Abstract
Targeting antigens directly to DCs through anti-DC receptor antibody fused to antigen proteins is a promising approach to vaccine development. However, not all antigens can be expressed as a recombinant antibody directly fused to a protein antigen. Here, we show that non-covalent assembly of antibody - antigen complexes, mediated by interaction between dockerin and cohesin domains from cellulose-degrading bacteria, can greatly expand the range of antigens for this DC-targeting vaccine technology. Recombinant antibodies with a dockerin domain fused to the antibody heavy chain C-terminus are efficiently secreted by mammalian cells, while many antigens not secreted as antibody fusion proteins are readily expressed as cohesin directly fused to antigen either via secretion from mammalian cells, or as soluble cytoplasmic E. coli products. These form very stable and homogeneous complexes with antibody fused to dockerin. In vitro, these complexes can efficiently bind to human DC receptors followed by presentation to antigen-specific CD4+ and CD8+ T cells. Low doses of the HA1 subunit of Influenza hemagglutinin conjugated through this means to anti-Langerin antibodies elicited Flu HA1-specific antibody and T cell responses in mice. Thus, the non-covalent assembly of antibody and antigen through dockerin and cohesin interaction provides a useful modular strategy for developing and testing prototype vaccines for eliciting antigen-specific T and B cell responses, particularly when direct antibody fusions to antigen cannot be expressed.
Introduction
Dendritic cells (DCs) are professional antigen presenting cells that play a key role in inducing and regulating antigen-specific T and B cell immune responses. DCs capture antigens, then process and present them to T cells as peptides bound to both major histocompatibility complex (MHC) class I and II (1–3). As the key players orchestrating immunity, DCs are a natural focus for immune therapy. A strategy to exploit DCs for vaccination is to deliver antigens directly to DCs using monoclonal antibodies (mAbs) directed against specific endocytic DC cell-surface receptors, resulting in processing and presentation of antigenic peptides to T cells. Studies in mice have demonstrated that such DC-targeting vaccines can result in potent antigen-specific CD4+ and CD8+ T cell responses. For example, low doses of anti-LOX-1 or anti-DEC-205 mAbs fused to protein or peptide antigens mediated presentation via MHC class I (4) and II (5) and induced protective T cell immunity (5). In another study, effective humoral responses to DC-targeted antigen via CLEC-9A were elicited without adjuvant (6). The ability to deliver antigens to specific DC subtypes and via specific receptors (7, 8) is a powerful new approach to dissecting their roles in immune responses, and this has exciting implications for vaccine development strategies. However, the full potential of this technology will only be realized when a wider range of both targeting antibodies and linked antigens become available. Studies in vitro with DC-targeting vaccines have used antigen chemically cross-linked to antibodies (9) or actual prototype vaccines -recombinant antibody directly fused to antigen, e.g., melanoma antigen pmel17 fused to the heavy (H) chain carboxyl (C) -terminus of a human mAb against mannose receptor (10) or HIV Gag p24 antigen similarly fused to a mouse antibody against human DEC-205 (11). In this work, we unfortunately find that many antigens, when fused to the mAb H chain C-terminus, prevent efficient secretion of the recombinant antibody from mammalian cells. We have circumvented this problem by developing separate recombinant antibody fused to dockerin and protein antigen fused to cohesin. Dockerin and cohesin are bacterial protein domains that interact non-covalently with high affinity and specificity and serve to assemble a cellulose-degrading macromolecular structure called the cellulosome (12). This supermolecular structure is formed via dockerin modules appended to cellulose-degrading catalytic subunits interacting with a protein called scaffoldin, which has multiple cohesin modules interspersed with linker sequences, and is, itself, anchored to cellulose via an integrated cellulose binding domain (13, 14). We show that stable and specific antibody-antigen complexes based on this interaction can be conveniently assembled for delivering antigen in vitro to DCs, permitting DCs to expand antigen-specific CD4+ and CD8+ T cells. Also, such antibody-antigen complexes are effective prototype vaccines in vivo for eliciting humoral and cellular responses in mice.
