Abstract
The genetic basis for the underlying individual susceptibility to chlorine-induced acute lung injury is unknown. To uncover the genetic basis and pathophysiological processes that could provide additional homeostatic capacities during lung injury, 40 inbred murine strains were exposed to chlorine, and haplotype association mapping was performed. The identified single-nucleotide polymorphism (SNP) associations were evaluated through transcriptomic and metabolomic profiling. Using ≥ 10% allelic frequency and ≥ 10% phenotype explained as threshold criteria, promoter SNPs that could eliminate putative transcriptional factor recognition sites in candidate genes were assessed by determining transcript levels through microarray and reverse real-time PCR during chlorine exposure. The mean survival time varied by approximately 5-fold among strains, and SNP associations were identified for 13 candidate genes on chromosomes 1, 4, 5, 9, and 15. Microarrays revealed several differentially enriched pathways, including protein transport (decreased more in the sensitive C57BLKS/J lung) and protein catabolic process (increased more in the resistant C57BL/10J lung). Lung metabolomic profiling revealed 95 of the 280 metabolites measured were altered by chlorine exposure, and included alanine, which decreased more in the C57BLKS/J than in the C57BL/10J strain, and glutamine, which increased more in the C57BL/10J than in the C57BLKS/J strain. Genetic associations from haplotype mapping were strengthened by an integrated assessment using transcriptomic and metabolomic profiling. The leading candidate genes associated with increased susceptibility to acute lung injury in mice included Klf4, Sema7a, Tns1, Aacs, and a gene that encodes an amino acid carrier, Slc38a4.
Keywords: ARDS, countermeasures, glutamine, genetics, metabolomics
Clinical Relevance
A major challenge to critical care involves the reliable prediction of survival in patients with acute lung injury. Because acute lung injury is a sporadic disease produced by heterogeneous precipitating factors, previous genetic analyses were mainly limited to case-control studies that evaluated candidate gene associations. This study functionally assesses single-nucleotide polymorphism associations linked with survival during acute lung injury in mice. Genetic associations from haplotype mapping were strengthened by an integrated assessment using transcriptomic and metabolomic profiling. The leading candidate genes associated with increased susceptibility to acute lung injury in mice included Klf4, Sema7a, Tns1, Aacs, and a gene that encodes an amino acid carrier, Slc38a4.
Accidental (e.g., railroad derailments) (1–3) or intentional (e.g., terrorism in Iraq) (4–6) chlorine exposures have led to acute lung injury. Over 10 million tons/year of chlorine are manufactured in the United States, and accidental releases have occurred in many industries (7, 8). Moreover, widespread use requires the transport of approximately 20,000 tank cars/year (340,000 L/car) (9). Rail accidents are rare but can be catastrophic, because a ruptured car can generate a lethal plume for several hours (10). Accidental chlorine releases are approximately five times more likely to produce casualties and evacuations compared with other chemical accidents (11).
A major consequence of chlorine exposure is acute lung injury (9–17). Acute lung injury, which comprises a heterogeneous syndrome caused by direct (chlorine) and indirect (sepsis) insults, involves decreased epithelial/endothelial integrity, fluid clearance, and surfactant function (18–21). Outcomes vary greatly, and survival is difficult to predict (22–26), which has stimulated investigation into individual susceptibility (27–48). However, genetic analyses are mainly limited to case-control studies because acute lung injury occurs sporadically. Nonetheless, investigations indicate that this syndrome is complex and governed by multiple genetic factors.
In a previous study of 16 murine strains, we reported that inbred mice varied approximately 3-fold in mean survival time, supporting the likelihood of a genetic basis of susceptibility (49). Haplotype mapping, using a large, genetically diverse panel of inbred murine strains (50–52), has emerged as a valuable tool to identify the genes responsible for complex traits (53–59). Recently, we used this method to identify the genetic determinants of acrolein-induced lung injury (59). In this study, we integrate haplotype mapping with transcriptomic and metabolomic profiling to identify candidate genes associated with delayed pulmonary edema resulting from chlorine-induced lung injury.
Materials and Methods
This study was performed with the approval of the Institutional Animal Care and Use Committee at the University of Pittsburgh, and mice (6–8-week-old females) were housed under specific pathogen-free conditions. At high concentrations, chlorine can produce rapid, often lethal, lung injury, whereas low concentrations may cause delayed pulmonary edema. To model the delayed form of lung injury, we previously exposed 16 inbred murine strains to 45 parts per million (ppm) chlorine for 24 hours, and monitored survival hourly (49). In this study, these data were combined with data from an additional 24 inbred strains (n = 334) for haplotype association mapping. After their 24-hour exposure, mice were returned to microisolator cages (filtered room air), and their survival was monitored hourly. To examine chlorine-induced changes in bronchoalveolar lavage, as well as lung histology and transcripts/metabolites, groups (n = 5–8 mice/group) of sensitive C57BLKS/J or resistant C57BL/10J mice were exposed to filtered air (0 hours) or chlorine (6 or 12 hours). Microarrays (n = 5 mice/strain/time) and quantitative RT-PCR (n = 8 mice/strain/time) were used to contrast transcript levels of candidate genes. Metabolomic profiling was performed as described previously (60–63), using lung tissue (n = 5 mice/strain/time) that was homogenized in deionized water containing recovery standards, extracted (80:20 methanol/water), and analyzed by positive or negative ultrahigh performance liquid chromatography–mass spectrometry/mass spectrometry (LC:Surveyor; ThermoFisher, Pittsburgh, PA) (61), or by gas chromatography–mass spectrometry (Thermo-Finnegan Mat-95XP; ThermoFisher) (61). In contrast to the accurate-mass and elution-time tags used in shotgun proteomics, our library-based approach combines accurate retention times and tandem mass spectrometric fragmentation patterns to unambiguously identify >2,400 biochemicals (63). LRpath (64) and CLEAN (65) were used to assess pathway enrichment in transcriptomic and metabolomic profiling. To contrast the strains, a difference in the mean response was considered significant at a threshold of −0.58 < X < 0.58 log 2 change (i.e., ±1.5-fold change; P < 0.05). Additional details are provided in the online supplement.
Results
Haplotype Mapping of Survival Times in 40 Murine Strains
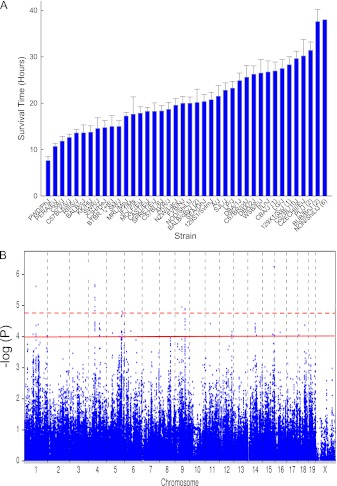
Mice developed varying signs of upper respiratory tract irritation and respiratory distress during exposure (45 ppm × 24 hours), or after return to filtered room air. During gross pathologic observations at death, the lung surface appeared red from hemorrhaging and coagulation consistent with severe lung injury (66). The mean survival time was distributed continuously among murine strains (supportive of a complex trait), with the polar strains varying by approximately 5-fold from 7.6 ± 0.8 (mean ± SEM) hours (PWD/PhJ) to 38.1 ± 0.5 hours (NON/ShiLtJ) (Figure 1A). A haplotype association map was obtained (Figure 1B), and significant single-nucleotide polymorphisms (SNPs) (n = 11 SNPs; −log(P) > 4.8) were identified on chromosomes 1, 4, 5, 9, and 15, with suggestive SNP associations (n = 56 SNPs; 4.8 ≥ −log(P) > 4.0) on chromosomes 1, 5, 9, 12, 14, 16, and 18 (please refer to Table E1 in the online supplement). Because haplotype association mapping identifies SNP associations in linkage with functional SNPs (67), we evaluated the nonsynonymous SNPs and promoter SNPs in genes ± 0.5 megabase pairs of the identified SNPs in 28 candidate genes.
Figure 1.
Haplotype association mapping of murine strains varying in sensitivity to chlorine-induced acute lung injury. (A) Acute lung injury survival time of 40 murine strains. Female mice were exposed to 45 parts per million (ppm) of chlorine for up to 24 hours, and survival times were recorded hourly. Values represent means ± SE (n = 5–22 mice/strain, 6–8 weeks old). Numbers in parenthesis represent the number of mice/strain imputed at 48 hours. The NON/ShiLtJ strain was imputed at 42 hours. (B) Haplotype association map for chlorine-induced acute lung injury in mice. The scatter (Manhattan) plot displays the corresponding −log(P) association probability for a single-nucleotide polymorphism (SNP) at the indicated chromosomal location. Dashed line, threshold of significant SNP associations of −log(P) > 4.8. Solid line, threshold of suggestive SNP associations of 4.8 ≥ −log(P) > 4.0.
