Abstract
We report for the first time the genetic and biological characterization of 10 HIV-1 primary isolates representing CRF28_BF and CRF29_BF together with additional unique BF recombinant forms (URFs) obtained by PBMC cocultivation. Recombination is an important factor promoting the increase in the genetic diversity of HIV-1. Notably, more than 20% of HIV-1 sequences worldwide were recombinants. Several recombinant viruses were reported in Brazil, and six circulating recombinant forms (CRFs) have been identified (CRF28_BF, CRF29_BF, CRF31_BC, CRF39_BF, CRF40_BF, and CRF46_BF). CRF28_BF and CRF29_BF were found to infect almost 30% of the patients in São Paulo State. The near full-length genomes of these 10 primary isolates were amplified by nested PCR in three overlapping segments, purified, and sequenced. Three samples were related to CRF28_BF, three to CRF29_BF, and four were unique recombinant forms (URFs), as determined by their breakpoint profile determined with the jpHMM program. Additionally, the coreceptor usage of these isolates was investigated in vitro using GHOST assays, which revealed three dual-tropic (X4/R5) viruses, four lymphotropic (X4) viruses, and three macrophage-tropic (R5) viruses with different V3-loop motifs, which challenges the notion that GWGR-carrying viruses are macrophage-tropic only. In sum, we report a much-anticipated well-characterized panel of viruses representing CRF28_BF, CRF29_BF, and URFs from São Paulo State, Brazil.
Introduction
The intense investigation of HIV-1 genetic diversity has led to the identification and description of different subtypes worldwide.1–5 It is believed that much of this variability results from the lack of proofreading activity of the viral reverse transcriptase,6,7 along with a high rate of viral replication.8,9 Additionally, recombination during reverse transcription is a factor promoting HIV diversity and adaptive change,10,11 by allowing advantageous mutations arising on different genomes to undergo linkage in the same progeny recombinant genome more frequently than what would be expected under random mutation alone.12–16 Moreover, intersubtype recombinants have been reported in increasing numbers in regions of the world where multiple subtypes do cocirculate to the extent that at least 20% of HIV-1 sequences worldwide were found to be recombinants.17–19 To date, 49 circulating recombinant forms (CRFs) have been identified (www.hiv.lanl.gov), and some of these were successful in causing regional epidemics, e.g., CRF01_AE in Asia20 and CRF02_AG in Africa.21,22 Likewise, several distinct BF recombinant viruses have been described in South America.23 Notably, full genome sequencing of these viruses revealed at least nine recombinant lineages that fulfilled the criteria of being recognized as CRFs (CRF12_BF, CRF17_BF, CRF28_BF, CRF29_BF, CRF38_BF, CRF39_BF, CRF40_BF, CRF44_BF, and CRF46_BF) (www.hiv.lanl.gov).
Such great diversity of successful CRFs BF (i.e., recombinants between subtypes B and F) could entail an adaptive advantage for these recombinants, since we can assume that the emerging recombinant must first overgrow its parental strain within the coinfected individual to be efficiently transmitted. Actually, two independent lines of evidence suggest that BF recombinants may exhibit different biological behaviors. First, a high rate of population growth was observed for both CRF12_BF and CRF38_BF in Argentina and Uruguay.24,25 Second, it was demonstrated that the BF recombinant LTR/Tat complex has higher transcriptional activity compared to the subtype B complex26 and that BF Vpu-harboring variants have increased fitness compared to subtype B in vitro.27 Nevertheless, it remains to be investigated to what extent these results could be generalized to other BF recombinants spreading in South America.
Therefore, it is paramount to try to obtain primary isolates for the most prevalent BF mosaics, such as CRF28_BF and CRF29_BF, which were already found to infect almost 30% of HIV-1 carriers in some places in São Paulo State.28,29 Herein, we report the genetic and biological characterization of 10 HIV-1 primary isolates representing CRF28_BF, CRF29_BF, and unique BF recombinant forms (URFs) in order to contribute to the construction of a panel of well-characterized BF recombinant viruses.
