Abstract
Objective To evaluate whether the prevalence of HIV-1 transmitted drug resistance has continued to decline in infections probably acquired within the United Kingdom.
Design Multicentre observational study.
Setting All UK public laboratories conducting tests for genotypic HIV resistance as a part of routine care.
Participants 14 584 patients infected with HIV-1 subtype B virus, who were first tested for resistance before receiving antiretroviral therapy between January 2002 and December 2009.
Main outcome measure Prevalence of transmitted drug resistance, defined as one or more resistance mutations from the surveillance list recommended by the World Health Organization.
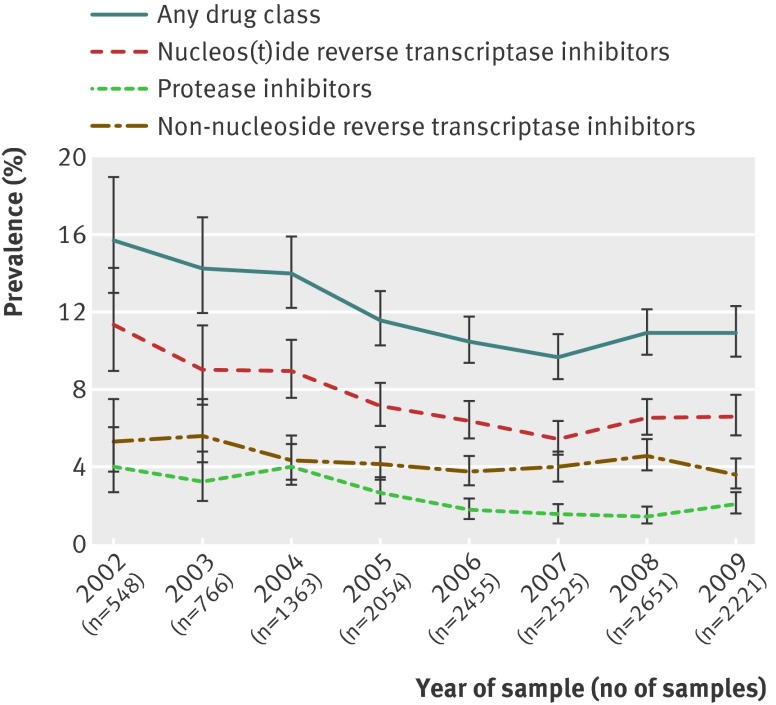
Results 1654 (11.3%, 95% confidence interval 10.8% to 11.9%) patients had one or more mutations associated with transmitted HIV-1 drug resistance; prevalence was found to decline from 15.5% in 2002 to 9.6% in 2007, followed by a slight increase to 10.9% in 2009 (P=0.21). This later rise was mainly a result of increases in resistance to nucleos(t)ide reverse transcriptase inhibitors (from 5.4% in 2007 to 6.6% in 2009, P=0.24) and protease inhibitors (1.5% to 2.1%, P=0.12). Thymidine analogue mutations, including T215 revertants, remained the most frequent mutations associated with nucleos(t)ide reverse transcriptase inhibitors, despite a considerable fall in stavudine and zidovudine use between 2002 and 2009 (from 29.4% of drug regimens in 2002 to 0.8% in 2009, from 47.9% to 8.8%, respectively).
Conclusions The previously observed decline in the prevalence of transmitted drug resistance in HIV-1 infections probably acquired in the UK seems to have stabilised. The continued high prevalence of thymidine analogue mutations suggests that the source of this resistance may be increasingly from patients who have not undergone antiretroviral therapy and who harbour resistant viruses. Testing of all newly diagnosed HIV-1 positive people should be continued.
Introduction
Combination antiretroviral therapy continues to be highly effective in treating HIV-1, and the introduction of new drugs and antiretroviral drug classes has notably improved patient prognosis. Nevertheless, resistance to antiretroviral drugs can develop in people on therapy and is associated with treatment failure.1 Against this backdrop, the rate of new HIV-1 infections within the United Kingdom continues to rise.2 In such new infections, transmitted drug resistance is sometimes assumed to reflect only direct infection from patients already receiving antiretroviral therapy. Concerns about an adverse effect of transmitted HIV-1 drug resistance on the success of antiretroviral therapy have led to national and international guidelines recommending that all newly diagnosed patients have resistance tests conducted to aid selection of first line regimens.3 4
A previous study showed a sharp decline in the prevalence of transmitted HIV-1 drug resistance in the UK between 2002 and 2005,5 which was mainly attributed to changes in testing guidelines and the wider use of regimens that suppress viral concentrations to below infectious levels. Since then, more potent and better tolerated antiretroviral drugs have been introduced, and the proportion of patients achieving viral suppression has continued to increase.6 This suggests that transmitted HIV-1 drug resistance may have declined even further and could eventually fall below levels in which universal testing before antiretroviral therapy is cost effective,7 an important issue at a time when the cost of HIV-1 management is being scrutinised.8 This paper examines recent time trends in transmitted drug resistance in HIV-1 infections probably acquired in the UK.
