes can be formed through inward budding of endosomal membranes, giving rise to intracellular multivesicular bodies (MVBs) that later fuse with the plasma membrane, releasing the exosomes to the exterior.95 Exosomes containing miRNAs were found not only in blood,96 but also in other types of body fluids such as saliva.97 Interestingly, one group of researchers has demonstrated the existence of tumor-derived exosomes98 and a miRNA signature for circulating ovarian cancer exosomes.99 This miRNA signature was significantly correlated with primary tumor-miRNA expression in women with cancer compared to women with benign disease and was not identified in normal controls. A similarity between miRNA signatures in circulating exosomal miRNA and originating tumor cells was also found in lung adenocarcinoma,100 with a significant difference in exosomal miRNA levels between cancer patients and controls.
Importantly, exosomes represent a newly discovered mechanism by which donor cells can communicate and influence the gene expression of recipient cells. These findings were first demonstrated by the same study that discovered miRNAs in exosomes, in which mouse mast cell exosomes were added to human mast cells, leading to a subsequent detection of mouse proteins in the human cells.92 Indeed, another study confirmed these findings and demonstrated that exosomes released by glioblastoma cells containing mRNA, miRNAs, and angiogenic proteins, such as EGFRvIII, are taken up by normal recipient cells, such as brain microvascular endothelial cells.101 In this study, it was shown that messages delivered by tumor-derived exosomes are translated by recipient cells to promote tumor progression by stimulating proliferation of a human glioma cell line and tubule formation by endothelial cells. In addition, the results indicated that cancer patients have elevated levels of tumor-derived exosomes in plasma compared with controls. A recent study, however, found that a known family of tumor-suppressor miRNAs, let-7, is abundant in exosomes produced by a metastatic gastric cancer cell line.102 The researchers suggested that the selective secretion of exosomal let-7 family members may be related to maintenance of an intercellular tumorigenic and metastatic state.
Nevertheless, little is known about the mechanisms by which miRNAs are generated in plasma and the biologic impact of these molecules in distant sites of the body.103 In a recent study, the investigators demonstrated that exosomal miRNAs promote gene silencing similar to cellular miRNAs and that exosomes with miRNAs are released through a ceramide-dependent secretory machinery.104 In this study, it was also demonstrated that the secretion of miRNAs is affected by ceramide levels regulated by neutral sphingomyelinase 2 (nSMase2) and that the inhibition of nSMase2 blocked the secretion of miRNAs and exosomes. In addition, by incubating the metastatic prostate cancer cell line PC-3M-luc with a conditioned medium from miR-146a-overexpressing HEK293 cells, the investigators showed an approximately 20% decrease in proliferation. Moreover, the addition of the conditioned medium of miR-146a-transduced COS-7 cells significantly knocked down the miR-146a target gene ROCK1 in PC-3M-luc cells. However, further studies are necessary to unveil how miRNAs are sorted into exosomes and whether there is a pathway in which specific miRNAs are chosen to be incorporated into exosomes.