Materials and Methods
Vectors for expression of recombinant antibody and antigen fusion proteins
Total RNA was prepared from hybridoma cells (RNeasy kit, Qiagen) and used for cDNA synthesis and PCR (SMART RACE kit, BD Biosciences) with supplied 5′ primers and gene-specific 3′ primers (mIgGκ, 5′-GGATGGTGGGAAGATGGATACAGTTGGTGCAGCATC-3′; mIgG1, 5′-GTCACTGGCTCAGGGAAATAGCCCTTGACCAGGCATC-3′; and mIgG2a, 5′-CCAGGCATCCTAGAGTCACCGAGGAGCCAGT-3′). PCR products were cloned (pCR2.1 TA kit, Invitrogen) and characterized by DNA sequencing (Molecular Cloning Laboratories). With the derived sequences for the mouse heavy (H) and light (L) chain variable (V) region cDNAs, specific primers were designed and used in PCR to amplify the signal peptide and V-regions while incorporating flanking restriction sites for cloning into expression vectors encoding downstream human IgGκ or IgG4H regions. The vector for expression of chimeric mVκ-hIgGκ was built by amplifying residues 401–731 of gi|63101937| flanked by Xho I and Not I sites and inserting this into the Xho I – Not I interval of the vector pIRES2-DsRed2 (BD Biosciences). PCR was used to amplify the mAb Vκ region from the initiator codon, appending a proximal Nhe I or Spe I site then CACC to the region encoding, e.g., residue 126 of gi|76779294|, while appending a distal in-frame Xho I site (the anti-DC receptor chimeric L and H chains sequences used in this study are GenBank entries HQ738667, HQ738666, HQ724328, HQ724329, HQ912690, HQ912691, HQ912692, HQ912693, JX002666, JX002667 http://www.ncbi.nlm.nih.gov/nucleotide). The PCR fragment was then cloned into the Nhe I – Not I interval of the above vector. The control hIgGκ sequence corresponds to gi|49257887| residues 26–85 and gi|21669402| residues 67–709. The hIgG4H vector corresponds to residues 12–1473 of gi|19684072| (with S229P and L236E substitutions to stabilize a labile disulphide bond and abrogate residual Fc receptor interaction (15)) inserted between the Bgl II and Not I sites of pIRES2-DsRed2 while adding the sequence 5′-GCTAGCTGATTAATTAA-3′ instead of the stop codon. PCR was used to amplify the mAb VH region from the initiator codon, appending CACC then a Bgl II site, to the region encoding residue 473 of gi|19684072|. The PCR fragment was then cloned into the Bgl II – Apa I interval of the above vector. GenBank entries for the L and H chain sequences for the anti-Langerin mIgG2b antibodies are JX002668, JX002669 for the anti-human Langerin 4C7 mAb, which cross-reacts with mouse Langerin (unpublished), as well as JX002670 and JX002671 for the mAb 2G3 (7). The anti-DCIR rAb is described in (16). A dockerin coding sequence (Doc) from Clostridium thermocellum (C. thermocellum) CelD (gi|40671| residues 1923–2150) flanked by a proximal Nhe I site and a distal Not I site following the stop codon was inserted into the Nhe I - Pac I - Not I interval of each H chain vector. The following antigen coding sequences were also inserted between the H chain vector Nhe I and Not I sites: Flu HA1-1 is CipA protein [C. thermocellum] gi|479126| residues 147–160 preceding gi|126599271| hemagglutinin HA [Influenza A virus (A/Puerto Rico/8/34(H1N1))] residues 18–331 with a P321L change and with 6 C-terminal His residues; Flu HA5-1 is CipA protein [C. thermocellum] gi|479126| residues 147–160 preceding gi|58618438| hemagglutinin HA [Influenza A virus (A/Viet Nam/1203/2004(H5N1))] residues 17–330 and with 6 C-terminal His residues; Flu HA5-0 is CipA protein [C. thermocellum] gi|479126| residues 147–160 preceding gi|58618438| hemagglutinin HA [Influenza A virus (A/Viet Nam/1203/2004(H5N1))] residues 17–519 with 6 C-terminal His residues; HIV Gag p24 is gi|119624034| major histocompatibility complex, class II, DR alpha residues 60–75 preceding gi|28872819| Gag p24 [Human immunodeficiency virus 1] residues 133–363; Flu M1 is gi|60458| matrix protein M1 [Influenza A virus (A/WSN/1933(H1N1))] residues 3–252 with V15I and N92S changes; Flex Flu M1 pep is CipA protein [C. thermocellum] gi|479126| residues 147–164 preceding gi|133754191| matrix protein M1 [Influenza A virus (A/Phila/1935(H1N1))] residues 50–72 and residues 58–67 joined by K and followed by C-terminal residues RKNGSGE; Flu M1 pep is gi|133754191| matrix protein M1 [Influenza A virus (A/Phila/1935(H1N1))] residues 50–72 and residues 58–67 joined by K; Flex V1 encoded ASQTPTNTISVTPTNNSTPTNNSNPKPNP; Gad B is NM_001134366.1 Homo sapiens glutamate decarboxylase 2 residues 513–2258; Cyclin B1 peptide 2 is NM_031966.2 residues 797–903; Cyclin B1-v1 is NM_031966.2 residues 475–1476 