Nonsynonymous SNPs
To prioritize SNPs associated with survival, candidate genes were evaluated based on whether the nucleotide sequence change could lead to nonsynonymous SNPs (i.e., an amino acid substitution, insertion, or deletion in the encoded protein). We identified 51 nonsynonymous SNPs in 21 genes (Table E2). Of these, 17 SNPs in 11 genes had a greater than 10% allelic frequency and could explain more than 10% of the phenotypic difference between polar strains (Table 1).
TABLE 1.
GENES WITH NONSYNONYMOUS SINGLE-NUCLEOTIDE POLYMORPHISMS ASSOCIATED WITH CHLORINE SURVIVAL TIME
| Name of Gene | Chromosome | Position (Base Pairs) | Association dbSNP | −log(P) | Nonsynonymous dbSNP | Exon | Amino Acid Substitution | Phenotype (%) | Allele Frequency |
| Tns1 | 1 | 72920639 | rs3021883 | 4.1 | rs40124058 | 19 | L944V | 20.1 | 30.5 |
| rs36340401 | 35 | N1882S | 20.3 | 30.5 | |||||
| Slco4c1 | 1 | 98701146 | rs13476000 | 5.6 | rs37471241 | 13 | T707A | 16.3 | 37.6 |
| Ikbkap | 4 | 56426555 | rs32078659 | 4.8 | rs27869828 | 11 | I345V | 11.3 | 24.5 |
| rs27869847 | 18 | G662V | 11.3 | 24.5 | |||||
| rs27835086 | 36 | A1309V | 17.7 | 23.4 | |||||
| Ncor2 | 5 | 125502713 | rs33262018 | 4.2 | rs36571333 | 44 | T2229A | 14.4 | 24.2 |
| Aacs | 5 | 125996775 | rs29541719 | 4.6 | rs38516850 | 1 | Q5R | 20.0 | 28.1 |
| rs36769124 | 9 | T321I | 20.0 | 28.1 | |||||
| Fry | 5 | 151155327 | rs3684936 | 4.1 | rs50343365 | 22 | T2568A | 13.5 | 19.1 |
| Cyp11a1 | 9 | 58075842 | rs3674363 | 5.0 | rs47078233 | 9 | N522D | 13.7 | 14.1 |
| Ttk | 9 | 84014785 | rs30022241 | 4.9 | rs33070516 | 9 | *302L | 11.6 | 47.8 |
| rs33070518 | 9 | T306P | 11.6 | 47.8 | |||||
| Tnfrsf19 | 14 | 61611931 | rs30388803 | 4.4 | rs37583013 | 9 | T342A | 14.4 | 40.0 |
| rs37057546 | 9 | N286D | 14.4 | 40.0 | |||||
| Slc38a4 | 15 | 96687641 | rs32255071 | 6.3 | rs46306226 | 16 | N544S | 12.6 | 17.9 |
| Slc35a5 | 16 | 45150868 | rs4180445 | 4.1 | rs32739736 | 6 | H403R | 12.0 | 48.1 |
Definition of abbreviations: Tns1, tensin 1; Slco4c1, solute carrier organic anion transporter family, member 4C1; IkbKap, inhibitor of kappa light polypeptide enhancer in B cells; Ncor2, nuclear receptor co-repressor 2; Aacs, acetoacetyl coenzyme A synthetase; Fry, furry homolog; Cyp11a1, cytochrome P450 family 11, subfamily A, polypeptide 1; Ttk, Ttk protein kinase; Tnfrsf19, tumor necrosis factor receptor superfamily, member 19; Slc38a4, solute carrier family 38, member 4; Slc35a5, solute carrier family 35, member 5; dbSNP, Single Nucleotide Polymorphism Database at NCBI; rs number, dbSNP reference sequence.
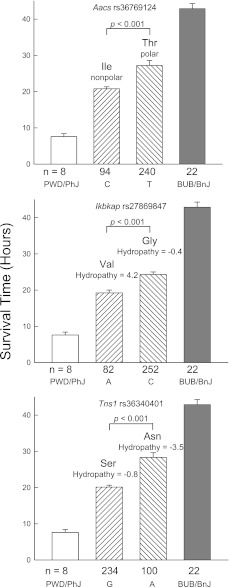
Three genes exhibited SNPs that could lead to amino acid substitutions in functional domains (Figure 2). Genes with predicted substitutions that could alter the protein hydropathy index or side chain polarity included Aacs Thr321Ile (domain, acyl-protein synthetase), Ikbkap Gly662Val (domain, the IKI3 family), and Tns1 Asn1882Ser (domain, pleckstrin homology–like).
Figure 2.
Assessment of the phenotypic difference in survival times between polar sensitive PWD/PhJ and resistant BUB/BnJ murine strains produced by nonsynonymous single-nucleotide polymorphism (SNP) associations in exemplary candidate genes. The mean survival time was determined for mice carrying either allele (n = number of mice with either allele, as indicated below the abscissas). The difference between these groups was then compared with the difference of the means of polar sensitive PWD/PhJ (n = 8 mice) and resistant BUB/BnJ (n = 22 mice) murine strains exposed to 45 ppm chlorine (total = 364 female mice). The SNP identification “rs” number is indicated in each histogram. The predicted amino acid is presented for either allele, with the consequences to side-chain polarity or hydropathy index. Value represent means ± standard errors, and P values indicate the significance of the difference between the allele means as determined by ANOVA, according to an all-pairwise multiple comparison procedure (the Holm-Sidak method). Aacs, acetoacetyl-coenzyme A synthetase; Ikbkap, inhibitor of κ light polypeptide enhancer in B cells, kinase complex–associated protein; Tns1, tensin 1. C = cytosine, T = thymidine, A = adenine, and G = guanine at the SNP position.
Histological and Lavage Protein Assessment
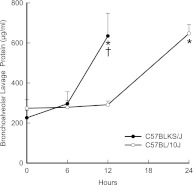
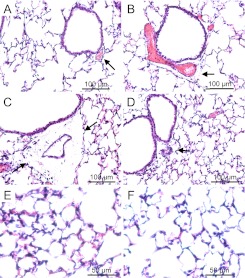
Based on the 40-strain analysis and our capability to obtain adequate numbers of mice, C57BLKS/J and C57BL10/J were selected for further analysis as the sensitive and resistant strains, respectively. At 12 hours, lavage protein increased in sensitive C57BLKS/J mice, but not in resistant C57BL/10J mice (P < 0.001) (Figure 3). At 24 hours, lavage protein increased in the C57BL/10J mice, compared with strain-matched control mice. The lung tissue from the sensitive C57BLKS/J strain demonstrated perivascular enlargement (Figure 4C) and alveolar wall thickening (Figure 4E) within 12 hours, compared with strain-matched control mice (Figure 4A). Neither perivascular enlargement nor alveolar wall thickening was evident in C57BL/10J mice after 12 hours (Figures 4D and 4F), compared with strain-matched control mice (Figure 4B).
Figure 3.
Chlorine-induced acute lung injury increased bronchoalveolar lavage protein. Female mice were exposed to 45 ppm chlorine for 0 (filtered air control), 6, 12, or 24 (C57BL/10J only) hours and anesthetized, and bronchoalveolar lavage was performed with Ca2+, Mg2+-free PBS. Bronchoalveolar lavage protein increased sooner in the sensitive C57BLKS/J murine strain than in the resistant C57BL/10J murine strain. Lavage fluid was centrifuged, and total protein in cell-free supernatants was measured according to a bicinchoninic acid assay. Values represent means ± SE (n = 6 mice/strain/time). *Significantly different (P < 0.05) from strain-matched control mice, as determined by ANOVA with an all-pairwise multiple comparison procedure (the Holm-Sidak method). †Significantly different (P < 0.05) between the sensitive C57BLKS/J and resistant C57BL/10J murine strain at indicated times, as determined by ANOVA with an all-pairwise multiple comparison procedure (the Holm-Sidak method).
Figure 4.