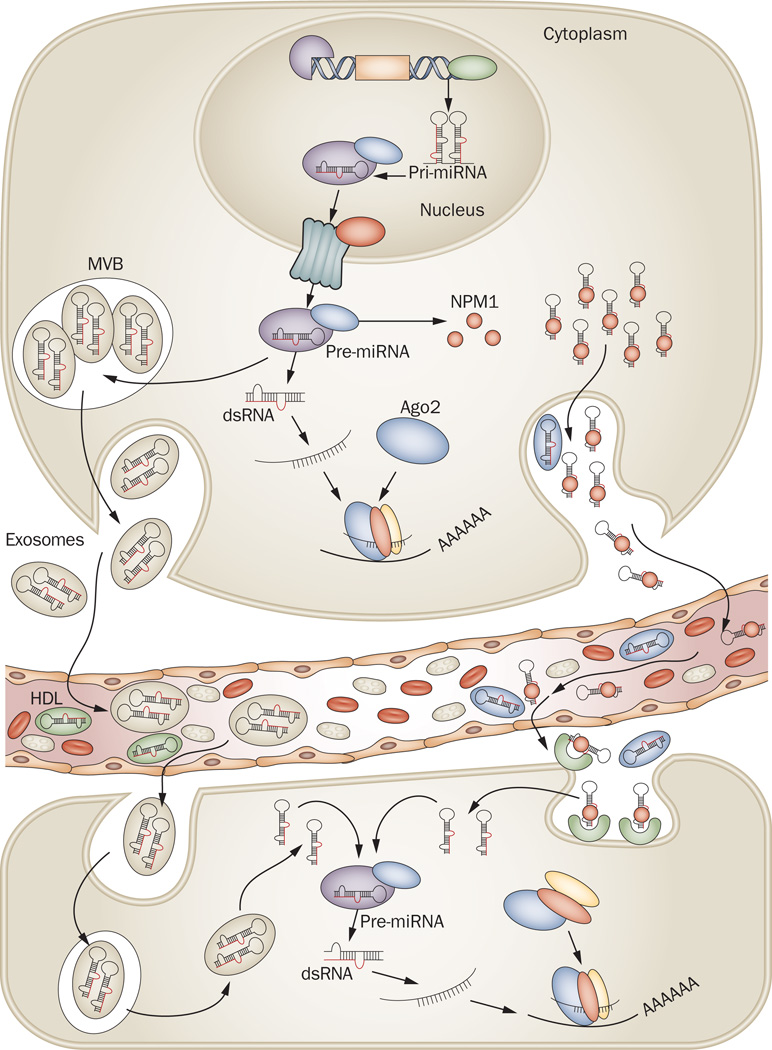
Growing evidence indicates that exosomal miRNA packaging occurs non-randomly based on differential expression of exosomal miRNA compared to that of donor cells.92 Indeed, studies have demonstrated that nearly 30% of the released miRNAs in vitro and in vivo do not reflect the expression profile found in donor cells, suggesting that specific miRNAs are selected to be intracellularly retained or released by exosomes.105 Taken together, these studies indicate that the secretion of miRNAs by tumor cells is associated with their ability to influence the surrounding microenvironment for their own benefit. After being transcribed in the nucleus and exported to the cytoplasm in donor cells, pre-miRNA molecules can bind to specific proteins responsible for their stability and association with MVBs and exosomes. After fusion with the plasma membrane, MVBs are able to release exosomes into the circulating compartments and bloodstream. These exosomes can donate their miRNAs to the recipient cells by the process of endocytosis. Exosomal miRNAs are processed by the same machinery used in miRNA biogenesis and thus promote widespread consequences within the cell and lead to an alteration in the physiologic state of the cell. Recently, another mechanism potentially involving the nSMase2 pathway was discovered showing that high-density lipoprotein (HDL) transports circulating miRNAs and can alter gene expression by transferring miRNAs to recipient cells (Figure 3).106 This is an exciting finding, but further study is necessary to determine the mechanism by which selective miRNAs are taken up from recipient cells and how they are released.
Figure 3.
Biogenesis and mechanism of action of circulating miRNAs. After being transcribed in the nucleus, pre-miRNA molecules can be processed further by Dicer in the cytoplasm. In addition, based on recent findings,92,104–108 there are at least two ways that pre-miRNAs can be packaged and transported using exosomes and MVBs or other (not fully explored) pathways together with RNA-binding proteins. After fusion with the plasma membrane, MVBs release exosomes into the circulating compartments and bloodstream. Likewise, pre-miRNA inside the donor cell can be stably exported in conjunction with RNA-binding proteins, such as NPM1,107 and Ago2,108 or by HDL.106 Circulating miRNAs enter the bloodstream and are taken up by the recipient cells by endocytosis or, hypothetically, by binding to receptors present at the recipient cellular membrane capable of recognizing RNA-binding proteins. More studies are necessary to elucidate how miRNAs are loaded into exosomes and how they can be internalized by recipient cells. Exosomal miRNAs are processed by the same machinery used in miRNA biogenesis and thus have widespread consequences within the cell by inhibiting the expression of target protein-coding genes. For processing machinery see Figure 1. Abbreviations: MVBs, multivesicular bodies; NPM1, nucleophosmin 1; Ago2, Argonaute2; HDL, high-density lipoprotein.