with 6 His codons appended; PE38KDL is gb|K01397.1| Pseudomonas aeruginosa exotoxin type A residues 856- 1195 1244–1926 followed by AAGGACGAGCTGTAA; Cyclin D1 is NM_053056.2 Homo sapiens Cyclin D1 residues 210–1009; a cohesin coding sequence (Coh), the 7th cohesin domain in CipA protein [C. thermocellum] gi|479126| residues 148–165 preceding gi|479126| residues 1–147 with R21N, E54K, E96K, and S109E changes and a C-terminal T residue. The DocVar1 derivative of the dockerin domain (gi|40671| 2085–2085, AAT to GAC) was made by site-specific mutagenesis (Quickchange kit, Stratagene). A mammalian expression vector for hIgG Fc fusion proteins was engineered as described (17). PCR was used to amplify human alkaline phosphatase (AP) gb|BC009647| residues 133–1581 while adding a proximal in-frame Xho I site and distal 6 C-terminal His residues followed by a TGA codon and a Not I site. This Xho I – Not I fragment replaced the hIgG Fc coding sequence in the above vector. A mammalian expression vector for G.AP fusion protein was constructed by inserting protein G precursor B2 domain gi|124267| residues 295–352 preceded by a Sal I site and followed by distal linker residues encoding GGSGGSGGS and an Xho I site into the Xho I site of the AP vector. A similar mammalian expression vector for the fusion of cohesin with AP (Coh.AP) was made by inserting a fragment (bound by a proximal Sal I site and distal Xho I site) encoding gi|479126| type I cohesin residues 165–312 then 148–169 into the Xho I site. The following antigen-coding sequences replaced AP in the above Xho I – Not I interval: Flu HA1-1 is gb|EF467821.1| residues 85–1025 (with a A290T change) with a proximal Xho I site and distal 5′-CACCATCACCATCACCATTGAGCGGCCGC-3′ sequence (encoding 6 C-terminal His residues, a stop codon, and a Not I site); Flu HA5-1 is gi|58618438| residues 17–330 with 6 C-terminal His residues; Gag p24 is Human immunodeficiency virus 1 gb|ABO61536.1| residues 45–256 with 6 C-terminal His residues. For expression of Coh.antigens in E. coli, PCR was used to amplify the ORF of gi|60458| Flu M1 protein [Influenza A virus (A/WSN/1933(H1N1))] with a V15I change while incorporating a Nhe I site distal to the initiator codon and a Not I site distal to the stop codon. The fragment was inserted into pET-28b (+) (Novagen), placing the Flu M1 ORF in-frame with 6 C-terminal His residues. A pET-28b (+) derivative encoding the N-terminal 169 residue cohesin domain from gi|479126| with R21N, E54K, E96K, and S109E changes was inserted between the Nco I and Nhe I sites and this encoded Coh fusion protein. For expression of Coh.Flu M1 protein, PCR was used to amplify the above Flu M1 residues while incorporating a Nhe I site proximal to the initiator codon and an Xho I site instead of the terminator codon. This fragment was inserted between the Nhe I and Xho I sites of the Coh.His vector. Similar strategies were used to generate expression vectors for His-tagged Coh.Gad B, Coh.Cyclin D1, Coh.Cyclin B1-v1, and Coh.PE38KDL, except with the latter the His-tag was N-terminal to the cohesin domain.
Expression and purification of recombinant antibodies and proteins
Recombinant antibodies and some fusionproteins were produced using the FreeStyle™ 293 Expression System (Invitrogen) according to the manufacturer’s protocol based on 1 mg total plasmid DNA with 1.3 ml 293 Fectin reagent/L of transfection. Production levels of rAb.antigen expression constructs were tested in 5 ml transiently transfected mammalian 293F cell cultures using ~2.5 μg each of the L chain and H chain constructs and the protocol described above. Culture supernatants were harvested after 3 days and analyzed by anti-hIgG Fc ELISA with AffiniPure goat anti-human IgG (H+L) as the capture agent and peroxidase-conjugated AffiniPure goat anti-human IgG, Fcγ fragment-specific antibody as the detection reagent (Jackson ImmunoResearch). Some measurements also used the anti-human Kappa ELISA kit (Bethyl Laboratories). In tests of this protocol, production of secreted rAb was independent of H chain and L chain vector concentrations over a ~2-fold range of each vector DNA concentration (i.e., the system was DNA saturated for each vector over this range). For rAb production, equal amounts of vector encoding the H and L chain were co-transfected. Transfected cells were cultured for 3 days, the culture supernatant was harvested and fresh 293 Freestyle™ media (Invitrogen) with 0.5% penicillin/streptomycin (Sigma) added with continued incubation for 2 days. The pooled supernatants were clarified by