Material and Methods
Ethics statements, clinical samples, and virus isolation
The study was submitted and approved by the Ethics Committee on Human Research of the University of São Paulo and informed consent terms were signed by all patients. Ten patients harboring BF recombinant viruses were selected from the Viral Genetic Diversity Network (VGDN) program,30 based on their available clinical and epidemiological information. HIV-1 isolation was performed by coculture of freshly harvested peripheral blood mononuclear cells (PBMCs) and uninfected PHA-stimulated PBMCs according to the ACTG Virology Manual for HIV Laboratories guidelines.31 Viral growth was monitored by p24 antigen production using the Vironostika HIV-1 antigen Microelisa kit (bioMérieux, Netherlands). Positive cultures were further expanded in PHA-stimulated PBMCs to produce viral stocks.
DNA extraction, polymerase chain reaction (PCR), and near full-length genome sequencing
DNA was extracted from infected PBMCs using the QIAamp DNA Blood kit (Qiagen, Germany), according to the manufacturer's instructions, and stored at −80°C until use. Three overlapping regions of the viral genome were amplified using nested PCR.32 PCR products were purified with the QIAquick PCR Purification Kit (Qiagen, Germany). Sequencing reactions were performed using BigDyeTerminator version 3.0 cycle sequencing (ABI Prism; PE Applied Biosystems, Foster City, CA), and the products were analyzed on ABI 3100 automated DNA sequencers (PE Applied Biosystem). Sequencing primers are available upon request. Sequence data were edited and assembled with CodonCode Aligner software (Gene Codes Corporation).
Recombination analysis
The available genome sequences were analyzed with the jumping profile hidden Markov model (jpHMM) program,33,34 which uses detailed information on the polymorphism of the putative parental populations rather than using individual parental strains and provides detailed information on the reliability of the predicted recombination breakpoints.35
Genotypic and phenotypic analysis of coreceptor usage
Despite the availability of different methods to predict coreceptor usage, the geno2pheno algorithm was found to be the more sensitive to detect CXCR4 variants.36 The V3 sequences were analyzed with the geno2pheno[coreceptor] tropism prediction algorithm using the clonal setting and a 20% false-positive rate (FPR) as the cutoff. This FPR value was chosen since it minimizes the number of falsely predicted R5-tropic viruses.36 The phenotypic evaluation of coreceptor usage was done on GHOST(3) indicator cell lines expressing CD4 and either CCR5 or CXCR4 (NIH AIDS Reagent Program).37 The GHOST cells were grown in DMEM (Invitrogen) supplemented with 10% FCS, penicillin (50 U/ml), streptomycin (50 U/ml), and l-glutamine (2 mM), and selected with G418 (500 μg/ml), hygromycin (100 μg/ml), and puromycin (1 μg/ml). Infection was carried out as previously described.37 Briefly, cells were seeded in 24-well plates 1 day before infection. The following day, the medium was replaced with 200 μl of fresh medium containing polybrene (2 μg/ml) and 300 μl of virus stock. Cultures were incubated overnight, washed with PBS, and further incubated with fresh medium. Infected cells expressing GFP were detected 2 or 3 days after infection by fluorescence microscopic observation. CXCR4 usage was confirmed using the antagonist AMD3100,38–40 added to the cells prior to infection at a concentration of 1 μM. JM-2987 (hydrobromide salt of AMD 3100) was obtained from the NIH AIDS Research and Reference Reagent Program.
Results
Clinical features and viral isolation
HIV-1 primary isolates were obtained from each sampled patient, as confirmed by the p24 antigen capture assay. Clinical information is summarized in Table 1. Most patients were men (7/10) and the predominant risk behavior for HIV infection was unprotected heterosexual intercourse. Six isolates were obtained from patients with CD4 count ranging from 10 to 693 cells/ml and viral load ranging from undetectable to 150,247 copies/ml (Table 1). The dates of the first HIV-1-positive test ranged from 1985 to 2003 (Table 1).
Table 1.
Clinical Data of Sampled Patients
| Patient ID | Sex | Risk behavior | Year of diagnosisa | CD4 (cells/ml) | Viral load (copies/ml) | Antiretroviral treatment |
|---|---|---|---|---|---|---|
| 0008SP | Female | HT | 1996 | 595 | 450 | NRTI+NRTI |
| 0063SP | Male | HT, IDU, BT | 1985 | 10 | 45,600 | NRTI+NRTI+PI |
| 0264RI | Male | MSM, IDU | 1995 | 632 | 22,000 | None |
| 0341RI | Female | HT, BT | 1995 | 554 | 54,500 | NRTI+NRTI |
| 0614SV | Male | HT | 1998 | 499 | <50 | NRTI+NRTI+PI |
| 0632SV | Male | HT | 2003 | 169 | 150,347 | NRTI+NRTI+PI |
| 0647SV | Male | HT | 1995 | 442 | 12,284 | None |
| 0679SV | Male | HT | 1995 | 202 | 5,665 | NRTI+NRTI+PI |
| 0736SV | Male | MSM, IDU | 1992 | 693 | 890 | NRTI+NRTI |
| 0744SV | Female | HT, BT | 2003 | na | 22,395 | NRTI+NRTI |
Patient reported date of first positive HIV test.