Although the presence of miRNAs in exosomes or HDL could explain their stability in serum, other possibilities include protection by chemical modifications or association with protein complexes. In this regard, recent findings showed that the RNA-binding protein nucleophosmin 1 (NPM1) may have a role in the exportation, packaging, and protection of extracellular miRNAs (Figure 3).107 Furthermore, recent studies demonstrated that potentially 90% of the plasma and serum miRNAs are not encapsulated by vesicles, but cofractionated with protein complexes. The results indicated that the association of Argonaute2 (Ago2; the effector of target mRNA silencing by miRNAs) with plasma and serum miRNAs influences their stability. Whether the Ago2–miRNA complex in plasma is capable of regulating the expression of recipient cells is not clear; however, these findings may be useful in the near future for establishing circulating miRNA as biomarkers.108
It was demonstrated that the cell-free miRNAs were probably derived from normal and/or tumor-lysed cells in body fluids.71,109 Therefore, in order to use miRNAs as biomarkers in cancer, it is important to determine the source of the tumor-specific miRNAs in body fluids and establish a signature capable of differentiating diseased from healthy states. Also, it is necessary to clarify whether the differential expression between tumor and normal tissues is related solely to the tumor or is a response mediated by the affected organ or system. There is evidence that circulating miRNAs in body fluids and extracellular fluid compartments have hormone-like effects, leading to widespread consequences within the cells at a distance from the ‘secreting’ cell (Box 1). Nonetheless, additional studies are necessary to elucidate the mechanism by which miRNAs reach the bloodstream and the physiologic impact of circulating miRNA in global cellular processes.
Box 1 | MiRNAs—the ‘oldest’ hormones.
After the seminal discovery of ribozymes, RNAs that perform catalytic functions in the absence of any DNA or protein molecule, the concept of the RNA world as the primordial world of ‘living’ organisms containing only RNA as genetic material is now widely accepted.126 This concept can be extended by considering that the first ‘signaling’ molecules between genomes were also RNAs, and such signals were short sequences and very stable, exactly the same as circulating microRNAs (miRNAs). As hormones, miRNAs should be released by a donor cell as exosomes or as ‘free’ molecules secreted by active mechanisms and spread signals that affect cells located in other parts of the organism that uptake the miRNAs either as exosomes or as ‘free’ RNAs (Figure 3). Consequently, ‘normal’ levels of circulating miRNAs vary widely among humans according to age, gender, physiologic events (such as menarche or pregnancy) and are influenced by various ‘extrinsic’ factors such as environmental temperature and stress. A practical consequence for this view, is the fact that for any study comparing the expression of miRNAs in any type of body fluid from normal individuals and cancer patients it is important to design the study in a ‘paired’ way at least for age, gender and race and use at least twice as many controls as patients. In this way, the ample expression variations in the normal population can be assessed and the comparisons with cancer groups will be more meaningful and reproducible in independent cohorts.
Detection of miRNAs in body fluids
Several techniques are currently available for establishing miRNA signatures in body fluids, such as miRNA microarrays,70 quantitative real-time PCR (qRT-PCR),68 and deep sequencing (next-generation sequencing). 71 Among these approaches, the most frequently used is qRT-PCR and its variations, such as stem-loop RT-PCR110,111 and poly(A)-tailed RT-PCR,112 which have improved the specificity and sensitivity of miRNA detection. 113,114 Nonetheless, most published studies present conflicting data and have limitations in their cross-comparison of miRNA-expression profiles due to differing methodologies, various reference genes being used to normalize the miRNA levels measured in body fluids, and differences in blood collection (for example, heparin contains an inhibitor of Taq polymerase).116 Frequently used reference genes, such as U6 small nuclear RNA (RNU6B) and 5S ribosomal RNA, were found to have a less-stable expression than others117 or degraded in serum samples.71 In addition, the considerable differences in choices of reference genes to use represents a major obstacle in comparing expression levels between normal tissue and tumors (Table 2). For example, in one study to identify stable controls for normalization, two of 21 miRNAs studied (miR-142-3p and miR-16) were identified as potential ‘normalizers’ given consistent expression across all patient and control samples.73 The addition of synthetic versions of miRNAs from other organisms such as C. elegans in serum and/or plasma samples has proven useful for normalizing the data obtained by qRT-PCR and also may represent an interesting approach to circumventing normalization issues.69 However, more studies are necessary for the identification of an accurate normalization protocol and empirical validation of stable endogenous control miRNAs for each type of body fluid. Moreover, specific methods have to be standardized for specimen collection, processing, and purification of total RNA and data analysis, similar to the standard operating procedures used routinely in laboratories.