filtration, loaded onto a 1 ml HiTrap MabSelect™ column (GE Healthcare), eluted with 0.1 M glycine pH 2.7, neutralized with Tris base and then dialyzed versus Dulbecco’s PBS (DPBS, GIBCO). Endotoxin levels ranged from 0.01 to 0.20 ng/mg for rAbs. Coh.antigens (Coh.Flu HA1-1, Coh.Flu HA5-1, and Coh.Gag p24) were also expressed using the system described above. The culture supernatant (1 L) was loaded onto a 20 ml Q Sepharose column (GE Healthcare) washed with PBS and then eluted with PBS + 1 M NaCl pH 7.4. The eluted fraction was purified by Ni++ chelating chromatography as described below, and then dialyzed versus DPBS, yielding 16.6 mg, 1.0 mg and 1.3 mg, respectively. Endotoxin level was 0.43 ng/mg for Coh. Flu HA1-1. All proteins were analyzed by SDS-PAGE gel and concentrations were based on theoretical extinction coefficient at 280 nm. Coh.Flu M1 was expressed in E. coli strain T7 Express (NEB) grown in Luria broth (Difco) at 37°C with selection for kanamycin resistance (40 μg/ml) and shaking at 200 rounds/min to mid-log growth phase. Then 120 mg/L isopropyl beta-D-1-thiogalactopyranoside (Bioline) was added and after 3 h, the cells were harvested by centrifugation and stored at −80°C. E. coli cells from each l L fermentation were resuspended in 50 ml ice-cold 50 mM Tris, 1 mM EDTA pH 8.0 with 0.2 ml of protease inhibitor Cocktail II (Calbiochem). The cells were sonicated twice on ice for 4 min at setting 18 (Fisher Sonic Dismembrator 60) with a 5 min rest period and then spun at 17,000 r.p.m. (Sorvall SA-600) for 20 min at 4°C. For Coh.Flu M1 purification, the 50 ml cell lysate supernatant was passed through 10 ml of Q Sepharose beads (GE Healthcare), then through a 1 ml S Sepharose column (GE Healthcare) and washed with 50 mM Tris pH 7.4. The flow-through was adjusted to binding buffer with 7.5 ml 160 mM Tris, 40 mM imidazole, 4 M NaCl pH 7.9 and loaded onto a 5 ml HiTrap chelating HP column (GE Healthcare) charged with Ni++. The bound protein was washed with 20 mM NaPO4, 300 mM NaCl, 10 mM imidazole pH 7.6 (buffer A) and eluted with a 10–500 mM imidazole gradient in buffer A. The peak fractions were analyzed by SDS-PAGE gel, pooled and dialyzed versus DPBS, yielding 6.4 mg. Endotoxin level was 2 ng/mg for Coh.Flu M1. Control cohesin protein was purified by breaking 2 L cells in 50 mM MES pH 5.5, then binding the soluble fraction onto a 20 ml QXL column (GE Healthcare) and eluting with a gradient to 1 M NaCl in pH 6.5 and the positive fractions were adjusted to binding buffer and run on a 10 ml Ni++ column, washed with 100 ml 0.5% ASB-14 (Calbiochem) in binding buffer, and then eluted as above. Positive fractions (second peak) were pooled and dialyzed vs. DPBS, yielding 33 mg. Coh.Gad B was isolated from the sonic extract pellet washed with 0.5% Triton-X100 then dissolved in binding buffer with 6 M urea and reacted with 30 mg MPEG-MAL-20K (Nektar) at R.T. o/n and run on a 1 ml Ni++ column as above. The eluted refolded PEG-derivatized protein was dialyzed into DPBS, yielding 9.6 mg/L cells. Coh.Cyclin D1 was isolated from the supernatant after passing it through a 5 ml ANX column (GE Healthcare) after two extractions with triton-X114 (Pierce) then loaded onto a 5 ml Ni++ column and eluted as described above. Fractions containing the protein (~16 mg total) were treated o/n with 10 mg MPEG-MAL-20K reagent at R.T. and dialyzed vs. DPBS yielding 16 mg. Coh.Cyclin B1-v1 was isolated from the sonic extract pellet washed in triton X-100 (Sigma) and dissolved in binding buffer with 6 M urea, reacted with 25 mg MPEG-MAL-20K at R.T. o/n, then diluted 10X in binding buffer and purified via a 5 ml Ni++ column, yielding 2.5 mg of refolded PEG-derivatized protein. Coh.PE38KDL was isolated from the supernatant fraction loaded onto a 20 ml Q XL column and eluted with a 5 volume gradient to 1 M NaCl. Pooled fractions at ~0.4 M NaCl were adjusted to binding buffer and further purified over a 5 ml Ni++ column yielding 7.5 mg. In each case, cohesin fusion proteins were confirmed to bind to rAb.Doc. Flu HA1-Cal04 protein was prepared from 1 L E. coli containing pET28 with gb|GQ117044.1| Influenza A virus (A/California/04/2009(H1N1)) residues 52–996 and appended CATCACCATCACCATCACTGA inserted between the Nhe I and Not I sites. The protein from the insoluble fraction was dissolved in 40 ml 7 M urea in 25 mM NaPO4 pH 6.3 with 5 mM DTT and then 140 mg MPEG-BTC-20K (Nektar) was added at R.T. o/n. This was purified via Ni++ affinity chromatography as described above and pooled fractions were dialyzed into PBS yielding 4.7 mg.