IDU, injecting drug user; MSM, men who have sex with men; HT, heterosexual; BT, blood transfusion; na, not available; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Genomic characterization and recombination patterns
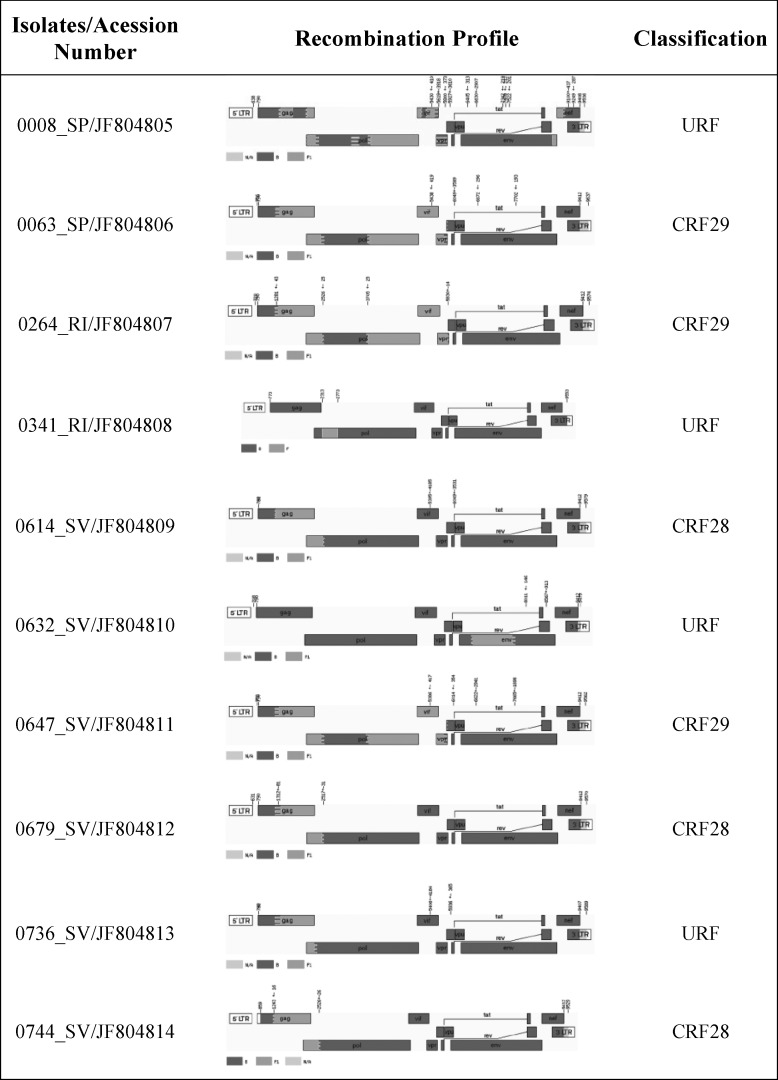
The near full-length genomes of all primary isolates were successfully amplified and sequenced. The sequences were 8.8 kb in length on average, with intact open reading frames. To allow the amplification of all open reading frames of each provirus, we had to avoid pairs of primers that would amplify only a single LTR. Therefore, 140 base pairs were not amplified by our strategy. It is worth mentioning that no other research group reports this stretch of proviral sequence, possibly for the same reason. Additionally, to check the possibility of contamination among samples, the sequenced genomes were compared using the Blast2Seq tool of NCBI,41 which revealed that no cross-contamination appeared to have occurred. Three samples were classified as CRF28_BF (0614SV, 0679SV, and 0744SV) and three as CRF29_BF (0063SP, 0264RI, and 0647SV), according to the breakpoints profile, summarized in Fig. 1 and Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/aid). The estimated breakpoints for these isolates were slightly different from those originally described for CRF28_BF (B: 832–1322, F1: 1323–2571, B: 2572–9432) and CRF29_BF (B: 823–1322, F1: 1323–2571, B: 2572–3682, F1: 3683–5462, B: 5463–9432).42 The remaining four isolates (0008SP, 0341RI, 0632SV, and 0736SV) were classified as URFs (Fig. 1). Interestingly, two of them (0008 and 0736) shared some breakpoints with CRF28_BF or CRF29_BF. The recombination pattern of isolate 0008SP was quite complex, sharing breakpoints with CRF29_BF at position 3748±45. The isolate 0736SV also had a breakpoint located at position 1300±78, resembling the profile of CRF28_BF, but had a second one at 2317±34, which is 200 bp away from the predicted,23 which indicates that this recombinant virus is possibly an example of a “second generation” recombinant (i.e., recombinant progeny involving at least one distinct parental CRFs). It was noteworthy that the isolate 0632SV had the same mosaic structure and breakpoints (6583±20–7659±30, Supplementary Fig. S1) as a previously published sequence (DQ358806/02BR005) sampled in São Paulo.43
FIG. 1.