Methods
Resistance data
The UK HIV Drug Resistance Database, described in detail elsewhere,9 was established in 2001 and collects the majority of genotypic resistance tests done within the UK as part of routine clinical care. The resistance tests analysed in this study used bulk sequencing of the pol gene, encoding at least codons 4-99 of the protease gene and 34-234 of the reverse transcriptase gene, using a variety of inhouse and commercial testing systems. Subtype was assigned centrally using the Rega algorithm.10
Clinical data
We acquired demographic and clinical information by linkage (using pseudonymised identifiers) to the UK Collaborative HIV Cohort Study (UK CHIC),9 which includes patients from 13 of the largest clinics within the UK, and to the HIV and AIDS Patient and the Survey of Prevalent HIV Infections Diagnosed databases, which are coordinated by the Health Protection Agency. When possible, we linked resistance tests done after 2007 to samples on which a recent infection testing algorithm had been conducted as part of a national health surveillance programme11; these tests use antibody avidity assays to classify infections as either recent (probably occurring in the previous five months) or non-recent;12 and clinical, laboratory, or historical information to reduce the false recent rate.
Tests included in analysis and definition of drug resistance
The UK has several parallel and largely non-overlapping HIV-1 epidemics with different levels and patterns of resistance to antiretroviral therapy.13 14 To simplify the understanding of temporal trends, we limited this analysis to subtype B viral infections, so it is not intended to generate nationally representative results. This epidemic was seeded by around six introductions to the UK in the early to mid-1980s15 and is largely confined to men infected through homosexual exposure, of whom 83% are estimated to have acquired infection within the UK.16
We identified the first resistance test for all patients older than 16 years who had not yet received antiretroviral therapy at the time of sampling, up to the end of 2009. Patients with an undetectable viral load (<50 copies/mL) were excluded; such levels may indicate unrecorded treatment use. Since guidelines in 2001 first recommended that resistance tests be performed for all patients who had not received antiretroviral therapy,3 we excluded tests conducted before 2002.
Transmitted HIV-1 drug resistance was defined as one or more mutations from the surveillance list recommended by the World Health Organization.17 We used the Stanford HIVdb algorithm 6.0.11 (29 Mar 2011) to examine susceptibility to antiretroviral drugs, and reported low level resistance or greater. Intermediate or high level resistance was considered to reflect a substantial loss in susceptibility. We assessed temporal trends in terms of the date of the resistance test sample rather than the date of the patient’s infection, which is generally not known.
Statistical methods
Confidence intervals for proportions were calculated using a 95% Wilson confidence interval for binomially distributed data. We analysed the patterns of trends over continuous time using both linear and piecewise linear logistic regression with a flexible choice of a single inflexion point calculated using least squares optimisation; we selected the model with the best fit according to Akaike’s information criterion. The trends for codons were reported if a mutation had an overall prevalence of more than 0.3% as well as other mutations for nucleos(t)ide reverse transcriptase with a strong effect on phenotype (K65R, K70E, L74I/V, Y115F). We examined differences between the prevalence of resistance in recent and non-recent infections tests using the χ2 test. All statistical analyses were conducted in Stata/IC 11.2 software.
Results
Population characteristics
We analysed 14 583 patients who were antiretroviral therapy naive, infected with a subtype B virus, and whose first drug resistance test was conducted between January 2002 and December 2009. Of these patients, 10 173 (70%) were white, 995 (7%) were black, 711 (5%) had a known other ethnicity, and 2704 (19%) had an unknown ethnicity. The median age at diagnosis was 36 years (interquartile range 30 to 42). Of the patients analysed, 10 288 (71%) were men who had sex with men, 1275 (9%) had a heterosexual exposure source, 313 (2%) had a known other exposure source, and 2707 (19%) had an unknown exposure source. The median number of days between HIV-1 diagnosis and resistance sample was 22 (interquartile range five to 358). The median CD4 count at the time of testing, available from 11 219 (76.9%) patients, was 408×106 cells/L (interquartile range 271×106 to 560×106).
The number of resistance tests conducted per year increased over time, reflecting a rise in the number of new diagnoses among men who have sex with men (fig 1). The decline between 2008 and 2009 is mainly due to an increase in the proportion of tests where the patient’s status regarding antiretroviral therapy exposure was uncertain (15.6% in 2008 to 32.0% in 2009), probably as a result of a reporting lag with demographic and clinical datasets.
Fig 1 Prevalence of transmitted drug resistance over time, by antiretroviral drug class. Bar=95% confidence interval
Trends in transmitted drug resistance
Samples from 1654 (11.3%, 95% confidence interval 10.8% to 11.9%) patients had one or more mutations associated with transmitted HIV-1 drug resistance. Of these samples, 1009 (6.9%, 6.5% to 7.3%), 604 (4.1%, 3.8% to 4.5%), and 319 (2.2%, 2.0% to 2.4%) had one or more mutations associated with nucleos(t)ide reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, and protease inhibitors, respectively. Of these samples, 1426 (9.8%, 9.3% to 10.3%) had single class resistance, 175 (1.2%, 1.0% to 1.4%) had dual class resistance, and 52 (0.4%, 0.3% to 0.5%) had triple class resistance; dual and triple class resistance have remained at a similar prevalence since 2005.
