Abstract
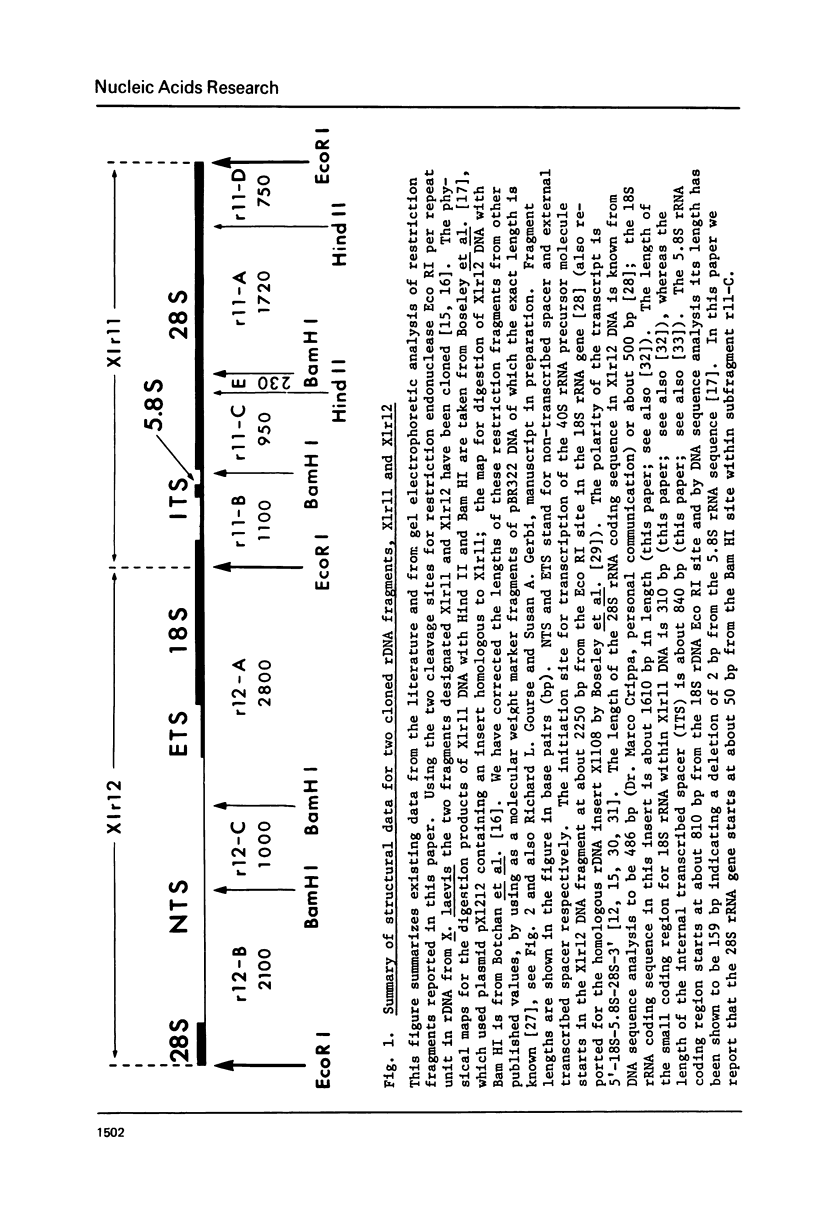
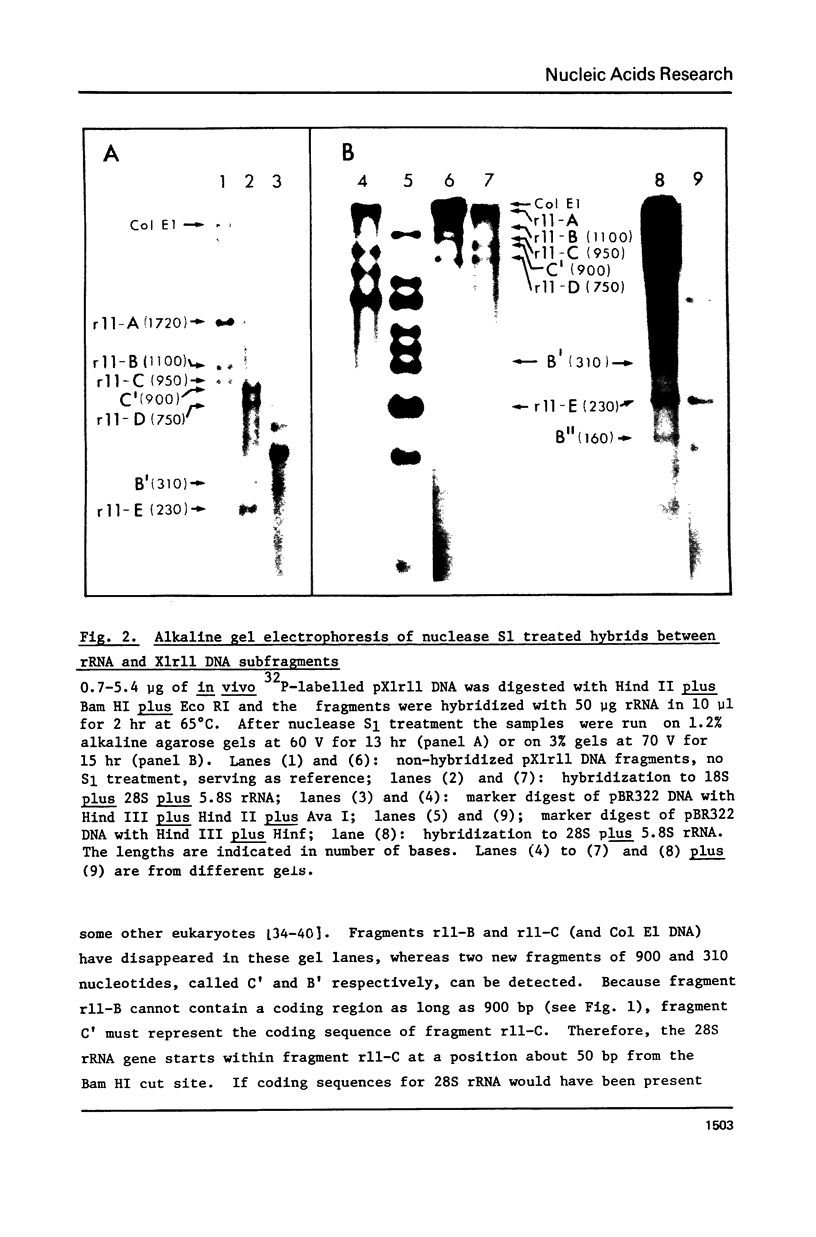
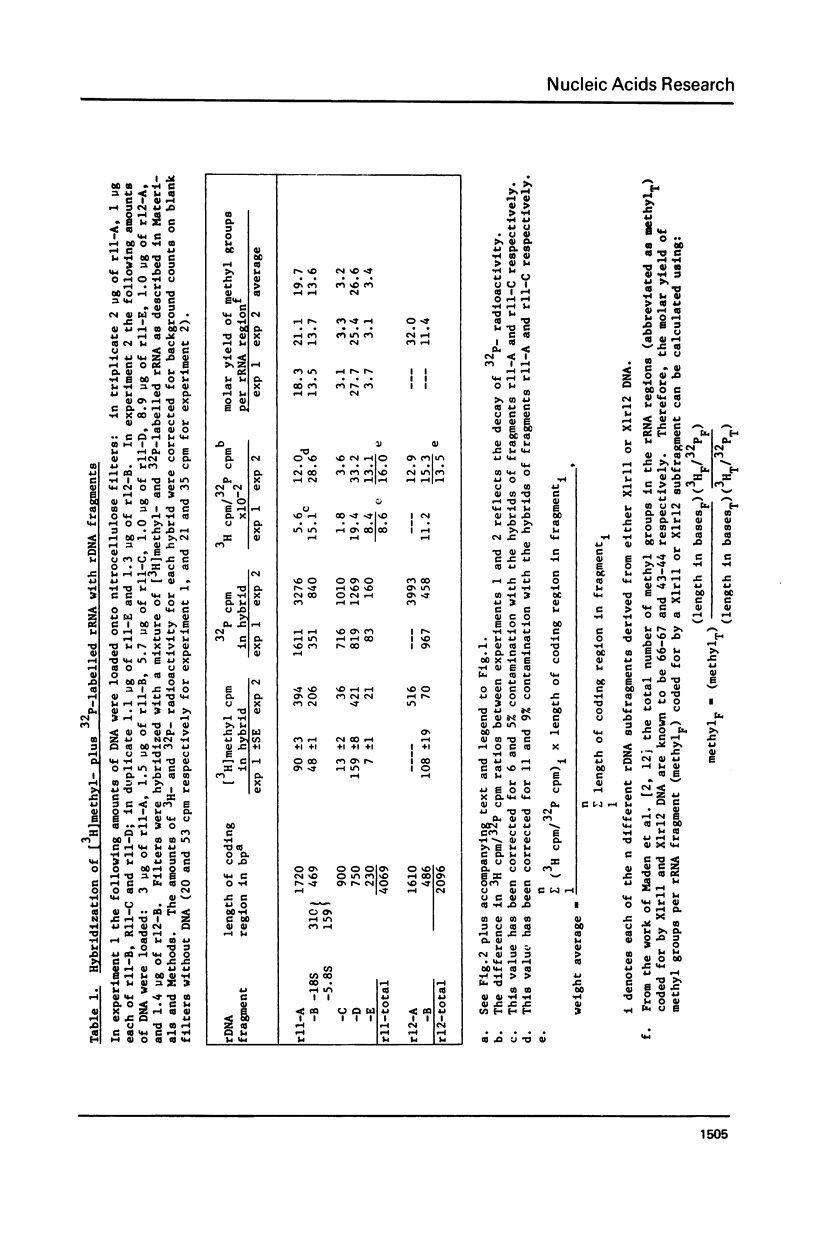
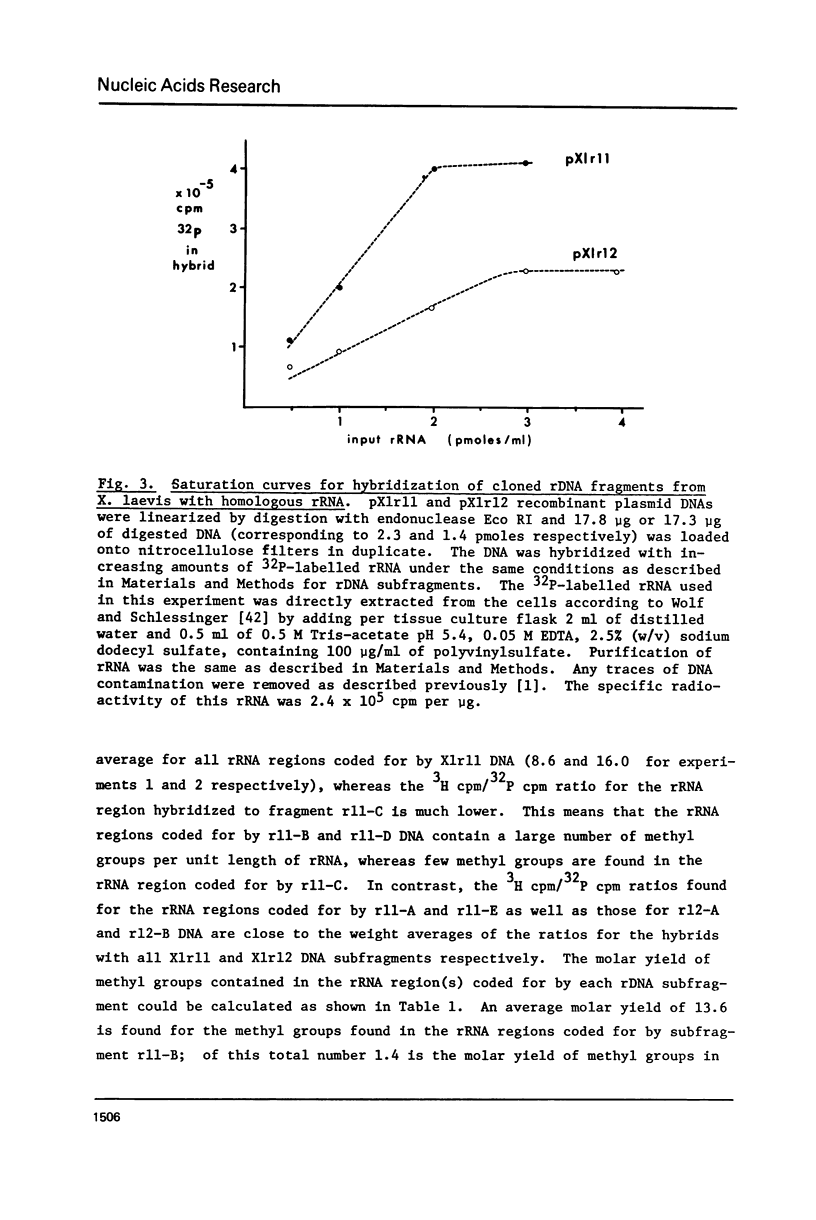
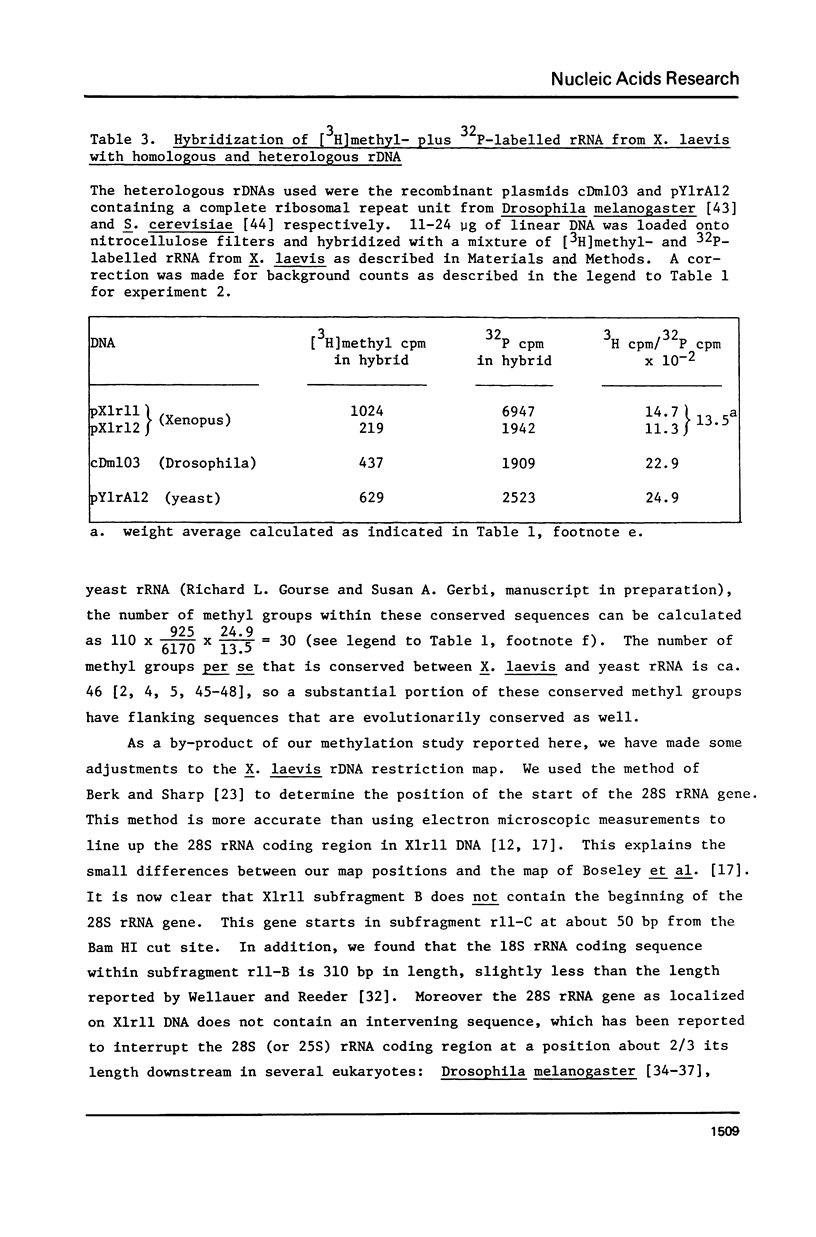
The distribution of methyl groups in rRNA from Xenopus laevis was analyzed by hybridization of rRNA to subfragments of either of two cloned rDNA fragments, X1r11 and X1r12, which together constitute a complete rDNA repeat unit. Using a mixture of 3H-methyl plus 32P-labelled rRNA as probe, the molar yield of methyl groups per rRNA region in hybrid could be calculated. For this calculation the length of the rRNA coding region in each DNA subfragment is needed, which was determined for X1r11 subfragments by the nuclease S1 mapping method of Berk and Sharp. The results show that both in 18S and 28S rRNA the methyl groups are nonrandomly distributed. For 18S rRNA, clustering was found within a 3' terminal fragment of 310 nucleotides. For 28S rRNA, clustering of methyl groups was found within a region of 750 nucleotides in length, which ends 500 nucleotides from the 3' end. In contrast, the 28S rRNA 5' terminal region of 900 nucleotides is clearly undermethylated. The general position of methyl groups in 28S rRNA correlates with the location of evolutionarily conserved sequences in this molecule, as recently determined in our laboratory.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberty H., Raba M., Gross H. J. Isolation from rat liver and sequence of a RNA fragment containing 32 nucleotides from position 5 to 36 from the 3' end of ribosomal 18S RNA. Nucleic Acids Res. 1978 Feb;5(2):425–434. doi: 10.1093/nar/5.