Histological assessment of lung tissue from (A) control C57BLKS/J mice, (B) control C57BL/10J mice, (C and E) chlorine-exposed C57BLKS/J mice, and (D and F) chlorine-exposed C57BL/10J mice. Consistent with acute lung injury, perivascular enlargement (C, arrows) and leukocyte infiltration (E) were more evident in the sensitive C57BLKS/J strain than in the resistant C57BL/10J strain (D and F). Female mice were exposed to filtered air (control) or chlorine (45 ppm, 12 hours) and anesthetized. Lung tissue was obtained and fixed in formaldehyde, and 5-μm sections were prepared with hematoxylin and eosin staining. Bars indicate magnification.
SNP Association in Promoters
In addition to nonsynonymous SNPs, the 28 candidate genes identified by haplotype mapping were evaluated for strain differences in lung transcript levels before (0 hours) or after (6 or 12 hours) chlorine exposure (n = 8 mice/strain/time) (Table E3). Baseline lung transcripts encoding acetoacetyl coenzyme A synthetase (log2 = 0.7 ± 0.1) and cytochrome P450 family 11, subfamily A, polypeptide 1 (log2 = 1.2 ± 0.2) increased, and Kruppel-like factor (gut)-4 (KLF-4) (log2 = −1.5 ± 0.3) decreased, in C57BLKS/J compared with C57BL/10J mice.
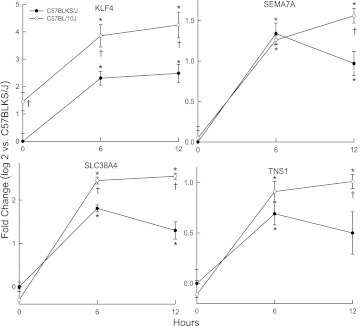
At 6 hours, KLF4 and solute carrier family 38, member 4 (SLC38A4) transcripts increased less in C57BLKS/J than in C57BL/10J mice (Figure 5). At 12 hours, KLF4, semaphorin-7A (SEMA7A), SLC38A4, and tensin 1 (TNS1) transcripts increased less in C57BLKS/J than in C57BL/10J mice. Transcripts for other candidate genes either decreased similarly in both murine strains (e.g., SLC35A5 or SLCO4C1; Figure E1 in the online supplement), or were not significantly different from control values after exposure. The interrogation of SNPs within the 5′ untranslated region (promoter) that could change putative DNA-binding sites was evident in six of the differentially expressed genes (Table 2). These SNPs (except those in Klf4) could explain approximately 14–35% of the phenotypic difference between polar strains. Using microarrays, we identified 41 increased and 10 decreased transcripts within 104 transcription factors that were related to the binding sites identified in the promoter SNPs (Table E4).
Figure 5.
Transcript levels of candidate genes that differed between the C57BL/10J and C57BLKS/J murine strains after chlorine exposure. Female mice were exposed to filtered air (control, 0 hours), or to chlorine (45 ppm) for 6 or 12 hours, lung mRNA was isolated, and transcript expression levels were determined by quantitative real-time polymerase chain reactions. KLF4, Kruppel-like factor–4 (gut); SEMA7A, sema domain, immunoglobulin domain, and glycophosphatidyl inositol membrane anchor (semaphorin)–7A; SLC38A4, solute carrier family 38, member 4; TNS1, tensin 1. Values represent means ± SE (n = 8 mice/strain/time), normalized to the sensitive C57BLKS/J control (filtered air, 0 hours). *Significantly different (P < 0.05) from strain-matched control mice, as determined by ANOVA with an all-pairwise multiple comparison procedure (the Holm-Sidak method). †Significantly different (P < 0.05) between the sensitive C57BLKS/J and resistant C57BL/10J murine strain at indicated times, as determined by ANOVA with an all-pairwise multiple comparison procedure (the Holm-Sidak method).
TABLE 2.
GENES WITH PROMOTER SINGLE-NUCLEOTIDE POLYMORPHISMS ASSOCIATED WITH CHLORINE SURVIVAL TIME
| Name of Gene | Chromosome | Position (Base Pairs) | Association dbSNP | −log(P) | Expression dbSNP | Substitution | DNA Binding Site | Phenotype (%) | Allele Frequency |
| Tns1 | 1 | 72920639 | rs3021883 | 4.1 | rs52269246 | A/G | Loss: AP2 | ND | ND |
| Gain: RAF and CTCF | |||||||||
| rs39579545 | A/G | Loss: AP2 and CACCC-binding factor | 35.1 | 17.7 | |||||
| Gain: SRF | |||||||||
| rs31543918 | T/C | Loss: LEF1/TCF | 27.8 | 30.2 | |||||
| Klf4 | 4 | 55986910 | rs27805492 | 5.0 | rs48027778 | C/T | Loss: RAF | ND | ND |
| Gain: NF-GMB | |||||||||
| rs46517881 | G/T | Loss: IPF1 | ND | ND | |||||
| Gain: AP-1, CEB/Pα | |||||||||
| Aacs | 5 | 125996775 | rs29541719 | 4.6 | rs36310016 | A/T | Loss: GAL4 and TFII-I | 18.1 | 28.1 |
| rs36610226 | A/C | Loss: GCF | 18.1 | 28.1 | |||||
| Gain: E2F + p107 | |||||||||
| Sema7a | 9 | 58075842 | rs3674363 | 5.0 | rs52190134 | C/T | gain: HES-1 and C/EBP-α | 11.8 | 16.5 |
| rs49278038 | G/A | Gain: AP-1,TF68, LCR-F1, GCR1 | 16.0 | 14.1 | |||||
| Cyp11a1 | 9 | 58075842 | rs3674363 | 5.0 | rs50082802 | A/G | Loss: IHF | 14.1 | 49.4 |
| Gain: GR and NF-E | |||||||||
| rs50603668 | G/T | Loss: AP-1 and RAR-β | 15.9 | 14.1 | |||||
| Gain: GAL4, T-Ag, and NFE2/CAC-bp | |||||||||
| rs46968424 | A/G | Gain: RAF, PEA3, E1A-F, and MAF | 13.9 | 45.8 | |||||
| Slc38a4 | 15 | 96687641 | rs32255071 | 6.3 | rs51436989 | C/T | Loss: NF-E and NF-S | 15.4 | 20.4 |
| Gain: CREB | |||||||||
| rs48402040 | A/G | Loss: ZP5 and T-Ag | 16.2 | 10.0 | |||||
| Gain: CACCC-binding factor |
Definition of abbreviations: Aacs, acetoacetyl coenzyme A synthetase; Cyp11a1, cytochrome P450, family 11, subfamily A, polypeptide 1; Klf4, Kruppel-like factor–4 (gut); ND, not done; Sema7a: sema domain, immunoglobulin domain, and glycophosphatidyl inositol membrane anchor (semaphorin)–7A; Slc38a4, solute carrier family 38, member 4; Tns1, tensin-1; dbSNP, Single Nucleotide Polymorphism Database; rs number, dbSNP reference sequence.
Transcriptomic Pathway Enrichment Analysis
The pathway enrichment of transcripts that differed between the sensitive and resistant strains before or during exposure (n = 6 female mice/strain/time) was assessed by microarray (Table E5). In general, the baseline lung transcriptome of C57BLKS/J mice was similar to that of C57BL10/J mice, with 161 increased and 106 decreased transcripts in the C57BLKS/J compared with the C57BL/10J strain. The only pathway with significant enrichment was that of natural killer–mediated cytotoxicity. Members in this pathway included nine transcripts that encoded killer cell lectin-like receptors (also known as inhibitory LY49-proteins), which decreased in C57BLKS/J mice compared with C57BL/10J mice.
After exposure, significantly increased lung transcripts in sensitive C57BLKS/J mice were enriched in pathways that included the Rous sarcoma oncogene (Src) homology–3 domain, transcription factor activity, and cell death (Figure E2A). After exposure, decreased transcripts in the sensitive C57BLKS/J mice were enriched in pathways that included protein transport, translation, and the development of vasculature (Figure E2B). After exposure, increased transcripts in resistant C57BL/10J mice were enriched in pathways that included cell adhesion, cytoskeletal organization, and the protein catabolic process (Figure E3A). Decreased transcripts in resistant C57BL/10J mice were enriched in pathways that included RNA binding, transcription, and mitochondria (Figure E3B).
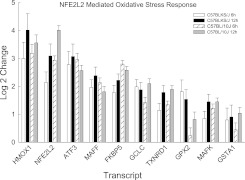
Noteworthy contrasts between strains in related pathways included the transcription factor activity pathway, which contained transcripts that increased more in sensitive C57BLKS/J lungs, and the transcription pathway, which contained transcripts that decreased more in resistant C57BL/10J lungs. Similarly, the protein transport pathway was enriched with decreased transcripts in sensitive C57BLKS/J lungs, whereas the protein catabolic process pathway was enriched with increased transcripts in resistant C57BL/10J lungs. Pathways that were altered nearly equally in both strains included the nuclear factor erythroid-derived–2–like–2 (NFE2L2, also known as NRF2)–mediated oxidative stress response (Figure 6).