The previously reported5 decline in the prevalence of transmitted HIV-1 drug resistance for any class of antiretroviral drug was observed to continue from 15.5% in 2002 until around January 2007 (95% confidence interval January 2006 to February 2008; odds ratio 0.88 per year (95% confidence interval 0.84 to 0.92); fig 1). However, between 2007 and 2009, we saw a non-significant increase from 9.6% to 10.9% (odds ratio 1.06, 0.97 to 1.17; P=0.21). We saw non-significant increases in resistance to nucleos(t)ide reverse transcriptase inhibitors (from 5.4% in 2007 to 6.6% in 2009; P=0.24) and protease inhibitors (1.5% to 2.1%; P=0.12). Inflection points were also identified, which gave a significantly better fit for mutations associated with nucleos(t)ide reverse transcriptase inhibitors (February 2007 (January 2006 to March 2008); second piece odds ratio 1.08 (0.95 to 1.22)) and protease inhibitors (June 2008 (August 2007 to June 2009); 1.69 (0.86 to 3.31)). The prevalence of mutations associated with non-nucleoside reverse transcriptase inhibitors remained stable at around 3.6% with no evidence of non-linearity (odds ratio 0.96 (0.92 to 1.00)).
The table displays the resistance trends over time for key individual codons with prevalence greater than 0.3%. The trends in HIV-1 drug resistance associated with nucleos(t)ide reverse transcriptase inhibitors largely reflect the most common mutations within this drug class, namely, the T215 revertant18 mutations (I/S/C/D/V/E), K219Q/E/N/R, and M41L. These mutations are either reversions or a subset of the thymidine analogue mutations that develop under regimens containing either stavudine or zidovudine. The finding of a levelling off in the prevalence of thymidine analogue mutations is paradoxical, in the light of the dramatic fall in the use of stavudine and zidovudine. For example, in the UK CHIC study, the proportion of treatment regimens that included stavudine dropped from 29.4% to 0.8% between 2002 and 2009, while the proportion that included zidovudine dropped from 47.9% to 8.8% over the same period. Stavudine and zidovudine have largely been replaced by tenofovir and abacavir,3 but signature mutations for these drugs (such as K65R) are still rare in patients with HIV-1. L90M mutations have increased in prevalence since 2007, despite the near cessation in the use of saquinavir, nelfinavir, and indinavir, which are first generation protease inhibitors that select for this mutation (their use in the UK as part of a drug regimen fell from 20.6% in 2002 to 4.6% in 2009), although L90M has broad cross resistance effects to the protease inhibitor class.19
Time trends of selected mutations with prevalence greater than 0.3%. Data are no (%) of samples with mutation unless stated otherwise
| Mutation | Year (no of samples) | |||||||