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averner M. J., Pace N. R. The nucleotide sequence of marsupial 5 S ribosomal ribonucleic acid. J Biol Chem. 1972 Jul 25;247(14):4491–4493. [PubMed] [Google Scholar]
- Barnett T., Rae P. M. A 9.6 kb intervening sequence in D. virilis rDNA, and sequence homology in rDNA interruptions of diverse species of Drosophila and other diptera. Cell. 1979 Apr;16(4):763–775. doi: 10.1016/0092-8674(79)90092-8. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boseley P., Moss T., Mächler M., Portmann R., Birnstiel M. Sequence organization of the spacer DNA in a ribosomal gene unit of Xenopus laevis. Cell. 1979 May;17(1):19–31. doi: 10.1016/0092-8674(79)90291-5. [DOI] [PubMed] [Google Scholar]
- Botchan P., Reeder R. H., Dawid I. B. Restriction analysis of the nontranscribed spacers of Xenopus laevis ribosomal DNA. Cell. 1977 Jul;11(3):599–607. doi: 10.1016/0092-8674(77)90077-0. [DOI] [PubMed] [Google Scholar]
- Brand R. C., Klootwijk J., Planta R. J., Maden B. E. Biosynthesis of a hypermodified nucleotide in Saccharomyces carlsbergensis 17S and HeLa-cell 18S ribosomal ribonucleic acid. Biochem J. 1978 Jan 1;169(1):71–77. doi: 10.1042/bj1690071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand R. C., Klootwijk J., Van Steenbergen T. J., De Kok A. J., Planta R. J. Secondary methylation of yeast ribosomal precursor RNA. Eur J Biochem. 