Figure 6.
Transcripts in the enriched nuclear factor erythroid-derived–2–like 2 (NFE2L2, also known as Nrf2)–mediated oxidative stress response pathway in the lungs of C57BLKS/J and C57BL/10J mice after chlorine-induced acute lung injury. Microarrays increased transcripts (significantly different, at P < 0.05, from strain-matched control mice) that were enriched as determined by the logistic regression approach LRpath. Transcripts in this pathway were similar between strains, and included heme oxygenase (decycling)–1 (HMOX1); nuclear factor, erythroid-derived–2–like 2 (NFE2L2); activating transcription factor–3 (ATF3); v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) (MAFF); FK506 binding protein–5 (FKBP5); glutamate–cysteine ligase, catalytic subunit (GCLC); thioredoxin reductase–1 (TXNRD1); glutathione peroxidase–2 (GPX2); v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) (MAFK); and glutathione S-transferase, α 1 (Ya) (GSTA1). Values represent means ± SE (n = 6 female mice/strain/time), normalized to strain-matched control mice (0 hours).
Metabolomic Profiling
Lung metabolomic profiling of these strains identified 280 compounds (Table E6). In general, basal lung metabolites were similar between the sensitive and resistant strains (i.e., only seven increased and six decreased molecules differed significantly between strains). Compared with resistant C57BL/10J mice, the basal metabolite in sensitive C57BLKS/J murine lungs that was reduced comprised cytosine, and metabolites that were elevated included phenylacetylglycine, S-methylglutathione, and ophthalmate.
Exposure markedly altered the lung metabolomic profile. At 6 hours and 12 hours of chlorine exposure, 95 metabolites were altered in at least one strain, compared with strain-matched control mice. LRpath analysis identified the amino acid pathway to be significantly different between strains and with treatment (Table E7).
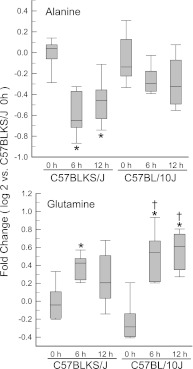
Noteworthy metabolites in the amino acid pathway that changed with exposure included alanine, which decreased more in C57BLKS/J than in C57BL/10J mice, and glutamine, which increased more in C57BL/10J than in C57BLKS/J mice (Figure 7). Subpathways enriched with exposure included (1) glycerolipid metabolism (e.g., decreased glycerophosphocholine and glycerol 3-phosphate), (2) medium chain fatty acids (e.g., increased caprylate [8:0] and laurate [12:0]), and (3) phenylalanine and tyrosine metabolism. The metabolites that changed in these subpathways did not differ between strains (except for phenol sulfate, which increased more in C57BL/10J mice). Other metabolites changed in both strains included increased 3-hydroxybutyric acid (BHBA), and decreased lactate, 1-palmitoleoylglycerophosphocholine, and 1-palmitoleoylglycerophosphoinositol. Small molecules that increased more in the sensitive C57BLKS/J than in the resistant C57BL/10J strain included two carbohydrates (sorbitol and fructose).
Figure 7.
Alanine decreased in sensitive C57BLKS/J murine lungs, but not in resistant C57BL/10J murine lungs, whereas glutamine increased more in the resistant C57BL10/J murine lungs, compared with the sensitive C57BLKS/J murine lungs. Female mice were exposed to filtered air (0 hours, control) or chlorine (45 ppm, 6 or 12 hours), and metabolome profiling was performed with lung tissue. Values are normalized to the sensitive C57BLKS/J control (filtered air, 0 hours) levels, and plots indicate the medians (lines within boxes) with 25% and 75% confidence intervals (borders of the boxes) and 95% confidence intervals (error bars). *Significantly different (P < 0.05) from strain-matched control mice, as determined by ANOVA with an all-pairwise multiple comparison procedure (the Holm-Sidak method). †Significantly different (P < 0.05) between the sensitive C57BLKS/J and resistant C57BL/10J murine strain at indicated times, as determined by ANOVA with an all-pairwise multiple comparison procedure (the Holm-Sidak method).
Discussion
In this study, chlorine produced an approximately 3-fold increase in lavage protein at 12 hours in the sensitive C57BLKS/J strain, or at 24 hours in the resistant C57BL/10J strain (Figure 3). These findings are similar to those of Zarogiannis and colleagues (17). Lung histology also indicated that the C57BLKS/J mice developed injury sooner than the C57BL/10J mice (Figure 4). The finding of perivascular enlargement should be evaluated with caution, because it can result from tissue processing. The histological samples obtained at 6 and 12 hours were selected mainly to coincide with transcriptomic and metabolomic analyses. These times may have been too early to uncover a great deal of lung injury or inflammatory infiltrate. Gross pathology revealed that lungs were marked by focal hemorrhages at the time of death. This similarity of this histological feature among strains suggests that the extent of injury was the same at the time of death, and that the phenotype being measured is survival time.
Ascorbate decreased slightly, but not to the extent of statistical significance. Both strains demonstrated nearly equivalent decreases in gulono-1,4-lactone (an ascorbate precursor) and dehydroascorbate (an ascorbate metabolite) (Table E6). Similarly, both strains exhibited a nearly equivalent enrichment of the NFE2L2-mediated oxidative stress pathway (Figure 4). Thus, although the response to oxidative stress was similar in both strains, resistance can be defined as the ability to prolong survival. The objective of this study was to uncover the genetic basis that could provide additional metabolic or other homeostatic capacities during chlorine-induced lung injury.
We identified 28 candidate genes with SNP associations. We prioritized these genes by several criteria, including the phenotypic difference associated with nonsynonymous SNPs in functional domains or with promoter SNPs matched with variable expression by transcriptomic analyses. In addition, we paired these relationships with altered metabolites identified by metabolomic profiling. This integrative approach revealed 13 candidate genes (Tables 1 and 2), and of these, Klf4, Sema7a, Tns1, Slc38a4, and Aacs were more noteworthy, and could be associated with survival in several ways.
For example, Krüppel-like factor (gut)–4 (KLF4) can protect against lung injury (68). KLF4, a transcription factor, regulates cadherin-5 expression in adherens junctions, and KLF4 knockdown augments LPS-induced lung injury in mice. KLF4 mRNA also can be induced by other stresses (69, 70). Here, lung KLF4 transcripts increased more in the resistant C57BL/10J mice than in the sensitive C57BLKS/J mice (Figure 5).
Another candidate, semaphorin-7A (SEMA7A), can be induced by transforming growth factor–β (TGFB1) and mediates TGFB1-induced alveolar apoptosis (71). SEMA7A polymorphisms are associated with abnormal bone mineral density in Korean women (72). During acute lung injury, TGFB1 can increase endothelial and epithelial permeability (73–75), and the inhibition of TGFB1 can diminish lung injury (66, 76–78). SEMA7A can mediate AKT phosphorylation (71), which is associated with increased cell survival (79) and is protective during lung injury (80). In this study, promoter Sema7a SNPs associated with 12–16% of the difference in survival phenotype (Table 2) and SEMA7A mRNA increased longer in the resistant C57BL/10J strain, compared with C57BLKS/J strain (Figure 5).
TNS1 polymorphisms have been associated with lung function and chronic obstructive lung disease (81, 82). TNS1, a scaffold protein, recruits and organizes enzymes at focal adhesions and mediates cell migration in wound healing (83). The C-terminal domain of TNS1 has a Src homology–2 domain that binds focal adhesion kinase, and a phosphotyrosine-binding domain that binds integrin-β. Osmotic stress alters the binding partners to the Src homology–2 domain (84). In this study, 12–16% of the difference in survival between polar strains associated with two Tns1 promoter SNPs (Table 2) and lungs from the resistant C57BL/10J strain demonstrated a prolonged increase in lung TNS1 mRNA after exposure to chlorine (Figure 5). In addition, we detected a nonsynonymous SNP (N1882S) in the integrin-β binding domain with an approximately 30% allelic frequency that associated with approximately 20% of the phenotypic difference (Figure 2 and Table 1).