Preparation of dendritic cells and T cells
Apheresis procedures were performed on healthy donors after informed consent was collected. This protocol was approved by the Baylor Research Institute Institutional Review Board. Monocyte-derived IFNα-DCs (IFNα-DCs) and monocyte-derived IL-4-DCs (IL-4-DCs) were prepared from frozen human monocytes from normal donors (HLA-A*0201+) cultured in CellGenix media (CellGenix) with GM-CSF and IFNα, or GM-CSF and IL-4 respectively and 1% penicillin/streptomycin as described previously (18). Autologous CD8+ T cells were negatively or positively selected from peripheral blood mononuclear cells using anti-CD8 magnetic beads (Miltenyi Biotec) or EasySep® enrichment cocktail (StemCell). Complexes between rAb.Doc and Coh.antigen were formed by mixing rAb.Doc fusion proteins with 2 molar equivalents of Coh.antigen fusion proteins in DPBS with Ca++/Mg++. 5×103 IFNα-DCs were loaded with various concentrations of pre-assembled complexes between rAb.Doc and Coh.Flu M1. After 16 h, 2×105 purified autologous CD8+ T cells in complete RPMI 1640 (Invitrogen) medium supplemented with 25 mM HEPES (GIBCO), 2 mM L-glutamine, 1 mM sodium pyruvate, 1% non-essential amino acids, 1% penicillin-streptomycin (all from Sigma) containing 10% heat-inactivated AB serum (Gemini Bioproducts), were added to the culture. IL-2 and IL-7 (20 and 10 units/ml, respectively, R&D) were added on day 2. The frequency of Flu M1-specific CD8+ T cells was measured by staining cells with anti-CD8, anti-CD3 mAbs (BD Biosciences), live/dead fixable aqua dye (Invitrogen) and with MHC class I Flu M1 tetramers (HLA-A*0201-GILGFVFTL)-PE (Beckman Coulter) on day 7 of the culture and then analyzed by flow cytometry. Autologous CD4+ T cells were negatively selected using the EasySep® enrichment cocktail. 5×103 IFNα-DCs were loaded with various concentrations of pre-assembled rAb.Doc-Coh.Flu HA1-1 complexes. After 16 h, 2×105 purified autologous CD4+ T cells labeled with 50 nM carboxyfluorescein succinimidyl ester (CFSE, Molecular Probes) were added to the culture. On day 10, cells were washed and restimulated with 5 μM of 17-mer peptides 43 (LEPDGTIIFEANGNLIA) and 65 (DQKSTQNAINGITNKVN) or 2 μM pools of overlapping (staggered by 4 aa) 15-mer peptides (Mimotopes) in 50% acetonitrile solution (Fluka) within the Flu HA1-1 protein and incubated for 48 h and then the secreted IFNγ was measured in the culture supernatant using BioPlex200 Luminex (BioRad). In parallel, intracellular IFNγ production was assessed after 6 h of restimulation with 5 μM of peptides 43 or 65 or 2 μM pools of overlapping 15-mer peptides within the Flu HA1-1 protein in the presence of brefeldin A (BD Biosciences). Cells were then fixed, permeabilized and stained with anti-CD4, anti-CD3, anti-CD8 and anti-IFNγ mAbs (all from BD Biosciences) and live/dead fixable aqua dye. The IFNγ production of the CFSElow CD4+ T viable cells was assessed by flow cytometry on a FACSCalibur instrument (BD Biosciences).
Dendritic cell activation
1–2×105 IFNα-DCs in complete RPMI containing 10% human AB serum were cultured in 96-well plates with premixed complexes or proteins (1.8 nM) or 100 ng/ml LPS from E. coli for 16 h. DCs were then stained with the indicated antibodies (BD Biosciences) and the expression levels of CD83 and CD86 were measured by flow cytometry on a FACSCantoII instrument (BD Biosciences).
Cohesin.antigen labeling and cell-surface staining
Serial dilutions of anti-CD40.antigen fusion proteins and pre-assembled anti-CD40.Doc-Coh.antigen complexes starting at 138 nM were incubated with 5×105 cells from a stable CHO cell line expressing CD40 receptor in DPBS, 2% BSA, on ice for 30 min. The cells were then washed and incubated with 4 μg/ml of PhycoLink® goat anti-human IgG (Fc-specific)-R-phycoerythrin (Prozyme). The cells were washed again and resuspended in DPBS, 1% PFA and then analyzed by flow cytometry on FACSArray flow cytometer (BD Biosciences). Coh.Flu M1 was treated with EZ-Link NHS-SS-PEO4-Biotin (Pierce) at a compound to protein ratio of 25 in a 100 μg/100 μl reaction in PBS for 1 h at R.T. with rotation. The derivatized proteins were dialyzed versus several changes of DPBS to remove unbound labeling reagent. For flow cytometry analysis, various concentrations of rAb.Doc and biotinylated Coh.Flu M1 were incubated in DBPS with Ca++/Mg++ for 1 h at R.T. Human IFNα-DCs (4.5×105/ml), IL4-DCs (2×105/ml) or PBMCs (107/ml) were resuspended in DBPS and 2% FCS (HyClone) and then rAb.Doc-Coh.Flu M1 complexes were added to the cells and kept on ice for 30 min. Cells were then washed with DBPS and incubated for 30 min at R.T. with streptavidin-phycoerythrin (SA-PE, 1:200 in 100 μl, BD Biosciences), anti-CD3, anti-CD14, CD19 mAbs (all from BD Biosciences) and live/dead fixable aqua dye; and then washed twice with DBPS and 2% FCS and analyzed by flow cytometry on FACSCalibur and FACSCantoII instruments (BD Biosciences).