1977 May 2;75(1):311–318. doi: 10.1111/j.1432-1033.1977.tb11531.x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Widada J. S., Krol A., Ebel J. P. Studies on the primary structure of the ribosomal 23S RNA of Escherichia coli: II. A characterisation and an alignment of 24 sections spanning the entire molecule and its application to the localisation of specific fragments. Nucleic Acids Res. 1977 Dec;4(12):4323–4345. doi: 10.1093/nar/4.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The sequence of Escherichia coli ribosomal 16 S RNA determined by new rapid gel methods. FEBS Lett. 1978 Oct 1;94(1):152–156. doi: 10.1016/0014-5793(78)80926-0. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K. A reinvestigation of 5' leads to 3' polarity in 40S ribosomal RNA precursor of Xenopus laevis. Cell. 1976 Jul;8(3):443–448. doi: 10.1016/0092-8674(76)90157-4. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Sequence of the 3'-terminal 21 nucleotides of yeast 17S ribosomal RNA. Nucleic Acids Res. 1977 Oct;4(10):3655–3663. doi: 10.1093/nar/4.10.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan J., Landy A. Structural analysis of the tRNA1Tyr gene of Escherichia coli. A 178 base pair sequence that is repeated 3.14 times. J Biol Chem. 1978 May 25;253(10):3607–3622. [PubMed] [Google Scholar]
- Forsheit A. B., Davidson N., Brown D. D. An electron microscope heteroduplex study of the ribosomal DNAs of Xenopus laevis and Xenopus mulleri. J Mol Biol. 1974 Dec 5;90(2):301–314. doi: 10.1016/0022-2836(74)90375-1. [DOI] [PubMed] [Google Scholar]