|---|---|---|---|---|---|---|---|---|
| 2002 (n=547) | 2003 (n=766) | 2004 (n=1364) | 2005 (n=2054) | 2006 (n=2455) | 2007 (n=2525) | 2008 (n=2651) | 2009 (n=2221) | |
| Protease inhibitors | ||||||||
| M46I/L | 12 (2.19) | 9 (1.17) | 13 (0.95) | 17 (0.83) | 14 (0.57) | 14 (0.55) | 15 (0.57) | 13 (0.59) |
| V82A/T/F/S/C/M | 9 (1.65) | 10 (1.31) | 19 (1.39) | 12 (0.58) | 7 (0.29) | 8 (0.32) | 3 (0.11) | 7 (0.32) |
| L90M | 8 (1.46) | 9 (1.17) | 20 (1.47) | 24 (1.17) | 18 (0.73) | 12 (0.48) | 16 (0.60) | 21 (0.95) |
| Non-nucleoside reverse transcriptase inhibitors | ||||||||
| K103N/S | 21 (3.84) | 28 (3.66) | 40 (2.93) | 57 (2.78) | 67 (2.73) | 76 (3.01) | 89 (3.36) | 62 (2.79) |
| Y181C/I/V | 5 (0.91) | 9 (1.17) | 10 (0.73) | 17 (0.83) | 11 (0.45) | 13 (0.51) | 12 (0.45) | 9 (0.41) |
| G190A/S/E | 7 (1.28) | 7 (0.91) | 10 (0.73) | 14 (0.68) | 9 (0.37) | 8 (0.32) | 8 (0.3) | 3 (0.14) |
| Nucleos(t)ide reverse transcriptase inhibitors | ||||||||
| M41L | 27 (4.94) | 17 (2.22) | 46 (3.37) | 49 (2.39) | 49 (2.00) | 41 (1.62) | 51 (1.92) | 33 (1.49) |
| K65R | 1 (0.18) | 3 (0.39) | 3 (0.22) | 1 (0.05) | 1 (0.04) | 3 (0.12) | 3 (0.11) | 0 |
| D67N/G/E | 12 (2.19) | 9 (1.17) | 20 (1.47) | 21 (1.02) | 21 (0.86) | 13 (0.51) | 13 (0.49) | 9 (0.41) |
| K70E* | 6 (1.10) | 9 (1.17) | 4 (0.29) | 5 (0.24) | 6 (0.24) | 2 (0.08) | 4 (0.15) | 3 (0.14) |
| L74I/V* | 5 (0.91) | 3 (0.39) | 3 (0.22) | 5 (0.24) | 4 (0.16) | 3 (0.12) | 2 (0.08) | 1 (0.05) |
| Y115F* | 0 | 2 (0.26) | 1 (0.07) | 1 (0.05) | 1 (0.04) | 0 | 1 (0.04) | 2 (0.09) |
| M184I/V | 11 (2.01) | 13 (1.70) | 15 (1.1) | 15 (0.73) | 13 (0.53) | 14 (0.55) | 9 (0.34) | 7 (0.32) |
| L210W | 12 (2.19) | 7 (0.91) | 15 (1.1) | 13 (0.63) | 12 (0.49) | 9 (0.36) | 12 (0.45) | 8 (0.36) |
| T215Y/F | 12 (2.19) | 9 (1.17) | 9 (0.66) | 17 (0.83) | 7 (0.29) | 3 (0.12) | 2 (0.08) | 2 (0.09) |
| T215I/S/C/D/V/E | 35 (6.40) | 35 (4.57) | 59 (4.33) | 82 (3.99) | 86 (3.50) | 82 (3.25) | 102 (3.85) | 87 (3.92) |
| K219Q/E/N/R | 10 (1.83) | 19 (2.48) | 31 (2.27) | 34 (1.66) | 34 (1.38) | 33 (1.31) | 41 (1.55) | 39 (1.76) |
*Included because these are major mutations to recommended first line antiretroviral drugs.
Drug susceptibility
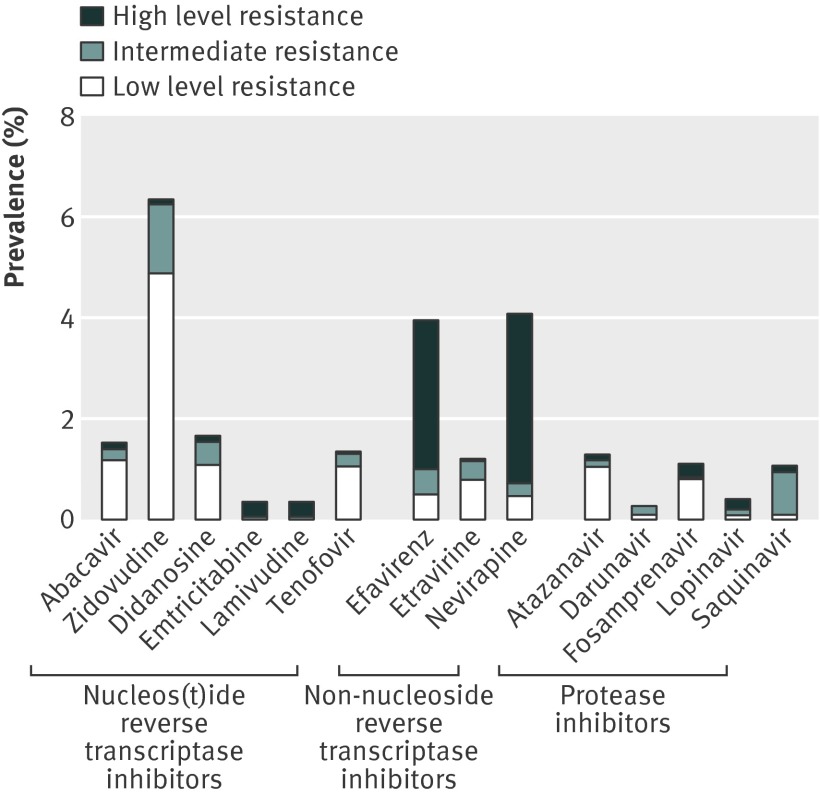
Figure 2 shows the predicted susceptibility of HIV-1 from samples collected in 2009 to currently recommended first line antiretroviral drugs3 and second generation antiretroviral drugs darunavir and etravirine. The association between genotypic mutations and the phenotypic susceptibility to antiretroviral drugs is complex, so although T215 revertants are considered by the Stanford HIVdb algorithm to result in only low level resistance to nucleos(t)ide reverse transcriptase inhibitors, they do confer a low genetic barrier to the development of high level resistance to this drug class.