Near full-length genome map of the 10 BF recombinants primary isolates. Breakpoints and subtype composition of isolates revealed mosaic structures related to CRF28_BF (3), CRF29_BF (3), and URFs (4). Recombination breakpoints were determined with the jpHMM algorithm available online at the GOBICS server (http://jphmm.gobics.de/jphmm.html). Fragments related to subtype B are shown in dark gray and to subtype F1 in light gray.
V3 loop characteristics and coreceptor usage
As shown in Fig. 1, the 0632SV had a subtype F1 V3 sequence, whereas the remaining sequences had a subtype B V3 loop. The predicted amino acid alignment showed a great diversity of tetrameric amino acid motifs at the tip of the V3 loop (Table 2). Overall, we found seven different motifs, with GWGR (3/10) and GPGR (2/10) being the most common. The GWGR motif was present in two out of three isolates of CRF28_BF and in one isolate of CRF29_BF. This contrasts with previous findings showing that the GWGR motif was not commonly present in CRF_BF isolates.44 Actually, this low frequency of GWGR variants was observed even when we analyzed all BF genomes available from the Los Alamos HIV database in which only 4 out of 137 have a GWG motif (data not shown). Furthermore, the length of the V3 loop was variable among our isolates (ranging from 34 to 37 aa). In particular, isolate 0008SP had a deletion at position 11, which is a wel- known determinant of cell tropism.45 To exclude the possibility of defective provirus amplification or sequencing artifact, both PCR and sequencing steps were repeated, confirming this was indeed a true deletion (Supplementary Fig. S2).
Table 2.
Genotypic and Phenotypic Characterization of Coreceptor Usage of Primary Isolates of HIV-1 BF Recombinants
| Sample | Acession number | Genome subtype | env subtype | V3 tetramer | Geno2pheno | GHOST assay | AMD sensitivity |
|---|---|---|---|---|---|---|---|
| 0008SP | JF804805 | URF | B | GPGR | X4 | X4 | Yes |
| 0063SP | JF804806 | CRF29 | B | GSGR | X4 | X4 | Yes |
| 0264RI | JF804807 | CRF29 | B | GRGR | X4 | X4 | Yes |
| 0341RI | JF804808 | URF | B | GLGR | X4 | R5/X4 | No |
| 0614SV | JF804809 | CRF28 | B | GPGR | X4 | R5/X4 | No |
| 0632SV | JF804810 | URF | F | GPGK | R5 | R5 | No |
| 0647SV | JF804811 | CRF29 | B | GWGR | X4 | R5/X4 | No |
| 0679SV | JF804812 | CRF28 | B | GWGR | R5 | R5 | No |
| 0736SV | JF804813 | URF | B | GVGR | X4 | X4 | Yes |
| 0744SV | JF804814 | CRF28 | B | GWGR | R5 | R5 | No |
We then investigated the coreceptor usage of these isolates, using both genotyping and phenotyping methods. Initially, we did a preliminary in silico analysis using the Geno2pheno algorithm. As shown in Table 2, seven isolates were predicted to use CXCR4 as their entry coreceptor, including the aforementioned isolate 0008SP, while the remaining three were predicted to use CCR5 exclusively. We then checked the isolates tropism in vitro using the GHOST(3) cell assay (Table 2). All three isolates predicted as CCR5-tropic replicated in GHOST(3)-CCR5 cells but did not replicate in cells expressing CXCR4, conforming that they were exclusively macrophage-tropic (i.e., used CCR5 as entry coreceptor). The remaining seven isolates that were predicted by Gene2pheno to use CXCR4 replicated in GHOST(3)–CXCR4 cells. Nevertheless, three of them (0341SV, 0614SV, and 0647SV) also replicated in GHOST(3)–CCR5 cells and were defined as dual-tropic viruses (Table 2). Since GHOST(3) cell lines express low levels of endogenous CXCR4, these results were further confirmed by blocking the CXCR4 coreceptor with the antagonist AMD3100. As expected, only those viruses previously classified as CCR5-tropic or dual-tropic were able to infect GHOST–CCR5. Despite the limited number of isolates described here, there was no relationship between clinical status and the presence of CXCR4-tropic variants.