Analysis of dockerin-cohesin interaction by gel filtration and surface plasmon resonance
Analysis of rAb.Dockerin and Coh.antigen protein interaction by size exclusion chromatography was on a Superdex 200 30/300 column (GE Healthcare) in DPBS with Ca++/Mg++ at R.T. and a 0.35 ml/min flow rate. The column was calibrated with gel filtration standards from Pierce. For determination of affinity and rate constants by surface plasmon resonance, analyses were performed using a SensiQ instrument (ICx Nomadics) at 25°C. A carboxyl sensor surface was modified to covalently couple protein G to both channels. Channel 1 was used to capture dockerin molecules and channel 2 was left as a reference protein G surface to subtract non-specific binding. A concentration of 2 nM anti-DC-ASGPR.Doc was used for capture and at 1 min of contact time the average capture was approximately 233 RU. A serial doubling dilution was prepared of Coh.Gag p24 at a top concentration of 20 nM down to 0.625 nM. The top concentration of 20 nM of Coh.Gag was near saturation of this interaction after 3 min of association. This concentration was allowed to dissociate for 30 min while the other concentrations were regenerated after 4 min. The data was fit in Qdat software (ICx Nomadics).
In vivo targeting of antibody.dockerin – cohesin.antigen complexes for detection of antigen-specific antibody and T cell responses
Human Langerin (huLangerin-DTR) transgenic mice have been previously described (7, 19). All experiments were performed on age- (7–12 week) and sex-matched mice (7). Littermate control, huLangerin-DTR and BALB/c mice were housed in in microisolator cages and fed irradiated food. The University of Minnesota and Baylor Research Institute Institutional Animal Care and Use Committees approved all mouse protocols. The anti-hLang.Doc antibody was mixed for conjugation with Coh.Flu HA1-1 in sterile Hank’s buffer (GIBCO) to a final concentration of 1 μg or 10 μg of conjugate or anti-CD40.Flu HA1-1 in 100 μl. For the control group in some experiments, anti-hLang antibody lacking the dockerin domain was mixed with Coh.Flu HA1-1. The mice were injected intra peritoneal (i.p.) in 250 μl with a 26 gauge needle at day 0 and day 14 and in some experiments at day 21 and at day 30. Blood samples were taken at day -2, day 14, day 21 and in some experiments at day 28. The blood was coagulated 1 h at R.T. and then centrifuged at 8,000 rpm for 8 minutes. The serum was stored at −80°C. To determine antigen-specific antibody titers, ELISA plates were coated with 1 μl/ml purified Influenza A PR8 virus (Charles River), or 0.25 μg hIgG4.Doc protein, or 2 μg/ml Flu HA1-Cal04 protein. Serial dilutions of serum in blocking buffer (TBS, Pierce) were incubated in the wells for o/n at 4°C. After washing, plates were incubated with HRP-conjugated goat anti mouse IgG (Jackson ImmunoResearch) in blocking solution for 2 h at 37°C, then washed and developed with HRP substrate and read at 405 nm. Antibody titer data are plotted on log scales. Assays for isotyping antigen-specific antibodies in the serum used biotinylated goat anti-mouse IgG1, IgG2a, IgG2b, and IgG3 polyclonal reagents and biotinylated anti-mouse IgE monoclonal 23G3 (SouthernBiotech) as detecting reagents, followed by development with NeutrAvidin HRP (Pierce). The capacity of Coh.Flu HA1-1 to present epitopes relevant to protective antibodies was verified by demonstrating that overnight incubation at 4°C with plate-bound Coh.Flu HA1-1, but not Coh.Gag p24, completely depleted hemagglutination inhibition (HAI) activity from sera with titers of 1:1280 and 1:2560. Also, addition of as little as 0.6 μg/ml Coh.Flu HA1-1, but not Coh.Gag p24 at levels as great as 20 μg/ml, completely inhibited HAI activity in sera with titer of 1:1280 against Influenza A PR8 virus. HAI assay was as described (20), except sera were treated with trypsin at 56°C for 30 min, then 0.01 M IKO4 at R.T. for 15 min, followed by addition of 1% glycerol for 15 min at R.T., then dilution into 85% saline with virus at 4°C for 30 min before addition of the red blood cells at 4°C for 45 min before scoring the assay. For the IFNγ ELISPOT assay, mouse splenocytes taken 4 days after the last boost and purified via Lympholyte®-M (Cedarlane) gradients were plated at 2.5×105 cells per assay point in precoated mouse IFNγ ELISPOT plates (PLUS kit, MabTech) with 1 μM peptides in 150 μl RPMI with 5% FCS, 10 mM HEPES, 2 mM GlutaMAX™ (Invitrogen), 50 μM mercaptoethanol and 1% penicillin/streptomycin. Controls were cells without peptide or protein with matching acetonitrile concentrations, as well as cells with 1 μg/ml phytohaemagglutinin. Plates were incubated for 36 h, developed with the supplied HRP reagent (MabTech), and read by ELISPOT reader (ZellNet Consulting).