Table 2.
Reference genes used to normalize miRNA expression in body fluids
| Reference gene | Cancer type | Body fluid source |
|---|---|---|
| miR-16 | Diffuse large B-cell lymphoma | Serum68 |
| Synthetic versions of the specific C. elegans microRNAs cel-miR-39, cel-miR-54, and cel-miR-238 | Prostate | Serum69 |
| miRs-142-3p and miR-16 | Ovarian | Plasma73 |
| U6 | Colorectal | Plasma74 |
| miR-16 | Gastric | Plasma76 |
| U6, 18S rRNA | Breast | Serum77 |
| miR-16 and cel-miR-54 | Pancreas | Plasma78,79 |
| miR-16 | Liver | Serum80 |
| miR-16 | Rhabdomyosarcoma | Serum81 |
| miR-16 | Tongue | Serum82 |
| miR-638 | Acute leukemia | Plasma84 |
| U6 and miR-16 | Oral squamous cell | Plasma,86 saliva87 |
| U6 | Bladder | Urine88 |
Predictors of therapy response
Clinical studies have demonstrated the potential for use of miRNAs as predictors of sensitivity to radiotherapy and anticancer agents.118 For instance, the loss of heterozygosity of miR-128b, an EGFR regulator, was correlated with response to the EGFR inhibitor gefitinib in relapsed patients with NSCLC.119 In colorectal cancer, it was demonstrated that let-7g and miR-181b may be indicators of chemotherapy response to 5-fluorouracil treatment.120 Likewise, upregulated and downregulated miRNAs were detected in sensitive or resistant cell lines and predicted patient response to anticancer agents. One study identified a miRNA chemosensitivity profile from a set of 59 human cancer cell lines derived from diverse tissues (NCI-60 cell lines). Downregulation of miR-34, miR-17, and let-7a was related to sensitivity to drugs commonly used in cancer treatment, such as 5-fluorouracil, adriamycin, and cyclophosphamide, respectively.121 These findings suggest that circulating miRNAs may be useful in predicting patterns of resistance and sensitivity to drugs used in cancer treatment.
Nonetheless, one study to date has demonstrated a correlation between circulating miRNA-expression levels and response to a given anticancer treatment. In this study, serum miR-21 levels were higher in patients with castration-resistant prostate cancer whose disease was resistant to docetaxel-based chemotherapy when compared to those with chemosensitive disease.122 Additional and more detailed investigations are needed to gauge the utility of circulating miRNAs in predicting resistance or sensitivity to specific treatments. Moreover, the study of circulating miRNAs would provide new insights regarding the identification of cancer patients responsive to a specific protocol of anticancer agents before treatment, circumventing unnecessary treatments and collateral side effects. Furthermore, by identifying specific miRNA signatures related to cancer progression in body fluids, it may be possible to better determine the efficacy of treatments and select patients for clinical trials.
Conclusions
MiRNA detection in body fluids is a ‘booming’ field in the world of biomarkers and, because more than 1,500 transcribed miRNAs have been identified in the human genome, miRNA detection could be a potential gold mine for identifying biomarkers, as well as for predicting response to cancer (and to other disease) therapy. However, important issues need to be addressed in order to establish circulating miRNAs as biomarkers for cancer. First, larger prospective clinical trials are needed to validate these results, since the majority of the published studies have small sample sizes and lack long-term outcome data. Second, as miRNAs upregulated or downregulated in body fluids are shared by several types of cancer, especially ones with common origins, further studies are necessary to establish a well-characterized panel of miRNAs specific to each type of tumor, early or advanced cancer stage, response to treatment, patient outcome, and recurrence.123,124 A combination of known biomarkers, such as cancer-related antigens (for example prostate-specific antigen), mutated genes (for example BRCA1, BRCA2, RB1, TP53, and PTEN), and chromosomal translocations, together with miRNAs, may also increase the specificity and sensitivity of cancer detection. Third, more studies are necessary to establish a standardized and robust method with universal parameters for tumor-specific miRNA detection in body fluids. Finally, such approaches can be used not only for cancer but for any type of human condition and disease, including highly lethal diseases such as septic shock.125,126 From the emerging studies, it is clear that it is only a matter of time until miRNA markers with widespread use and marketability will be identified and confirmed in large cohorts of patients not only with cancer, but with many other disorders.