During injury, lung epithelial cells are likely to be challenged by energetic stress (60). In general, cell survival can depend, in part, on limiting energy expenditure through many defense strategies (85, 86). Alternately, the activation of energy-yielding pathways for ATP production may be required for energetic needs incurred upon injury. Lactate and alanine decreased after chlorine exposure, and these metabolites are the precursors for pyruvate and subsequently acetyl-coenzyme A (acetyl-CoA). Thus, this response implicates an increased utilization of aerobic metabolism via the Krebs cycle. Interestingly, both strains exhibited a marked elevation in Krebs-cycle intermediates (citrate and cis-aconitate in C57BLKS/J, and fumarate in C57BL/10J).
Restricting energy-dependent solute carriers can conserve energy, but this may be counterproductive because they are critical for energy substrate uptake and fluid absorption. Thus, cellular stress may modulate the array of solute carriers. Several pathways and candidate genes identified in this study include solute carrier (SLC) proteins. In particular, SLC35A5, SLCO4C1, and SLC38A4 were associated with increased susceptibility to chlorine-induced lung injury. Little is known about SLC35A5, which is a putative nucleotide–sugar transporter, based on a shared homology with SLC35A1 (87). SLCO4C1, an organic anion transporter, can transport eicosanoids, thyroid hormone, and steroids (88). Although the SLCO4C1 transcript and protein are present in the lung (Figures E4 and E5), the role of SLOC4C1 in lung injury remains unclear. SLCO4C1 is protective in kidney disease (89), and a human SLCO4C1 SNP was associated with preeclampsia (90). In this study, nonsynonymous SNPs were identified (Table 1), but these SNPs did not occur in known functional domains. In addition, lung SLCO4C1 and SLC35A1 transcripts decreased nearly equivalently in the resistant and sensitive strains during chlorine injury (Figure E1).
In contrast, lung SLC38A4 transcripts increased more in the resistant compared with the sensitive mice (Figure 5). SLC38A4 is a sodium-coupled neutral amino acid (including alanine and glutamine) transporter. The tagSNP (rs32255071) on chromosome 15 was associated with SLC38A4 (−log(P) = 6.25). Promoter Slc38a4 SNPs were associated with 13–16% of the difference in survival (Table 2).
The transcriptomic profiling of lung transcripts that decreased more in sensitive C57BLKS/J compared with resistant C57BL/10J mice identified enrichment in the protein transport pathway (Figure E2B). In contrast, the protein catabolic process pathway contained transcripts that increased more in the resistant C57BL/10J strain than in the sensitive C57BLKS/J strain (Figure E3A). Similarly, metabolomic profiling indicated enrichment in the amino acid pathway, and individual amino acids, including glutamine, increased more in the resistant C57BL/10J than in the sensitive C57BLKS/J strain. Alanine decreased more in the sensitive C57BLKS/J than in the resistant C57BL/10J strain. Alanine can be used during energetic stress to generate pyruvate and glutamate (91). Moreover, glutamine can attenuate acute lung injury by inducing heat-shock proteins (92–94). Thus, of the three solute carrier proteins identified, SLC38A4 is worthy of additional investigation.
The ability to increase lung glutamine also may be important in the improved survival of the resistant C57BL/10J strain through additional roles in metabolism. Although glucose is generally thought to be the primary substrate for energy metabolism in most tissues, energetics in the lung are complex, as manifested by multiple substrate usage. Here, glucose was unchanged, whereas lactate decreased equally between strains. Interestingly, a fatty acid β-oxidation ketone body, BHBA, increased in both strains initially, but was maintained longer in resistant C57BL/10J mice, possibly reflecting greater fatty-acid β-oxidation in the resistant strain. Previously, Fox and colleagues (95) measured oxidation rates of glucose, glutamine, lactate, and BHBA in alveolar Type II cells from fetal rats. The CO2 formation from lactate was greater than from glutamine, which in turn was greater than from BHBA. The rate of glucose oxidation was lower than in all these substrates (∼ 5 times less than that of glutamine). In addition, glucose, but not lactate, inhibited the oxidation of glutamine. Similarly, alanine is also a substrate for energy production in alveolar Type II cells (96). Thus, glutamine, alanine, and other substrates can be oxidized for added energy in alveolar epithelial cells.
The alveolar Type II cell is a critical target during lung injury because it generates pulmonary surfactant, which maintains alveolar patency. Pulmonary surfactant consists of phospholipids (mainly dipalmitoylglycerophosphocholine), surfactant proteins, electrolytes, and other biomolecules. Surfactant-associated protein B (SFTPB) mRNA decreased in the sensitive C57BLKS/J mice more rapidly than in the C57BL/10J mice (i.e., log2 = −1.2 versus −0.3 at 6 hours, respectively). This is relevant because maintaining SFTPB is critical to survival during acute lung injury in mice (19, 27, 39).
Surfactant lipid production uses glucose-dependent fatty-acid synthesis, but fatty acids can also be generated from lactate or ketone bodies (97). In the lung, ketone metabolism also can serve as an energy source. Alternately, acetoacetate can be used in the synthesis of phospholipids, including palmitoylglycerophosphocholines, and thus have a potential role in supplying adequate surfactant lipids. Cytosolic lipid synthesis from acetoacetate can conserve energy by bypassing the pathway involving the ATP-dependent supply of acetyl units from the mitochondria to cytosol (98, 99). Nonsynonymous SNPs were identified in Aacs that could lead to Thr321Ile substitution in the acyl-protein synthetase domain (Table 1). AACS, a cytosolic acetoacetate (ketone bodies)–specific ligase, catalyzes the formation of short-chain acyl-CoA from acetoacetate, thereby providing acetyl-CoA for fatty-acid synthesis (100). Two lysophospholipids, 1-palmitoleoylglycerophosphocholine and 1-palmitoleoylglycerophosphoinositol, were decreased in the lungs of the sensitive C57BLKS/J murine strain (Table E6). Overall, the use of similar substrates for energy production and surfactant synthesis could create competition between these pathways, especially in times of stress and injury. Therefore, to better define these relationships in future mechanistic studies is imperative.
Another candidate gene is the inhibitor of κB kinase complex–associated protein (Ikbkap, also known as IKAP). Human IKBKAP polymorphisms produce a truncated protein that has been associated with familial dysautonomia, a recessive disease that affects the nervous system (101, 102). Neuronal dysfunction leads to several defects, including abnormal respiratory hypoxic responses and pneumonia (103). IKBKAP was named on the basis of its reported ability to bind to and assemble IκB kinases into an active complex (104). Later studies, however, failed to confirm a role in NF-κB activation, and instead reported IKBKAP to be involved in transcription elongation (105), Jun N-terminal kinase-mediated stress signaling (106), and cell migration (107). Here, we identified three nonsynonymous SNPs in murine Ikbkap that could lead to amino acid substitutions in exons 11, 18, and 36, associated with a phenotypic difference in survival (Table 1). The Gly662Val substitution is in the IKI3 domain likely to be involved in IKBKAP's transcriptional elongation function.
This study provides an integrative strategy that combines haplotype mapping, transcriptomics, and metabolomics to assess chlorine-induced acute lung injury in mice. However, as with any investigation, each approach has limitations that cannot be fully overcome by our combined approach. First, we acknowledge that each approach is essentially descriptive, and that our experimental design did not provide information on a mechanistic basis for acute lung injury. Rather, this study was designed to provide high-content information that screened potential candidate genes, transcriptional responses, and metabolic pathways related to strain-specific differences in lung injury that can be followed up in mechanistic studies.
Second, analyses of transcripts and metabolites detected in lung tissue involve limitations. Lung homeostasis is complex and requires the consideration of contributions from other tissues such as liver, kidney, adipose tissue, and blood. The steady-state level of transcripts or metabolites in a pathway is limited, and could reflect either the activation of an upstream process or the inhibition of a downstream process. Full interpretation of these results will require further assessments of precursor/product relationships, biochemical sites of regulation, and combinatorial rate-limiting steps in multienzymatic pathways.
Third, although chlorine-induced acute lung injury has relevance to accidental human exposures, numerous other agents can produce acute lung injury (18). The genetic and metabolomic findings here may have been limited by the use of a single agent. We recently identified candidate genes associated with acrolein-induced acute lung injury (58) and these genes differed from those identified with chlorine. Until several types of chemically induced acute lung injury have been evaluated, generalizations to other forms may not be warranted. Nonetheless, a major candidate identified previously with acrolein was Acvr1 (Activin A receptor, type 1), which implicated TGFB1 signaling. In the present study, TGFB1 signaling was also implicated with chlorine by the identification of Sema7a.