Discussion
We performed for the first time the isolation and characterization of CRF28_BF and CRF29_BF viruses, which are highly prevalent among our HIV-1-infected population. It is noteworthy that clinical data showed that one of the sampled patients, harboring a CRF29_BF (0063SP) virus, was first diagnosed in 1985. This suggests that this CRF has been cryptically circulating at least 14 years before its description, in patients sampled between 1999 and 2002,42 even before the first description of an HIV-1 recombinant in Brazil in 1992.46 Alternatively, this patient could have been infected with another subtype at the time of the first HIV-1-positive diagnosis and later reinfected by the recombinant form isolated here. However, sequencing several distinct PCR products obtained after limiting dilution (which effect mimics cloning) did not reveal any evidence of dual infection in PBMCs from this patient (data not shown). Also, the observed date of 1985 agrees with an independent estimate for the origin of Brazilian BF recombinants, ranging from 1984 to 1991.47 Taken together, these results provide an independent estimate for a lower boundary for the origin of CRF 29 at around 1985.
We have also identified two URFs sharing breakpoints with CRF28/29, which may constitute evidence for a so-called second generation of BF recombinants. Alternatively, these URFs may have convergent breakpoints as a consequence of shared recombination hotspots. Nevertheless, the existence of these URFs may constitute important evidence that superinfection may be a common event, which agrees with reports on the high frequency of reinfection among HIV carriers.48 Indeed, the large number of unique BF recombinants found in South America23 strongly supports the notion that different genotypes may be circulating and reinfecting hosts. Together these findings stress a pivotal role of cocirculation and superinfection in the emergence of new recombinant forms. This could be the case of isolate 0632SV, which may be a new CRF_BF circulating in São Paulo State, since it shares all the recombination breakpoints with a previously published sequence from a nearby location.49
Most previous in silico analyses characterizing HIV recombinants have been done with bootscanning methods. Even though this methodology is efficient in recombination detection, it is largely dependent on the choice of parental strains, which affect the accuracy of the breakpoint prediction. In our case, the use of jpHMM provided a better resolution of breakpoints, which is crucial for the proper identification of CRFs (Supplementary Table S1). Most likely, this was a consequence of an improved accuracy in breakpoint prediction available in more recent versions of the jpHMM algorithm and the increasing availability of near full-length genome sequences used as references.35 Recently, an extensive reassessment of public database HIV sequences using jpHMM also described such discordant breakpoints, and reassigned two reference sequences, CRF28_BF (DQ085874) and CRF29_BF (AY771590),23 as URFs.
The V3 loop in the gp120 protein is considered the major viral determinant for cellular tropism45 and also a preferential target of humoral50 and cellular51 responses. Accordingly, we observed among our isolates considerable variability at the tetrapeptide motif at the V3 loop. Furthermore, we did not find any relationship between tetrapeptide motif and coreceptor usage, in line with a previous work that also described GWGR isolates as being capable of using CXCR4 as an entry coreceptor.52 We would argue that our finding was relevant, since it provided empirical evidence against the belief that variants carrying the GWGR tetramer would exclusively use the CCR5 coreceptor.53 Crucially, these findings provide further biological evidence that this motif (GWGR) alone does not determine the coreceptor usage.54 Moreover, CXCR4-tropic viruses were more common than CCR5-tropic viruses (7 and 3, respectively), regardless of the recombination profile and patient clinical status. Furthermore, despite our limited sample size but because both in vitro and in silico procedures converged on CXCR4 usage determination, it is worthwhile to try to determine in greater detail whether the Geno2pheno algorithm may indeed increase accuracy in coreceptor usage prediction. This information is of relevance for patient treatment with CCR5 antagonists, which is a drug class recently approved for extensive use in Brazil.