Key points.
-
▪
A single microRNA (miRNA) can target and regulate hundreds or thousands of mRNAs; aberrant miRNA expression is involved in the initiation of many diseases, including cancer
-
▪
MiRNAs are potentially useful as biomarkers in cancer diagnosis, prognosis and response to treatment owing to the unique expression profile of each tumor and limited complex transcriptional and translational modifications
-
▪
The discovery of miRNAs in body fluids opens up the possibility of using them as non-invasive biomarkers in cancer detection and as predictors of therapy response in clinical trials
-
▪
Standardized methods with well-established parameters for miRNA detection are necessary to indicate cancer stage, response to treatment, outcome and cancer recurrence
Acknowledgments
G. A. Calin is supported as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar and by the CLL Global Research Foundation. Work in G. A. Calin’s laboratory is supported in part by the National Institutes of Health, a Department of Defense Breast Cancer Idea Award, Developmental Research Awards in MD Anderson’s Breast Cancer, Ovarian Cancer, Brain Cancer, Multiple Myeloma and Leukemia SPOREs, a CTT/3I-TD grant, a 2009 Seena Magowitz–Pancreatic Cancer Action Network AACR Pilot Grant, and MD Anderson’s Cancer Center Support Grant CA016672 and the Arnold Foundation. This work was also supported, in part, by U54 CA151668. We would like to thank Maude Veech (MD Anderson Cancer Center) for help with the editing of this manuscript. We apologize to all colleagues whose work was not cited because of space limitations.
C. P. Vega, University of California, Irvine, CA, is the author of and is solely responsible for the content of the learning objectives, questions and answers of the Medscape, LLC-accredited continuing medical education activity associated with this article.
Footnotes
Competing interests
The authors, the journal Chief Editor L. Hutchinson and the CME questions author C. P. Vega declare no competing interests.
Author contributions
M. A. Cortez and G. A. Calin devised, wrote and edited the article, J. Ferdin contributed to writing and C. Bueso-Ramos, G. Lopez-Berestein, and A. K. Sood contributed substantially to the content of the article through in-depth discussions and editing the manuscript.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of premicroRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 15.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol. Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 16.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let 7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA binding sites in the 5' UTR as in the 3' UTR. Proc. Natl Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee I, et al. New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merritt WM, et al. Dicer, Drosha, and outcome in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melo SA, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Melo SA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 24.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNAbinding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, et al. Multiple microRNAs modulate p21 Cip1/Waf1 expression by directly targeting its 3' untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 27.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Mathonnet G, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 31.Calin GA, et al. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 33.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 34.Ciafre SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 35.Murakami Y, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 36.He H, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 38.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenfeld N, et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 40.Bloomston M, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 41.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asangani IA, et al. MicroRNA-21 (miR-21) posttranscriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 43.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 44.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bommer GT, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 47.Visone R, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 48.Nam EJ, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin. Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 49.Le MT, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 51.Fabbri M, Ivan M, Cimmino A, Negrini M, Calin GA. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin. Biol. Ther. 2007;7:1009–1019. doi: 10.1517/14712598.7.7.1009. [DOI] [PubMed] [Google Scholar]
- 52.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ota A, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 54.Garzon R, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fazi F, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 56.Saunders MA, Liang H, Li WH. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl Acad. Sci. USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chin LJ, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang H, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 59.Ye Y, et al. Genetic variations in microRNArelated genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. (Phila.) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman AE, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–5977. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int. J. Cancer. 2010;127:589–597. doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- 62.Jazdzewski K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc. Natl Acad. Sci. USA. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 64.Schetter AJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chung GE, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol. Rep. 2010;23:113–119. [PubMed] [Google Scholar]