Fig 2 Predicted susceptibility to antiretroviral drugs
The prevalence of intermediate or high level resistance to nucleos(t)ide reverse transcriptase inhibitors other than zidovudine was less than 0.9%, reflecting the low frequency of multiple mutations in thymidine analogues. There was a comparatively high level of reduced susceptibility to recommended first line regimens of the non-nucleoside reverse transcriptase inhibitors efavirenz and nevirapine (3.7%). However, etravirine, a non-nucleoside reverse transcriptase inhibitor, displayed very low levels of resistance (0.4%), reflecting the lack of predicted effect of K103N. The potency of modern drugs from the protease inhibitor class was high (only 25 (1.1%) patients had substantially reduced susceptibility to any protease inhibitor), owing to the rarity of multiple protease mutations. These findings indicate that potent first line regimens can still be constructed from the original three drug classes for almost all patients.
Prevalence of resistance in recently infected patients
Between 2007 and 2009, 742 samples were linked to a recent infection test result, of which 171 (23.0%) were classified as recent. The overall prevalence of resistance did not differ significantly (P=0.66) between recent samples (20, 11.7%) or non-recent samples (60/571, 10.5%). Furthermore, we detected no significant differences in the prevalence of resistance by individual drug class (results not shown).
Discussion
Interpretation
The previously observed decline in the prevalence of transmitted drug resistance in subtype B viruses seems to have been reversed for mutations associated with nucleos(t)ide reverse transcriptase inhibitors and protease inhibitors, despite an increase in the proportion of patients on antiretroviral therapy who are virologically suppressed (from 62% in 2000 to 84% in 20076). The most frequent mutations were T215 revertants, which may be transmitted as such or evolve from a virus harbouring a T215F or T215Y mutation.19 These and other mutations are associated with a significantly higher risk of virological failure than wild type genotypes.1 17 The prevalence of transmitted thymidine analogue mutations remained moderately high, despite a marked shift away from the prescribing in the UK of drugs that select for these mutations.
One plausible explanation for this paradox is the onward transmission of resistant viruses from people who have not received antiretroviral therapy before and who were themselves infected with a resistant virus. This hypothesis is supported by a previous phylogenetic analysis of UK subtype B sequences, which described five transmission clusters comprised exclusively of patients with resistance mutations who are antiretroviral therapy naive.20 It was postulated that an increasingly greater proportion of transmitted HIV-1 drug resistance could originate from antiretroviral therapy naive lineages, and that there could ultimately be a limit in the decline of transmitted HIV-1 drug resistance.
The first factor contributing to this possible limit is that people with undiagnosed HIV could disproportionally21 spread the epidemic, since they are more infectious in the period immediately after infection.22 Some studies23 24 (but not all25) have also shown that undiagnosed patients with HIV have more sexual partners than diagnosed patients. Furthermore, evidence is emerging that transmitted resistant viruses are more persistent than originally thought. The fitness cost (relative to wild type virus) of certain mutations, such as the T215 revertants and K103N, has been shown to be marginal in laboratory studies.26 27 Jain and colleagues provided clinical confirmation of this finding in a series of patients infected with resistant virus and who had two or more resistance tests before they started antiretroviral therapy.28 With the exception of the M184V mutation, which is highly replicatively deficient, all groups of transmitted mutations persisted beyond at least three years in the majority of patients.
A second possible explanation for the continued prevalence of thymidine analogue mutations is that the use of tenofovir and abacavir are maintaining the prevalence of such mutations in patients who have received antiretroviral therapy despite the decline in the use of zidovudine and stavudine. Further phylogenetic research could shed light on the transmission dynamics of these mutations.
Comparison with other studies
Two recent studies have reported on time trends in transmitted HIV drug resistance. In a study conducted in 20 European countries between 2002 and 2006,29 Vercauteren and colleagues found a small, linear decline in levels of nucleos(t)ide reverse transcriptase inhibitor and protease inhibitor resistance; non-nucleoside reverse transcriptase inhibitor resistance was observed to increase followed by a decrease between 2004 and 2006. Bartmeyer and colleagues30 performed a similar analysis of a German seroconverter cohort between 1996 and 2007. In more recent years, resistance to nucleos(t)ide reverse transcriptase inhibitors seemed to be stable and resistance to non-nucleoside reverse transcriptase inhibitors seemed to increase, although clear patterns are difficult to discern due to the relatively small sample size.
Study limitations
Our study has several limitations. Firstly, the analysis is based on resistance found at date of sample rather than date of infection. Since viral quasi-species harbouring resistance mutations may revert to, or be overgrown by, virus without the mutations,19 29 the true level of transmitted HIV drug resistance may have been under-estimated in this analysis. Also, the degree of this bias will be affected by the average time between HIV-1 infection and diagnosis, which may have changed over time. Furthermore, the diagnosis delay could mask the underlying trend in the prevalence of transmitted resistance by date of infection. However, we found no difference in the prevalence of resistance between recent and non-recent infections, in the subset of patients in which this analysis was possible.
Secondly, the genotypic data analysed were generated by population sequencing with a limit of sensitivity of approximately 15 to 25%.31 32 Our estimates of the prevalence of transmitted HIV drug resistance may therefore be biased downwards. This will have also biased the type of mutations observed, with persistent mutations appearing to be more prevalent than those which rapidly become undetectable such as K65R or M184V. We do not consider this limitation to be a major concern, because the main objective of our study was to examine changes in the prevalence of transmitted HIV drug resistance over time.