We report a unique and well-characterized panel of viruses representing CRF28_BF, CRF29_BF, and URFs BF. We also present evidence that at least CRF29_BF may have been circulating around 1985, at the beginning of the AIDS epidemic in Brazil, which dates at least 20 years before its detection in samples from 2002.42 Moreover, we found a high number of URFs (4:10) compared with that of CRFs (6:10), which highlights the fact that superinfection may be frequent enough to merit in-depth studies of its intrahost and epidemiological relevant factors. In sum, we believe that the isolates we present could be useful to advance our knowledge of well-established BF recombinants of growing epidemiological importance. It is quite relevant to consider that BF recombinant viruses constitute the majority (11 out of 49) of the CRFs described to date (Los Alamos). Moreover, BF viruses that originated in Brazil may have a far reaching effect on HIV pandemics, since they have already been isolated in Asia and Europe.55
Sequence Data
Sequences are deposited in GenBank under accession numbers: JF804805–JF804814.
Supplementary Material
Acknowledgments
This research was made possible by the Viral Genetic Diversity (VGDN) program funded by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (www.fapesp.br) project number 00/04205-6. F.L.M. holds a FAPESP doctorate scholarship (process number 07/01554-9). P.M.A.Z. holds a CNPq PQ research scholarship. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: bicyclam JM-2987 (hydrobromide salt of AMD-3100) and GHOST cells from Dr. Vineet N. Kewal Ramani and Dr. Dan R. Littman. We are also grateful to those patients who kindly agreed to collaborate in this study.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Taylor BS. Sobieszczyk ME. McCutchan FE. Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358(15):1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papathanasopoulos MA. Hunt GM. Tiemessen CT. Evolution and diversity of HIV-1 in Africa—a review. Virus Genes. 2003;26(2):151–163. doi: 10.1023/a:1023435429841. [DOI] [PubMed] [Google Scholar]
- 3.McCutchan FE. Understanding the genetic diversity of HIV-1. AIDS. 2000;14(Suppl 3):S31–S44. [PubMed] [Google Scholar]
- 4.Cock KM. Weiss HA. The global epidemiology of HIV/AIDS. Trop Med Int Health. 2000;5(7):A3–A9. doi: 10.1046/j.1365-3156.2000.00590.x. [DOI] [PubMed] [Google Scholar]
- 5.Hamers FF. Downs AM. Infuso A. Brunet JB. Diversity of the HIV/AIDS epidemic in Europe. AIDS. 1998;12(Suppl A):S63–S70. [PubMed] [Google Scholar]
- 6.Preston BD. Poiesz BJ. Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242(4882):1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 7.Roberts JD. Bebenek K. Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242(4882):1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 8.Perelson AS. Neumann AU. Markowitz M. Leonard JM. Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271(5255):1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 9.Ho DD. Neumann AU. Perelson AS. Chen W. Leonard JM. Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373(6510):123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 10.Neher RA. Leitner T. Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput Biol. 2010;6(1):e1000660. doi: 10.1371/journal.pcbi.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez BC. Simon-Loriere E. Galetto R. Negroni M. Implications of recombination for HIV diversity. Virus Res. 2008;134(1–2):64–73. doi: 10.1016/j.virusres.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Arora P. Dixit NM. Timing the emergence of resistance to anti-HIV drugs with large genetic barriers. PLoS Comput Biol. 2009;5(3):e1000305. doi: 10.1371/journal.pcbi.1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streeck H. Li B. Poon AF, et al. Immune-driven recombination and loss of control after HIV superinfection. J Exp Med. 2008;205(8):1789–1796. doi: 10.1084/jem.20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nora T. Charpentier C. Tenaillon O. Hoede C. Clavel F. Hance AJ. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81(14):7620–7628. doi: 10.1128/JVI.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charpentier C. Nora T. Tenaillon O. Clavel F. Hance AJ. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J Virol. 2006;80(5):2472–2482. doi: 10.1128/JVI.80.5.2472-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moutouh L. Corbeil J. Richman DD. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci USA. 1996;93(12):6106–6111. doi: 10.1073/pnas.93.12.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osmanov S. Pattou C. Walker N. Schwardlander B. Esparza J. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 2002;29(2):184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- 18.Hemelaar J. Gouws E. Ghys P. Osmanov S. Characterisation WUNfHIa. Global trends in molecular epidemiology of HIV-1 during 2000–2007. IAS, ed. XVIII International AIDS Conference; Vienna Austria. 2010. Vol Abstract no. THAA0201. [Google Scholar]