Results
Production of recombinant antibodies fused to protein antigens
A limited number of DC targeting vaccines based on recombinant anti-DC receptor antibodies with antigen fused to the heavy (H) chain C-terminus have been described. These include a model hen egg lysozyme peptide (21), melanoma antigen peptide pmel17 (10), HIV Gag p24 (5), Leishmania LACK (8), and they all employed mammalian cell secretion systems which typically yield correctly folded and glycosylated functional antibody products.
Our purpose was the production of prototype antigen-targeting vaccines composed of recombinant anti-DC receptor mAbs (rAbs) fused to several other desired antigens (rAb.antigen, throughout, we use a period to indicate direct fusion between rAb and antigen). Thus variable regions from light (L) and H chains of anti-DC receptor mAbs with different specificities (DC-ASGPR, DC-SIGN/L, Langerin, and CD40) were cDNA cloned, characterized by DNA sequence analysis, and engineered into mammalian expression vectors bearing either human Igκ or human IgG4H constant regions. Coding sequences for HIV Gag p24, used as a positive control, or Influenza A antigens were then placed in-frame with each H chain C-terminus. Antibodies fused to the 249 residue HIV Gag p24 domain, or to the 336 residue HA-1 subunit of Influenza A PR8 subtype H1N1 hemagglutinin (Flu HA1-1), were secreted at levels between 25–75% compared to cells transfected with expression vectors encoding the same antibodies without the C-terminal antigen fusion (rAb) (Fig. 1A). Surprisingly, expression constructs encoding the 336 residue HA-1 domain from the H5N1 strain (Flu HA5-1) were very poorly secreted at levels >10-fold reduced compared to cells transfected with matching control Flu HA1-1 constructs (Fig. 1A). This is despite their structural homology and 57% amino acid identity. Prototype DC-targeting vaccines bearing the full HA ectodomain from either the H5N1 (Flu HA5-0) or the H1N1 strain (Flu HA1-0, data not shown), or the entire 336 residue Influenza A matrix protein 1 (Flu M1) were also very poorly secreted (Fig. 1A). Vectors encoding two copies of a region of Flu M1 containing a known immunodominant epitope either directly fused to the H chain C-terminus (Flu M1 pep), or via a flexible linker sequence (Flex Flu M1 pep), also failed to direct secretion of significant amounts of the fusion proteins (Fig. 1A). There was no significant difference in the secretion properties between such rAb.antigen fusion proteins based on the three different mouse variable regions. Similar data showing extremely poor or no secretion from 293F cells was obtained for constructs linking Gad B, Cyclin B1, Cyclin D1 and PE38 directly to the C-terminus of anti-DCIR rAb or control hIgG4 H chain (see Methods, data not shown). Thus, many antigens are recalcitrant to production as DC-targeting prototype vaccines when linked to the rAb H chain C-terminus, and this seems independent of differences in the vaccine variable region sequences and the size of antigens tested in this study.
FIGURE 1.
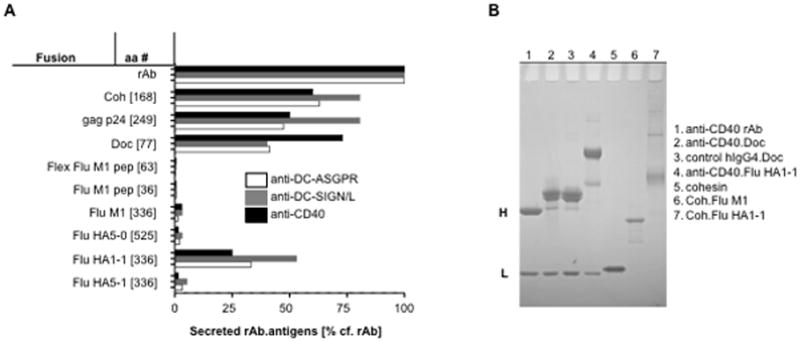
Recombinant antibody expression varies based on the nature of the heavy chain C-terminal antigen. A, Expression constructs for anti-DC-ASGPR, anti-DC-SIGN/L and anti-CD40 rAbs were engineered with various H chain C-terminal antigen-coding regions (antigen total residue number are shown in brackets). These constructs were transiently co-transfected with matching L chain constructs into 293F cells and then tested for expression of secreted rAbs.antigen fusion proteins by anti-hIgG Fc ELISA, compared to rAb alone (100%, ~0.5 – 1 μg/ml). Similar data were obtained when L chain-specific ELISA reagents were used (data not shown). B, Reduced SDS-PAGE analysis of purified rAb, rAb.Doc, rAbs.antigen and Coh.antigen fusion proteins resolved by SDS-PAGE under reducing conditions and stained with Coomassie Blue. Some products contain minor degradation or contaminant proteins. H and L indicate the positions of the IgG4 heavy (~50 kDa) and light (~25 kDa) chains.