In conclusion, mean survival times varied by approximately 5-fold among strains, and haplotype mapping identified SNP associations on chromosomes 1, 4, 5, 9, and 15. Microarrays revealed several enriched pathways, including protein transport, which decreased more in sensitive C57BLKS/J lungs, and the protein catabolic process, which increased more in resistant C57BL/10J lungs. Lung metabolomic profiling revealed that 95 of the 280 metabolites measured were altered by chlorine exposures, and included alanine, which decreased more in the C57BLKS/J strain, and glutamine, which increased more in the C57BL/10J than in the C57BLKS/J strain. The results from haplotype mapping were evaluated by an integrated assessment using transcriptomic and metabolomic profiling. The identified candidate genes associated with increased susceptibility to acute lung injury in mice included Klf4, Sema7a, Tns1, Aacs, and an amino acid carrier, Slc38a4. These genes or genes in related pathways may help direct future human genetic studies to evaluate such pathways.
Supplementary Material
Footnotes
Author Contributions: G.D.L., K.B., A.B., M.M., N.K., M.Y., D.R.P., and D.L.K. were responsible for this study's conception and design. H.P.-V., V.J.C., P.L., K.B., A.B., T.M.M., K.G., A.S.J., K.A.B., R.A.D., S.U., Y.P.P.D., Q.L., Z.H., L.J.V., M.M., N.K., M.Y., D.C.A., J.E.M., D.R.P., G.D.L., and J.P.F. were responsible for the analyses and interpretation in this study. G.D.L., K.B., D.R.P., and J.P.F. were responsible for drafting the manuscript for important intellectual content.
This study was supported by National Institutes of Health grants ES015675, HL077763, and HL085655 (G.D.L.), HL091938 (Y.P.P.D.), HG003749 and LM009662 (M.M.), HL084932 and HL095397 (N.K.), AT003203, AT005522, CA113793, and CA134433 (M.Y.), HL075562 (D.R.P.), and HL086981 (D.L.K.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2012-0026OC on March 23, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Jones RN, Hughes JM, Glindmeyer H, Weill H. Lung function after acute chlorine exposure. Am Rev Respir Dis 1986;134:1190–1195 [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention Public health consequences from hazardous substances acutely released during rail transit—South Carolina, 2005: selected states, 1999–2004. MMWR Morb Mortal Wkly Rep 2005;54:64–67 [PubMed] [Google Scholar]
- 3.Van Sickle D, Wenck MA, Belflower A, Drociuk D, Ferdinands J, Holguin F, Svendsen E, Bretous L, Jankelevich S, Gibson JJ, et al. Acute health effects after exposure to chlorine gas released after a train derailment. Am J Emerg Med 2009;27:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szinicz L. History of chemical and biological warfare agents. Toxicology 2005;214:167–181 [DOI] [PubMed] [Google Scholar]
- 5.Fitzgerald GJ. Chemical warfare and medical response during World War I. Am J Public Health 2008;98:611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones R, Wills B, Kang C. Chlorine gas: an evolving hazardous material threat and unconventional weapon. West J Emerg Med. 2010;11:151–156 [PMC free article] [PubMed] [Google Scholar]
- 7.Kales SN, Polyhronopoulos GN, Castro MJ, Goldman RH, Christiani DC. Mechanisms of and facility types involved in hazardous materials incidents. Environ Health Perspect 1997;105:998–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruckart PZ, Wattigney WA, Kaye WE. Risk factors for acute chemical releases with public health consequences: hazardous substances emergency events surveillance in the US, 1996–2001. Environ Health 2004;3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans RB. Chlorine: state of the art. Lung 2005;183:151–167 [DOI] [PubMed] [Google Scholar]
- 10.Buckley RL, Hunter CH, Addis RP, Parker MJ. Modeling dispersion from toxic gas released after a train collision in Graniteville, SC. J Air Waste Manage Assoc 2007;57:268–278 [DOI] [PubMed] [Google Scholar]
- 11.Horton DK, Berkowitz Z, Kaye WE. The public health consequences from acute chlorine releases, 1993–2000. J Occup Environ Med 2002;44:906–913 [DOI] [PubMed] [Google Scholar]
- 12.Leustik M, Doran S, Bracher A, Williams S, Squadrito GL, Schoeb TR, Postlethwait E, Matalon S. Mitigation of chlorine-induced lung injury by low-molecular-weight antioxidants. Am J Physiol Lung Cell Mol Physiol 2008;295:L733–L743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Squadrito GL, Postlethwait EM, Matalon S. Elucidating mechanisms of chlorine toxicity: reaction kinetics, thermodynamics, and physiological implications. Am J Physiol Lung Cell Mol Physiol 2010;299:L289–L300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song W, Wei S, Shou Y, Lazrak A, Liu G, Londino JD, Squadrito GL, Matalon S. Inhibition of lung fluid clearance and epithelial Na+ channels by chlorine, hypochlorous acid and chloramines. J Biol Chem 2010;285:9716–9728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyle GW. Mitigation of chlorine lung injury by increasing cyclic AMP levels. Proc Am Thorac Soc 2010;7:284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGovern T, Day BJ, White CW, Powell WS, Martin JG. AEOL10150: a novel therapeutic for rescue treatment after toxic gas lung injury. Free Radic Biol Med 2011;50:602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarogiannis SG, Jurkuvenaite A, Fernandez S, Doran SF, Yadav AK, Squadrito GL, Postlethwait EM, Bowen L, Matalon S. Ascorbate and deferoxamine administration post chlorine exposure decrease mortality and lung injury in mice. Am J Respir Cell Mol Biol 2011;45:386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349 [DOI] [PubMed] [Google Scholar]
- 19.Bein K, Wesselkamper SC, Liu X, Dietsch M, Majumder N, Concel VJ, Medvedovic M, Sartor MA, Henning LN, Venditto C, et al. Surfactant-associated protein B is critical to survival in nickel-induced injury in mice. Am J Respir Cell Mol Biol 2009;41:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsushima K, King LS, Aggarwal NR, De Gorordo A, D'Alessio FR, Kubo K. Acute lung injury review. Intern Med 2009;48:621–630 [DOI] [PubMed] [Google Scholar]
- 21.Bein K, Leight H, Leikauf GD. JUN–CCAAT/enhancer binding protein (C/EBP) complexes inhibit surfactant associated protein B promoter activity. Am J Respir Cell Mol Biol 2011;45:436–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson SE, Martin GS, Davis JL, Matthay MA, Eisner MD, NIH NHLBI ARDS Network Recent trends in acute lung injury mortality: 1996–2005. Crit Care Med 2009;37:1574–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siempos II, Vardakas KZ, Kyriakopoulos CE, Ntaidou TK, Falagas ME. Predictors of mortality in adult patients with ventilator-associated pneumonia: a meta-analysis. Shock 2010;33:590–601 [DOI] [PubMed] [Google Scholar]
- 24.Ware LB, Koyama T, Billheimer DD, Wu W, Bernard GR, Thompson BT, Brower RG, Standiford TJ, Martin TR, Matthay MA, et al. Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest 2010;17:288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gajic O, Dabbagh O, Park PK, Adesanya A, Chang SY, Hou P, Anderson H, III, Hoth JJ, Mikkelsen ME, Gentile NT, et al. , Lung Injury Prevention Study Investigators (USCIITG-LIPS): early identification of patients at risk of acute lung injury: evaluation of lung injury prediction score in a multicenter cohort study. Am J Respir Crit Care Med 2011;183:462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damluji A, Colantuoni E, Mendez-Tellez PA, Sevransky JE, Fan E, Shanholtz C, Wojnar M, Pronovost PJ, Needham DM. Short-term mortality prediction for acute lung injury patients: external validation of the ARDSNet prediction model. Crit Care Med 2011;39:1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein–B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest 2004;125:203–211 [DOI] [PubMed] [Google Scholar]
- 28.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Boyce P, Christiani DC. −308GA and TNFB polymorphisms in acute respiratory distress syndrome. Eur Respir J 2005;26:382–389 [DOI] [PubMed] [Google Scholar]
- 29.Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, et al. Pre–B-cell colony–enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 2005;171:361–370 [DOI] [PubMed] [Google Scholar]