Another limitation was that our method of classifying treatment status could have resulted in misclassification bias if some patients who had received antiretroviral therapy were included in the analysis. Previous research by the UK HIV Drug Resistance Database has suggested that this effect could distort trends if there is misclassification in 4% or more of the samples analysed.33 Finally, as the prevalence of transmitted HIV drug resistance in subtype B viruses, the focus of this analysis, is known to be higher than that observed in other subtypes,34 our findings are not generalisable to the UK epidemic as a whole, although coverage of patients infected with subtype B virus is high.
Conclusions and policy implications
Finally, we consider the clinical implications of our main conclusion that resistant lineages may have become fixed in the circulating viral pool. This concept, if confirmed to be correct, will apply universally, particularly in countries where first generation nucleos(t)ide reverse transcriptase inhibitors continue to be used, and underscores the importance of sentinel surveillance. In terms of the UK (and probably other well resourced countries), the detectable mutations that tend to be transmitted should have little effect on nucleos(t)ide reverse transcriptase inhibitors currently used in first line regimens, that is, abacavir, tenofovir, lamivudine, and emtricitabine. Non-nucleoside reverse transcriptase inhibitors used in first line regimens are of greater concern, with approximately 4% of patients being infected with viruses with reduced susceptibility to efavirenz and nevirapine. Previous models7 have suggested that baseline resistance testing remains cost effective at the levels observed in this study. Therefore, our findings argue that testing at HIV diagnosis and continued monitoring should remain the standard of care.
What is already known on this topic
Transmitted HIV drug resistance can affect therapy success
Some resistance mutations could persist more than others in the absence of selective drug pressure
A 2007 paper has shown a reduction in transmitted drug resistance in the UK since 2005
What this study adds
Transmitted drug resistance is no longer declining in UK, and evidence suggests a sustained epidemic that is resistant to nucleos(t)ide reverse transcriptase inhibitors, irrespective of previous antiretroviral therapy use
Susceptibility to antiretroviral therapy remains relatively high, and potent first line regimens can still be constructed from the original three drug classes for almost all patients
The UK Collaborative Group on HIV Drug Resistance is a collaboration between the UK HIV Drug Resistance Database; UK CHIC; Health Protection Agency HARS; and participating academic centres, clinics, and laboratories.
Analysis/writing group: David Dolling, Caroline Sabin, Valerie Delpech, Erasmus Smit, Anton Pozniak, David Asboe, Andrew Leigh Brown, Duncan Churchill, Ian Williams, Anna Maria Geretti, Andrew Phillips, Nicola Mackie, Gary Murphy, Hannah Castro, Deenan Pillay, Patricia Cane, David Dunn. David Dolling is the guarantor.
Steering Committee: Celia Aitken, Gartnavel General Hospital, Glasgow; David Asboe, Anton Pozniak, Chelsea and Westminster Hospital, London; Clare Booth, Royal Free NHS Trust, London; Patricia Cane, Health Protection Agency, Porton Down; Hannah Castro, Jonathan Crofts, David Dunn (co-chair), David Dolling, Esther Fearnhill, Kholoud Porter, MRC Clinical Trials Unit, London; David Chadwick, South Tees Hospitals NHS Trust, Middlesbrough; Duncan Churchill, Brighton and Sussex University Hospitals NHS Trust; Duncan Clark, St Bartholomew’s and The London NHS Trust; Simon Collins, HIV i-Base, London; Valerie Delpech, Health Protection Agency, Centre for Infections, London; Anna Maria Geretti, University of Liverpool; David Goldberg, Health Protection Scotland, Glasgow; Antony Hale, Leeds Teaching Hospitals NHS Trust; Stéphane Hué, University College London; Steve Kaye, Imperial College London; Paul Kellam, Wellcome Trust Sanger Institute and UCL Medical School; Linda Lazarus, Expert Advisory Group on AIDS Secretariat, Health Protection Agency, London; Andrew Leigh-Brown, University of Edinburgh; Nicola Mackie, Imperial NHS Trust; Chloe Orkin, St. Bartholomew’s Hospital, London; Philip Rice, St George’s Healthcare Trust, London; Deenan Pillay (co-chair), Andrew Phillips, Caroline Sabin, University College London Medical School; Erasmus Smit, Health Protection Agency, Birmingham Heartlands Hospital; Kate Templeton, Royal Infirmary of Edinburgh; Peter Tilston, Manchester Royal Infirmary; William Tong, Guy’s and St. Thomas’ NHS Foundation Trust, London; Ian Williams, Mortimer Market Centre, London; Hongyi Zhang, Addenbrooke’s Hospital, Cambridge; Mark Zuckerman, King’s College Hospital, London.