- 19.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 20.Gao F. Robertson DL. Morrison SG, et al. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70(10):7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montavon C. Toure-Kane C. Liegeois F, et al. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype AG recombinant virus IBNG. J Acquir Immune Defic Syndr. 2000;23(5):363–374. doi: 10.1097/00126334-200004150-00001. [DOI] [PubMed] [Google Scholar]
- 22.Andersson S. Norrgren H. Dias F. Biberfeld G. Albert J. Molecular characterization of human immunodeficiency virus (HIV)-1 and -2 in individuals from Guinea-Bissau with single or dual infections: Predominance of a distinct HIV-1 subtype A/G recombinant in West Africa. Virology. 1999;262(2):312–320. doi: 10.1006/viro.1999.9867. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M. Foley B. Schultz AK, et al. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology. 2010;7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bello G. Aulicino PC. Ruchansky D, et al. Phylodynamics of HIV-1 circulating recombinant forms 12_BF and 38_BF in Argentina and Uruguay. Retrovirology. 2010;7:22. doi: 10.1186/1742-4690-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aulicino PC. Holmes EC. Rocco C. Mangano A. Sen L. Extremely rapid spread of human immunodeficiency virus type 1 BF recombinants in Argentina. J Virol. 2007;81(1):427–429. doi: 10.1128/JVI.01403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turk G. Carobene M. Monczor A. Rubio AE. Gomez-Carrillo M. Salomon H. Higher transactivation activity associated with LTR and Tat elements from HIV-1 BF intersubtype recombinant variants. Retrovirology. 2006;3:14. doi: 10.1186/1742-4690-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Candia C. Espada C. Duette G, et al. Viral replication is enhanced by an HIV-1 intersubtype recombination-derived Vpu protein. Virol J. 2010;7(1):259. doi: 10.1186/1743-422X-7-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Souza AC. de Oliveira CM. Rodrigues CL. Silva SA. Levi JE. Molecular characterization of HIV type 1 BF pol recombinants from Sao Paulo, Brazil. AIDS Res Hum Retroviruses. 2008;24(12):1521–1525. doi: 10.1089/aid.2008.0089. [DOI] [PubMed] [Google Scholar]
- 29.Teixeira D. Munerato P. Komninakis SC, et al. The detection of in vivo and in vitro HIV type 1 B/F profiles in Brazil using a real-time PCR assay for five HIV type 1 genomic regions. AIDS Res Hum Retroviruses. 2010;26(9):981–990. doi: 10.1089/aid.2010.0023. [DOI] [PubMed] [Google Scholar]
- 30.Pardini MI. Jamal LF. Durigon EL, et al. Boosting virology in Brazil. Plos Biol. 2008;6(3):428–429. doi: 10.1371/journal.pbio.0060057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollinger FB. Bremer JW. Myers LE. Gold JW. McQuay L. Standardization of sensitive human immunodeficiency virus coculture procedures and establishment of a multicenter quality assurance program for the AIDS Clinical Trials Group. The NIH/NIAID/DAIDS/ACTG Virology Laboratories. J Clin Microbiol. 1992;30(7):1787–1794. doi: 10.1128/jcm.30.7.1787-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadai Y. Eyzaguirre LM. Constantine NT, et al. Protocol for nearly full-length sequencing of HIV-1 RNA from plasma. Plos One. 2008;3(1):e1420. doi: 10.1371/journal.pone.0001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M. Schultz AK. Calef C, et al. jpHMM at GOBICS: A web server to detect genomic recombinations in HIV-1. Nucleic Acids Res. 2006;34:W463–W465. doi: 10.1093/nar/gkl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz AK. Zhang M. Leitner T, et al. A jumping profile hidden Markov model and applications to recombination sites in HIV and HCV genomes. BMC Bioinformatics. 2006;7:265. doi: 10.1186/1471-2105-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz AK. Zhang M. Bulla I, et al. jpHMM: Improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 2009;37:W647–W651. doi: 10.1093/nar/gkp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez V. Masia M. Robledano C. Padilla S. Ramos JM. Gutierrez F. Performance of genotypic algorithms for predicting HIV-1 tropism measured against the enhanced-sensitivity Trofile coreceptor tropism assay. J Clin Microbiol. 