- 30.Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, et al. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol 2006;34:487–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong MN, Thompson BT, Williams PL, Zhou W, Wang MZ, Pothier L, Christiani DC. Interleukin-10 polymorphism in position −1082 and acute respiratory distress syndrome. Eur Respir J 2006;27:674–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 2007;21:2237–2246 [DOI] [PubMed] [Google Scholar]
- 33.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Christiani DC. Polymorphisms in the mannose binding lectin–2 gene and acute respiratory distress syndrome. Crit Care Med 2007;35:48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai R, Zhou W, Gong MN, Thompson BT, Su L, Yu C, Kraft P, Christiani DC. Inhibitor kappaB–alpha haplotype GTC is associated with susceptibility to acute respiratory distress syndrome in Caucasians. Crit Care Med 2007;35:893–898 [DOI] [PubMed] [Google Scholar]
- 35.Bajwa EK, Yu CL, Gong MN, Thompson BT, Christiani DC. Pre–B-cell colony–enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit Care Med 2007;35:1290–1295 [DOI] [PubMed] [Google Scholar]
- 36.Arcaroli J, Sankoff J, Liu N, Allison DB, Maloney J, Abraham E. Association between urokinase haplotypes and outcome from infection-associated acute lung injury. Intensive Care Med 2008;34:300–307 [DOI] [PubMed] [Google Scholar]
- 37.Flores C, Ma SF, Maresso K, Wade MS, Villar J, Garcia JG. IL6 gene-wide haplotype is associated with susceptibility to acute lung injury. Transl Res 2008;52:11–17 [DOI] [PubMed] [Google Scholar]
- 38.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, Garcia JG. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med 2008;36:2794–2800 [DOI] [PubMed] [Google Scholar]
- 39.Currier PF, Gong MN, Zhai R, Pothier LJ, Boyce PD, Xu L, Yu CL, Thompson BT, Christiani DC. Surfactant protein–B polymorphisms and mortality in the acute respiratory distress syndrome. Crit Care Med 2008;36:2511–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su L, Zhai R, Sheu CC, Gallagher DC, Gong MN, Tejera P, Thompson BT, Christiani DC. Genetic variants in the angiopoietin-2 gene are associated with increased risk of ARDS. Intensive Care Med 2009;35:1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arcaroli JJ, Hokanson JE, Abraham E, Geraci M, Murphy JR, Bowler RP, Dinarello CA, Silveira L, Sankoff J, Heyland D, et al. Extracellular superoxide dismutase haplotypes are associated with acute lung injury and mortality. Am J Respir Crit Care Med 2009;179:105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheu CC, Zhai R, Wang Z, Gong MN, Tejera P, Chen F, Su L, Thompson BT, Christiani DC. Heme oxygenase–1 microsatellite polymorphism and haplotypes are associated with the development of acute respiratory distress syndrome. Intensive Care Med 2009;35:1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tejera P, Wang Z, Zhai R, Su L, Sheu CC, Taylor DM, Chen F, Gong MN, Thompson BT, Christiani DC. Genetic polymorphisms of peptidase inhibitor 3 (ELAFIN) are associated with acute respiratory distress syndrome. Am J Respir Cell Mol Biol 2009;41:696–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Z, Tong C, Sun Z, Shen Y, Yao C, Jiang J, Yin J, Gao L, Song Y, Bai C. Genetic variants in the TIRAP gene are associated with increased risk of sepsis-associated acute lung injury. BMC Med Genet 2010;11:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Z, Jin X, Kang Y, Liu C, Zhou Y, Wu X, Liu J, Zhong M, Luo C, Deng L, et al. Angiotensin-converting enzyme insertion/deletion polymorphism associated with acute respiratory distress syndrome among Caucasians. J Int Med Res 2010;38:415–422 [DOI] [PubMed] [Google Scholar]
- 46.Glavan BJ, Holden TD, Goss CH, Black RA, Neff MJ, Nathens AB, Martin TR, Wurfel MM. ARDSnet Investigators: genetic variation in the FAS gene and associations with acute lung injury. Am J Respir Crit Care Med 2011;183:356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bajwa EK, Cremer PC, Gong MN, Zhai R, Su L, Thompson BT, Christiani DC. An NFKB1 promoter insertion/deletion polymorphism influences risk and outcome in acute respiratory distress syndrome among Caucasians. PLoS ONE 2011;6:e19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christie JD, Wurfel MM, Feng R, O'Keefe GE, Bradfield J, Ware LB, Christiani DC, Calfee CS, Cohen MJ, Matthay M, et al. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS ONE 2012;7:e28268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leikauf GD, Pope-Varsalona H, Concel VJ, Liu P, Bein K, Brant KA, Dopico RA, Di YP, Jang AS, Dietsch M, et al. Functional genomics of chlorine-induced acute lung injury in mice. Proc Am Thorac Soc 2010;7:294–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol 2004;2:e393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, Liu Y, You M. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet 2006;38:888–895 [DOI] [PubMed] [Google Scholar]
- 52.Burgess-Herbert SL, Tsaih SW, Stylianou IM, Walsh K, Cox AJ, Paigen B. An experimental assessment of in silico haplotype association mapping in laboratory mice. BMC Genet 2009;10:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prows DR, Shertzer HG, Daly MJ, Sidman CL, Leikauf GD. Genetic analysis of ozone-induced acute lung injury in sensitive and resistant strains of mice. Nat Genet 1997;17:471–474 [DOI] [PubMed] [Google Scholar]
- 54.Wesselkamper SC, Prows DR, Biswas P, Willeke K, Bingham E, Leikauf GD. Genetic susceptibility to irritant-induced acute lung injury in mice. Am J Physiol Lung Cell Mol Physiol 2000;279:L575–L582 [DOI] [PubMed] [Google Scholar]
- 55.Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D, et al. In silico genetics: identification of a functional element regulating H2-Ealpha gene expression. Science 2004;306:690–695 [DOI] [PubMed] [Google Scholar]
- 56.Liu P, Vikis H, Lu Y, Wang D, You M. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS ONE 2007;2:e651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prows DR, Hafertepen AP, Winterberg AV, Gibbons WJ, Jr, Wesselkamper SC, Singer JB, Hill AE, Nadeau JH, Leikauf GD. Reciprocal congenic lines of mice capture the ALIQ1 effect on acute lung injury survival time. Am J Respir Cell Mol Biol 2008;38:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leikauf GD, Concel VJ, Liu P, Bein K, Berndt A, Ganguly K, Jang AS, Brant KA, Dietsch M, Pope-Varsalona H, et al. Haplotype association mapping of acute lung injury in mice implicates activin a receptor, type 1. Am J Respir Crit Care Med 2011;183:1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fabisiak JP, Medvedovic M, Alexander DC, McDunn JE, Concel VJ, Bein K, Jang AS, Berndt A, Vuga LJ, Brant KA, et al. Integrative metabolome and transcriptome profiling reveals discordant energetic stress between mouse strains with differential sensitivity to acrolein-induced acute lung injury. Mol Nutr Food Res 2011;55:1423–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 2009;81:6656–6657 [DOI] [PubMed] [Google Scholar]
- 62.Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA, Milburn MV. Analysis of the adult plasma metabolome. Pharmacogenomics 2008;9:383–397 [DOI] [PubMed] [Google Scholar]
- 63.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics 2009;25:211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freudenberg JM, Joshi VK, Hu Z, Medvedovic M. CLEAN: clustering enrichment analysis. BMC Bioinformatics 2009;10:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wesselkamper SC, Case LM, Henning LN, Borchers MT, Tichelaar JW, Mason JM, Dragin N, Medvedovic M, Sartor MA, Tomlinson CR, et al. Gene expression changes during the development of acute lung injury: role of transforming growth factor beta. Am J Respir Crit Care Med 2005;172:1399–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variant create synthetic genome-wide associations. PLoS Biol 2010;8:e1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cowan CE, Kohler EE, Dugan TA, Mirza MK, Malik AB, Wary KK. Kruppel-like factor–4 transcriptionally regulates VE-cadherin expression and endothelial barrier function. Circ Res 2010;107:959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem 2005;280:38247–38258 [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, Wang J, Yi Y, Zhang H, Liu J, Liu M, Yuan C, Tang D, Benjamin IJ, Xiao X. Induction of KLF4 in response to heat stress. Cell Stress Chaperones 2006;11:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang HR, Lee CG, Homer RJ, Elias JA. Semaphorin 7A plays a critical role in TGF-beta1–induced pulmonary fibrosis. J Exp Med 2007;204:1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koh JM, Oh B, Lee JY, Lee JK, Kimm K, Kim GS, Park BL, Cheong HS, Shin HD, Hong JM, et al. Association study of semaphorin 7a (SEMA7a) polymorphisms with bone mineral density and fracture risk in postmenopausal Korean women. J Hum Genet 2006;51:112–117 [DOI] [PubMed] [Google Scholar]