Centres contributing data: Clinical Microbiology and Public Health Laboratory, Addenbrooke’s Hospital, Cambridge (Jane Greatorex); HIV/GUM Research Laboratory, Chelsea and Westminster Hospital, London (Adrian Wildfire); Guy’s and St. Thomas’ NHS Foundation Trust, London (Siobhan O’Shea, Jane Mullen); HPA – Public Health Laboratory, Birmingham Heartlands Hospital, Birmingham (Erasmus Smit); HPA London (Tamyo Mbisa); Imperial College Health NHS Trust, London (Alison Cox); King’s College Hospital, London (Richard Tandy); Medical Microbiology Laboratory, Leeds Teaching Hospitals NHS Trust (Tony Hale, Tracy Fawcett); Specialist Virology Centre, Liverpool (Mark Hopkins, Lynn Ashton); Department of Clinical Virology, Manchester Royal Infirmary, Manchester (Peter Tilston); Department of Virology, Royal Free Hospital, London (Clare Booth, Ana Garcia-Diaz); Edinburgh Specialist Virology Centre, Royal Infirmary of Edinburgh (Jill Shepherd); Department of Infection and Tropical Medicine, Royal Victoria Infirmary, Newcastle (Matthias L Schmid, Brendan Payne); South Tees Hospitals NHS Trust, Middlesbrough (David Chadwick); St George’s Hospital, London (Phillip Hay, Phillip Rice, Mary Paynter); Department of Virology, St Bartholomew’s and The London NHS Trust (Duncan Clark, David Bibby); Molecular Diagnostic Unit, Imperial College, London (Steve Kaye); University College London Hospitals (Stuart Kirk); West of Scotland Specialist Virology Lab Gartnavel, Glasgow (Alasdair MacLean, Celia Aitken, Rory Gunson).
Coordinating centre: Medical Research Council Clinical Trials Unit, London (Kate Coughlin, Jonathan Crofts, David Dolling, David Dunn, Esther Fearnhill, Kholoud Porter).
Funding: This work was supported by the UK Medical Research Council (grant G0900274) and the European Community’s 7th framework programme (FP7/2007-2013) under the Collaborative HIV and Anti-HIV Drug Resistance Network (CHAIN; project 223131).
Competing interests: All authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare: support from the UK Medical Research Council and the European Community’s 7th framework programme; no financial relationships with any organisations that might have an interest in the submitted work in the previous 3 years; and no other relationships or activities that could appear to have influenced the submitted work.
Ethical approval: This study was approved by the UK multicentre research ethics committee and relevant local research ethic committees.
Data sharing: No additional data available.
Cite this as: BMJ 2012;345:e5253
References
- 1.Wittkop L, Günthard HF, de Wolf F, Dunn D, Cozzi-Lepri A, de Luca A, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis 2011;11:363-71. [DOI] [PubMed] [Google Scholar]
- 2.Health Protection Agency. HIV in the United Kingdom: 2010 report. Health Protection Report 2010;4:47. [Google Scholar]
- 3.Gazzard BG on behalf of the BHIVA Treatment Guidelines Writing Group. British HIV Association guidelines for the treatment of HIV-1-infected adults with antiretroviral therapy 2008. HIV Med 2008;9:563-608. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch MS, Günthard HF, Schapiro JM, Brun-Vézinet F, Clotet B, et al. Antiretroviral drug resistance testing in adult HIV-1 infection—2008 recommendations of an IAS-USA panel. Clin Infect Dis 2008;47:266-85. [DOI] [PubMed] [Google Scholar]
- 5.UK Collaborative Group on HIV Drug Resistance, UK Collaborative HIV Cohort Study, UK Register of HIV Seroconverters. Evidence of a decline in transmitted HIV-1 drug resistance in the United Kingdom. AIDS 2007;21:1035-9. [DOI] [PubMed] [Google Scholar]
- 6.Bansi L, Sabin C, Delpech V, Hill T, Fisher M, Walsh J, et al. Trends over calendar time in antiretroviral treatment success and failure in HIV clinic populations. HIV Med 2010;11:432-8. [DOI] [PubMed] [Google Scholar]
- 7.Sax PE, Islam R, Walensky RP, Losina E, Weinstein MC, Goldie SJ, et al. Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin Infect Dis 2005;41:1316-23. [DOI] [PubMed] [Google Scholar]
- 8.Cairns G. UK HIV treatment guidelines: “thrifty changes.”. 2011. www.natap.org/2011/newsUpdates/052611_06.htm.
- 9.The UK Collaborative HIV Cohort (UK CHIC) Study Group. The creation of a large UK-based multicentre cohort of HIV-infected individuals: the UK Collaborative HIV Cohort (UK CHIC) study. HIV Med 2004;5:115-24. [DOI] [PubMed] [Google Scholar]
- 10.De Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 2005;21:3797-800. [DOI] [PubMed] [Google Scholar]
- 11.Health Protection Agency. Recent Infection Testing Algorithm (RITA)/HIV incidence. 2012. www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/HIV/HIVIncidence/.