- Gerbi S. A. Fine structure of ribosomal RNA. I. Conservation of homologous regions within ribosomal RNA of eukaryotes. J Mol Biol. 1976 Sep 25;106(3):791–816. doi: 10.1016/0022-2836(76)90265-5. [DOI] [PubMed] [Google Scholar]
- Glover D. M. Cloned segment of Drosophila melanogaster rDNA containing new types of sequence insertion. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4932–4936. doi: 10.1073/pnas.74.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D. M., Hogness D. S. A novel arrangement of the 18S and 28S sequences in a repeating unit of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):167–176. doi: 10.1016/0092-8674(77)90212-4. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Santer M., Steitz J. A., Mans R. J. Conservation of the primary structure at the 3' end of 18S rRNA from eucaryotic cells. Cell. 1978 Mar;13(3):551–563. doi: 10.1016/0092-8674(78)90328-8. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Salim M., Maden B. E. Extensive homologies between the methylated nucleotide sequences in several vertebrate ribosomal ribonucleic acids. Biochem J. 1978 Mar 1;169(3):531–542. doi: 10.1042/bj1690531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., Planta R. J. Analysis of the methylation sites in yeast ribosomal RNA. Eur J Biochem. 1973 Nov 15;39(2):325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- Klootwijk J., van den Bos R. C., Planta R. J. Secondary methylation of yeast ribosomal RNA. FEBS Lett. 1972 Oct 15;27(1):102–106. doi: 10.1016/0014-5793(72)80419-8. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The determination of the molecular weight of ribonucleic acid by polyacrylamide-gel electrophresis. The effects of changes in conformation. Biochem J. 1969 Jun;113(1):131–138. doi: 10.1042/bj1130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Forbes J., de Jonge P., Klootwijk J. Presence of a hypermodified nucleotide in HeLa cell 18 S and Saccharomyces carlsbergensis 17 S ribosomal RNAs. FEBS Lett. 1975 Nov 1;59(1):60–63. doi: 10.1016/0014-5793(75)80341-3. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Reeder R. H. Partial mapping of methylated sequences in Xenopus laevis ribosomal RNA by preparative hybridization to cloned fragments of ribosomal DNA. Nucleic Acids Res. 1979 Mar;6(3):817–830. doi: 10.1093/nar/6.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Salim M. The methylated nucleotide sequences in HELA cell ribosomal RNA and its precursors. J Mol Biol. 1974 Sep 5;88(1):133–152. doi: 10.1016/0022-2836(74)90299-x. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Manning J., Davidson N. Sequence arrangement of the rDNA of Drosophila melanogaster. Cell. 1977 Feb;10(2):213–214. doi: 10.1016/0092-8674(77)90215-x. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Hereford L. M., Skryabin K. G. Characterization of two types of yeast ribosomal DNA genes. J Bacteriol. 