- 66.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties . Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawrie CH, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 69.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lodes MJ, et al. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 72.Hu Z, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 73.Resnick KE, et al. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 74.Ng EK, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 75.Huang Z, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer. 2010;127:118–126. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 76.Tsujiura M, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heneghan HM, et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 78.Wang J, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. (Phila.) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho AS, et al. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl. Oncol. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamamoto Y, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009;14:529–538. doi: 10.3109/13547500903150771. [DOI] [PubMed] [Google Scholar]
- 81.Miyachi M, et al. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem. Biophys. Res. Commun. 2010;400:89–93. doi: 10.1016/j.bbrc.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 82.Wong TS, et al. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 83.Boeri M, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl Acad. Sci. USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanaka M, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS ONE. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moussay E, et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2011;108:6573–6578. doi: 10.1073/pnas.1019557108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park NJ, et al. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu CJ, et al. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16:360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 88.Hanke M, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 89.Weber JA, et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res. Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Patnaik SK, Mallick R, Yendamuri S. Detection of microRNAs in dried serum blots. Anal. Biochem. 2010;407:147–149. doi: 10.1016/j.ab.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 93.El-Hefnawy T, et al. Characterization of amplifiable, circulating RNA in plasma and its potential as a tool for cancer diagnostics. Clin. Chem. 2004;50:564–573. doi: 10.1373/clinchem.2003.028506. [DOI] [PubMed] [Google Scholar]
- 94.Smalheiser NR. Exosomal transfer of proteins and RNAs at synapses in the nervous system. Biol. Direct. 2007;2:35–49. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 96.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michael A, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor DD, Homesley HD, Doellgast GJ. Binding of specific peroxidase-labeled antibody to placental-type phosphatase on tumor-derived membrane fragments. Cancer Res. 1980;40:4064–4069. [PubMed] [Google Scholar]
- 99.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 100.Rabinowits G, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 101.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohshima K, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin. Biol. Ther. 2009;9:703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 104.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pigati L. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vickers KC, et al. MicroRNAs are transported in plasma and delivered to recipient cells by highdensity lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang K, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chim SS, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 110.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kroh EM, et al. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fu HJ, et al. A novel method to monitor the expression of microRNAs. Mol. Biotechnol. 2006;32:197–204. doi: 10.1385/MB:32:3:197. [DOI] [PubMed] [Google Scholar]
- 113.Rossi S, et al. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm. Genome. 2008;19:526–540. doi: 10.1007/s00335-008-9119-8. [DOI] [PubMed] [Google Scholar]
- 114.Ferdin J, Kunej T, Calin GA. Non-coding RNAs: identification of cancer-associated microRNAs by gene profiling. Technol. Cancer Res. Treat. 2010;9:123–138. doi: 10.1177/153303461000900202. [DOI] [PubMed] [Google Scholar]
- 115.Beutler E, Gelbart T, Kuhl W. Interference of heparin with the polymerase chain reaction. Biotechniques. 1990;9:166. [PubMed] [Google Scholar]
- 116.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur. J. Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 118.Weiss GJ, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann. Oncol. 2008;19:1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 119.Nakajima G, et al. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics. 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- 120.Salter KH, et al. An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PLoS ONE. 2008;3:e1908. doi: 10.1371/journal.pone.0001908. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121.Zhang HL, et al. Serum miRNA-21: Elevated levels in patients with metastatic hormonerefractory prostate cancer and potential predictive factor for the efficacy of docetaxelbased chemotherapy. Prostate. 2011;71:326–331. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 122.Li A, et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70:5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheng H, et al. Circulating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vasilescu C, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS ONE. 2009;4:e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang JF, et al. Serum miR-1467a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 126.Cech TR. The efficiency and versatility of catalytic RNA: implications for an RNA world. Gene. 1993;135:33–36. doi: 10.1016/0378-1119(93)90046-6. [DOI] [PubMed] [Google Scholar]