- 12.Murphy G, Parry JV. Assays for the detection of recent infection with human immunodeficiency virus type 1. Eurosurveillance 2008;13:4-10. [PubMed] [Google Scholar]
- 13.Presanis AM, Gill ON, Chadborn TR, Hill C, Hope V, Logan L, et al. Insights into the rise in HIV infections, 2001 to 2008: a Bayesian synthesis of prevalence evidence. AIDS 2010;24:2849-58. [DOI] [PubMed] [Google Scholar]
- 14.Chilton DN, Castro H, Lattimore S, Harrison LJ, Fearnhill E. HIV type-1 drug resistance in antiretroviral treatment-naive adults infected with non-B subtype virus in the United Kingdom. Antiviral Therapy 2010;15:985-91. [DOI] [PubMed] [Google Scholar]
- 15.Hué S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc Natl Acad Sci U S A 2005;102;4425-9. [DOI] [PMC free article] [PubMed]
- 16.Health Protection Agency. Testing times; HIV and other sexually transmitted infections in the United Kingdom. Health Protection Agency, 2007.
- 17.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE 2009;4:e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pingen M, Nijhuis M, de Bruijn JA, Boucher CAB, Wensing AMJ. Evolutionary pathways of transmitted drug-resistant HIV-1. J Antimicrob Chemother 2011;66:1467-80. [DOI] [PubMed] [Google Scholar]
- 19.Hertogs K, Bloor S, Kemp SD, Van den Eynde C, Alcorn TM, Pauwels R, et al. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 2000;14:1203-10. [DOI] [PubMed] [Google Scholar]
- 20.Hué S, Gifford R, Dunn D, Fearnhill E, Pillay D. Demonstration of sustained drug-resistant HIV-1 lineages circulating amongst treatment-naïve individuals. J Virol 2009;83:2645-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner BG, Roger M, Moisi DD, Oliveira M, Hardy I, Turgel R, et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS 2008;22:2509-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilcher CD, Joaki G, Hoffman IF, Martinson FEA, Mapanje C, Stewart PW, et al. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS 2007;21:1723-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heijman RLJ, Geskus G, Davidovich U, Coutinho R, Prins M, Stolte I. Changes in sexual behaviour after HIV diagnosis among MSM who seroconverted before and after the introduction of ART. Eighteenth Conference on Retroviruses and Opportunistic Infections, 2011.
- 24.Vallabhaneni S, Loeb L, Bragg L, McConnell J, Hartogensis W, Grant R, et al. Seroadaptive tactics adopted by HIV-positive MSM can contribute to profound and sustained reductions in HIV transmission risk following HIV diagnosis. Eighteenth Conference on Retroviruses and Opportunistic Infections, 2011.
- 25.Williamson LM, Dodds JP, Mercey DE, Hart GJ, Johnson AM. Sexual risk behaviour and knowledge of HIV status among community samples of gay men in the UK. AIDS 2008;22:1063-70. [DOI] [PubMed] [Google Scholar]
- 26.García-Lerma JG, Nidtha S, Blumoff K, Weinstock H, Heneine W. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. PNAS 2001;98:13907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkarni R, Miller MD, White K. The contribution of HIV-1 reverse transcriptase resistance mutations to antiviral synergy and replication fitness in vitro. Sixth Conference on HIV Pathogenesis, Treatment and Prevention, 2011. http://pag.ias2011.org/Abstracts.aspx?AID=4454.
- 28.Jain V, Sucupira MC, Bacchetti P, Hartogensis W, Diaz RS, Kallas EG. Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis 2011;203:1174-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vercauteren J, Wensing AM, van de Vijver DA, Albert J, Barlotta C, et al. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J Infect Dis 2009;200:1503-8. [DOI] [PubMed] [Google Scholar]
- 30.Bartmeyer B, Kuecherer C, Houareau C, Werning J, Keeren K, Somogyi S, et al. Prevalence of transmitted drug resistance and impact of transmitted resistance on treatment success in the German HIV-1 seroconverter cohort. PLoS ONE 2010;5:e12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant RM, Kuritzkes DR, Johnson VA, Mellors JW, Sullivan JL, Swanstrom R, et al. Accuracy of the TRUGENE HIV-1 genotyping kit. J Clin Microbiol 2003;41:1586-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuurman R, Brambilla D, de Groot T, Huang D, Land S, Bremer J, et al. Underestimation of HIV type 1 drug resistance mutations: results from the ENVA-2 genotyping proficiency program. AIDS Res Hum Retrovir 2002;18:243-8. [DOI] [PubMed] [Google Scholar]
- 33.Castro H, Pillay D, Sabin C, Dunn DT. Effect of misclassification of antiretroviral treatment status on the prevalence of transmitted HIV-1 drug resistance. BMC Med Res Methodol 2012. [forthcoming]. [DOI] [PMC free article] [PubMed]
- 34.Chilton DN, Castro H, Lattimore S, Harrison LJ, Fearnhill E. HIV type-1 drug resistance in antiretroviral treatment-naive adults infected with non-B subtype virus in the United Kingdom. Antivir Ther 2010;15:985-91. [DOI] [PubMed] [Google Scholar]