1978 Apr;134(1):295–305. doi: 10.1128/jb.134.1.295-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R. H., Higashinakagawa T., Miller O., Jr The 5' leads to 3' polarity of the Xenopus Ribosomal RNA precursor molecule. Cell. 1976 Jul;8(3):449–454. doi: 10.1016/0092-8674(76)90158-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Sollner-Webb B., Wahn H. L. Sites of transcription initiation in vivo on Xenopus laevis ribosomal DNA. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5402–5406. doi: 10.1073/pnas.74.12.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz R., Gerbi S. A., Glätzer K. H. Ribosomal DNA of fly Sciara coprophila has a very small and homogeneous repeat unit. Mol Gen Genet. 1979 May 23;173(1):1–13. doi: 10.1007/BF00267685. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. The structural organization of ribosomal DNA in Drosophila melanogaster. Cell. 1977 Feb;10(2):193–212. doi: 10.1016/0092-8674(77)90214-8. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Reeder R. H. A comparison of the structural organization of amplified ribosomal DNA from Xenopus mulleri and Xenopus laevis. J Mol Biol. 1975 May 15;94(2):151–161. doi: 10.1016/0022-2836(75)90074-1. [DOI] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- Wild M. A., Gall J. G. An intervening sequence in the gene coding for 25S ribosomal RNA of Tetrahymena pigmentosa. Cell. 1979 Mar;16(3):565–573. doi: 10.1016/0092-8674(79)90030-8. [DOI] [PubMed] [Google Scholar]
- Wolf S. F., Schlessinger D. Nuclear metabolism of ribosomal RNA in growing, methionine-limited, and ethionine-treated HeLa cells. Biochemistry. 1977 Jun 14;16(12):2783–2791. doi: 10.1021/bi00631a031. [DOI] [PubMed] [Google Scholar]