2010;48(11):4135–4139. doi: 10.1128/JCM.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morner A. Bjorndal A. Albert J, et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol. 1999;73(3):2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrix CW. Flexner C. MacFarland RT, et al. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob Agents Chemother. 2000;44(6):1667–1673. doi: 10.1128/aac.44.6.1667-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Clercq E. Yamamoto N. Pauwels R, et al. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob Agents Chemother. 1994;38(4):668–674. doi: 10.1128/aac.38.4.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bridger GJ. Skerlj RT. Thornton D, et al. Synthesis and structure-activity relationships of phenylenebis(methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication. Effects of macrocyclic ring size and substituents on the aromatic linker. J Med Chem. 1995;38(2):366–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF. Madden TL. Schaffer AA, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Sa Filho DJ. Sucupira MC. Caseiro MM. Sabino EC. Diaz RS. Janini LM. Identification of two HIV type 1 circulating recombinant forms in Brazil. AIDS Res Hum Retroviruses. 2006;22(1):1–13. doi: 10.1089/aid.2006.22.1. [DOI] [PubMed] [Google Scholar]
- 43.Sanabani S. Kleine Neto W. Kalmar EM. Diaz RS. Janini LM. Sabino EC. Analysis of the near full length genomes of HIV-1 subtypes B, F and BF recombinant from a cohort of 14 patients in Sao Paulo, Brazil. Infect Genet Evol. 2006;6(5):368–377. doi: 10.1016/j.meegid.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Leal E. Villanova FE. Diversity of HIV-1 subtype B: Implications to the origin of BF recombinants. PLoS One. 2010;5(7):e11833. doi: 10.1371/journal.pone.0011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fouchier RA. Groenink M. Kootstra NA, et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66(5):3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabino EC. Shpaer EG. Morgado MG, et al. Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil. J Virol. 1994;68(10):6340–6346. doi: 10.1128/jvi.68.10.6340-6346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leal E. Martins LO. Janini LM. Diaz RS. Evolutionary dynamics of HIV-1 BF and CB recombinants and its parental counterparts in South America. Retrovirol: Res Treat. 2008 2008(RRT-1-Leal-et-al):1. [Google Scholar]
- 48.Piantadosi A. Chohan B. Chohan V. McClelland RS. Overbaugh J. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 2007;3(11):e177. doi: 10.1371/journal.ppat.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanabani S. Pastena E. Neto W. Martinez V. Sabino E. Characterization and frequency of a newly identified HIV-1 BF1 intersubtype circulating recombinant form in Sao Paulo, Brazil. Virol J. 2010;7:74. doi: 10.1186/1743-422X-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Javaherian K. Langlois AJ. LaRosa GJ, et al. Broadly neutralizing antibodies elicited by the hypervariable neutralizing determinant of HIV-1. Science. 1990;250(4987):1590–1593. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- 51.Kmieciak D. Wasik TJ. Teppler H, et al. The effect of deletion of the V3 loop of gp120 on cytotoxic T cell responses and HIV gp120-mediated pathogenesis. J Immunol. 1998;160(11):5676–5683. [PubMed] [Google Scholar]
- 52.Ferraro GA. Mello MAG. Sutmoller F, et al. Biological characterization and chemokine receptor usage of HIV type 1 isolates prevalent in Brazil. Aids Res Hum Retroviruses. 2001;17(13):1241–1247. doi: 10.1089/088922201750461294. [DOI] [PubMed] [Google Scholar]
- 53.Leal E. Silva WP. Sucupira MC. Janini LM. Diaz RS. Molecular and structural characterization of HIV-1 subtype B Brazilian isolates with GWGR tetramer at the tip of the V3-loop. Virology. 2008;381(2):222–229. doi: 10.1016/j.virol.2008.08.029. [DOI] [PubMed] [Google Scholar]
- 54.Tomasini-Grotto R-M. Montes B. Triglia D, et al. Variability of the conserved V3 loop tip motif in HIV-1 subtype B isolates collected from Brazilian and French patients. Brazilian J Microbiol. 2010;41:720–728. doi: 10.1590/S1517-83822010000300024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruselles A. Rozera G. Bartolini B, et al. Use of massive parallel pyrosequencing for near full-length characterization of a unique HIV Type 1 BF recombinant associated with a fatal primary infection. AIDS Res Hum Retroviruses. 2009;25(9):937–942. doi: 10.1089/aid.2009.0083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.