- 73.Fahy RJ, Lichtenberger F, McKeegan CB, Nuovo GJ, Marsh CB, Wewers MD. The acute respiratory distress syndrome: a role for transforming growth factor–beta1. Am J Respir Cell Mol Biol 2003;28:499–503 [DOI] [PubMed] [Google Scholar]
- 74.Birukova AA, Birukov KG, Adyshev D, Usatyuk P, Natarajan V, Garcia JG, Verin AD. Involvement of microtubules and Rho pathway in TGF-beta1–induced lung vascular barrier dysfunction. J Cell Physiol 2005;204:934–947 [DOI] [PubMed] [Google Scholar]
- 75.Clements RT, Minnear FL, Singer HA, Keller RS, Vincent PA. RhoA and Rhokinase dependent and independent signals mediate TGF-beta–induced pulmonary endothelial cytoskeletal reorganization and permeability. Am J Physiol Lung Cell Mol Physiol 2005;288:L294–L306 [DOI] [PubMed] [Google Scholar]
- 76.Shenkar R, Coulson WF, Abraham E. Anti–transforming growth factor–beta monoclonal antibodies prevent lung injury in hemorrhaged mice. Am J Respir Cell Mol Biol 1994;11:351–357 [DOI] [PubMed] [Google Scholar]
- 77.Pittet JF, Griffiths MJ, Geiser T, Kaminski N, Dalton SL, Huang X, Brown LA, Gotwals PJ, Koteliansky VE, Matthay MA, et al. TGF-beta is a critical mediator of acute lung injury. J Clin Invest 2001;107:1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao J, Zhao WX, Xue FS, Zhou LJ, Xu SQ, Ding N. Early administration of propofol protects against endotoxin-induced acute lung injury in rats by inhibiting the TGF-beta1–Smad2 dependent pathway. Inflamm Res 2010;59:491–500 [DOI] [PubMed] [Google Scholar]
- 79.Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med 2005;9:59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lai JP, Bao S, Davis IC, Knoell DL. Inhibition of the phosphatase PTEN protects mice against oleic acid–induced acute lung injury. Br J Pharmacol 2009;156:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, Zhao JH, Ramasamy A, Zhai G, Vitart V, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet 2010;42:36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soler Artigas M, Wain LV, Repapi E, Obeidat M, Sayers I, Burton PR, Johnson T, Zhao JH, Albrecht E, Dominiczak AF, et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med 2011;184:786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lo SH. Tensin. Int J Biochem Cell Biol 2004;36:31–34 [DOI] [PubMed] [Google Scholar]
- 84.Hall EH, Balsbaugh JL, Rose KL, Shabanowitz J, Hunt DF, Brautigan DL. Comprehensive analysis of phosphorylation sites in Tensin1 reveals regulation by p38MAPK. Mol Cell Proteomics 2010;9:2853–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boutilier RG. Mechanisms of cell survival in hypoxia and hypothermia. J Exp Biol 2001;204:3171–3181 [DOI] [PubMed] [Google Scholar]
- 86.Hochachka PW. Defense strategies against hypoxia and hypothermia. Science 1986;231:234–241 [DOI] [PubMed] [Google Scholar]
- 87.Handford M, Rodriguez-Furlán C, Orellana A. Nucleotide–sugar transporters: structure, function and roles in vivo. Braz J Med Biol Res 2006;39:1149–1158 [DOI] [PubMed] [Google Scholar]
- 88.Mikkaichi T, Suzuki T, Onogawa T, Tanemoto M, Mizutamari H, Okada M, Chaki T, Masuda S, Tokui T, Eto N, et al. Isolation and characterization of a digoxin transporter and its rat homologue expressed in the kidney. Proc Natl Acad Sci USA 2004;101:3569–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toyohara T, Suzuki T, Morimoto R, Akiyama Y, Souma T, Shiwaku HO, Takeuchi Y, Mishima E, Abe M, Tanemoto M, et al. SLCO4C1 transporter eliminates uremic toxins and attenuates hypertension and renal inflammation. J Am Soc Nephrol 2009;20:2546–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morrison AC, Srinivas SK, Elovitz MA, Puschett JB. Genetic variation in solute carrier genes is associated with preeclampsia. Am J Obstet Gynecol 2010;203:491. [DOI] [PubMed] [Google Scholar]
- 91.Matthews CC, Zielke HR, Wollack JB, Fishman PS. Enzymatic degradation protects neurons from glutamate excitotoxicity. J Neurochem 2000;75:1045–1052 [DOI] [PubMed] [Google Scholar]
- 92.Oliveira GP, Oliveira MB, Santos RS, Lima LD, Dias CM, Ab'Saber AM, Teodoro WR, Capelozzi VL, Gomes RN, Bozza PT, et al. Intravenous glutamine decreases lung and distal organ injury in an experimental model of abdominal sepsis. Crit Care 2009;13:R74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weitzel LR, Wischmeyer PE. Glutamine in critical illness: the time has come, the time is now. Crit Care Clin 2010;26:515–525 [DOI] [PubMed] [Google Scholar]
- 94.Kwon WY, Suh GJ, Kim KS, Jo YH, Lee JH, Kim K, Jung SK. Glutamine attenuates acute lung injury by inhibition of high mobility group box protein–1 expression during sepsis. Br J Nutr 2010;103:890–898 [DOI] [PubMed] [Google Scholar]
- 95.Fox RE, Hopkins IB, Cabacungan ET, Tildon JT. The role of glutamine and other alternate substrates as energy sources in the fetal rat lung Type II cell. Pediatr Res 1996;40:135–141 [DOI] [PubMed] [Google Scholar]
- 96.Greenleaf RD. Characteristics of amino acid metabolism by isolated alveolar Type II cells. Exp Lung Res 1984;7:85–91 [DOI] [PubMed] [Google Scholar]
- 97.Sheehan PM, Yeh YY. Pulmonary surfactant lipid synthesis from ketone bodies, lactate and glucose in newborn rats. Lipids 1985;20:835–841 [DOI] [PubMed] [Google Scholar]
- 98.Endemann G, Goetz PG, Edmond J, Brunengraber H. Lipogenesis from ketone bodies in the isolated perfused rat liver: evidence for the cytosolic activation of acetoacetate. J Biol Chem 1982;257:3434–3440 [PubMed] [Google Scholar]
- 99.Yamasaki M, Hasegawa S, Suzuki H, Hidai K, Saitoh Y, Fukui T. Acetoacetyl-CoA synthetase gene is abundant in rat adipose, and related with fatty acid synthesis in mature adipocytes. Biochem Biophys Res Commun 2005;335:215–219 [DOI] [PubMed] [Google Scholar]
- 100.Bergstrom JD, Wong GA, Edwards PA, Edmond J. The regulation of acetoacetyl-CoA synthetase activity by modulators of cholesterol synthesis in vivo and the utilization of acetoacetate for cholesterogenesis. J Biol Chem 1984;259:14548–14553 [PubMed] [Google Scholar]
- 101.Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet 2001;68:753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet 2001;68:598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Axelrod FB. Familial dysautonomia: a review of the current pharmacological treatments. Expert Opin Pharmacother 2005;6:561–567 [DOI] [PubMed] [Google Scholar]
- 104.Cohen L, Henzel WJ, Baeuerle PA. IKAP is a scaffold protein of the IkappaB kinase complex. Nature 1998;395:292–296 [DOI] [PubMed] [Google Scholar]
- 105.Krappmann D, Hatada EN, Tegethoff S, Li J, Klippel A, Giese K, Baeuerle PA, Scheidereit C. The I kappa B kinase (IKK) complex is tripartite and contains IKK gamma but not IKAP as a regular component. J Biol Chem 2000;275:29779–29787 [DOI] [PubMed] [Google Scholar]
- 106.Holmberg C, Katz S, Lerdrup M, Herdegen T, Jäättelä M, Aronheim A, Kallunki T. A novel specific role for I kappa B kinase complex–associated protein in cytosolic stress signaling. J Biol Chem 2002;277:31918–31928 [DOI] [PubMed] [Google Scholar]
- 107.Johansen LD, Naumanen T, Knudsen A, Westerlund N, Gromova I, Junttila M, Nielsen C, Bøttzauw T, Tolkovsky A, Westermarck J, et al. IKAP localizes to membrane ruffles with filamin A and regulates actin cytoskeleton organization and cell migration. J Cell Sci 2008;121:854–864 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.