For experimental studies, the antigen component can also be made independently and chemically cross-linked to the antibody (4), but manufacture of such a product for human use has significant quality control issues unless linking the antigen to the antibody is controlled (22). The specific protein A or G interaction with the Ig constant region (23) provides a conceptual framework for an alternate strategy of formulating such prototype vaccines by the controlled assembly of the antibody and antigen via non-covalent protein-protein interactions, however this method is not suited to applications where other antibodies are present. As an alternative, we explored the use of the well-characterized dockerin and cohesin interaction (12–14) for this purpose.
Antibody.dockerin and cohesin.antigen fusion proteins form stable and specific complexes in vitro
Fusion proteins composed of Clostridium thermocellum dockerin (Doc) and cohesin (Coh) domains linked to antibody H chain C-termini (respectively, rAb.Doc and rAb.Coh) were efficiently secreted by mammalian cells (Fig. 1A,B). Some antigens of interest that were not well secreted as rAb.antigen, e.g., Flu M1 and Flu HA5-1 (Fig. 1A), were readily purified from E. coli bearing expression vectors encoding cohesin fused to antigen (Coh.antigen) expression vectors (e.g., Coh.Flu M1, Fig. 1B), or were efficiently secreted by 293F cells transfected with Coh.antigen expression vectors (e.g., Flu HA5-1, not shown).
We used cohesin fused to alkaline phosphatase (Coh.AP) as a model antigen. Coh.AP interacted specifically with rAb.Doc (Fig. 2A), but not control rAb (Fig. 2B) in ELISA and the binding was efficient since ~ 2 mol equivalents of Coh.AP (0.5 μg, 72 kDa) saturated the rAb.Doc (0.25 μg, 160 kDa) surface (Fig. 2A). Preformed dockerin-cohesin complexes appeared to be very stable. In a competition study, only ~20% of the rAb.Doc-Coh.AP complex (we use a dash throughout to indicate this non-covalent interaction) dissociated within 300 min in the presence of a 20-fold excess of free rAb.Doc, compared to antibody-protein G.AP complexes, where ~90% dissociated within 300 min when a 20-fold excess free rAb was added (Fig. 2C). The interaction between dockerin and cohesin was also very stable in pure human serum, where virtually no dissociation was observed in 4 h (total Ig in serum is ~15 mg/ml), in contrast to rAb interaction with protein G, where ~1 mg/ml total Ig displaced all the bound protein G in 4 h (Fig. 2D). Thus the rAb.Doc-Coh.antigen interaction is specific and stable and this is a key attribute for potential use in in vitro and in vivo biological situations, which typically contain serum.
FIGURE 2.
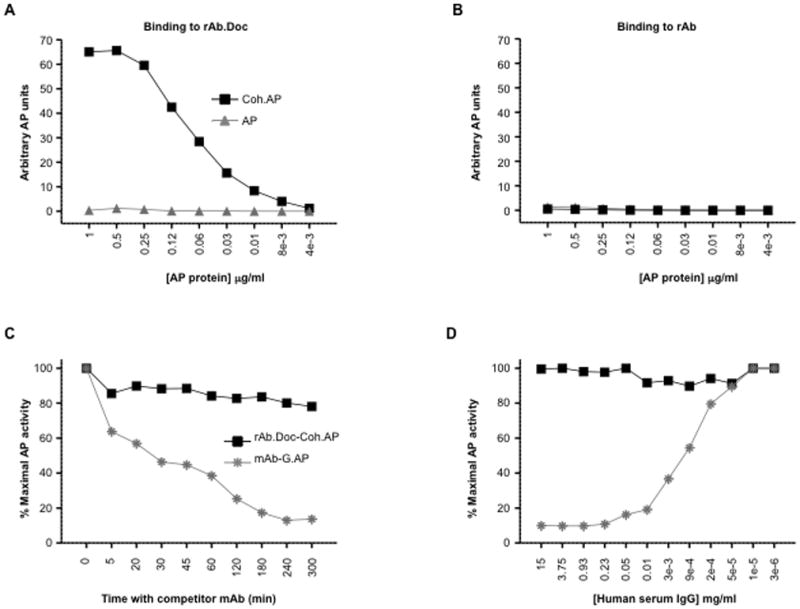
Cohesin fused to alkaline phosphatase binds specifically and stably to recombinant antibody fused to dockerin. A, 2-fold dilutions starting at 1 μg/ml of secreted alkaline phosphatase (AP) or cohesin fused to alkaline phosphatase (Coh.AP) were incubated for 1 h in microtiter wells containing 0.25 μg of immobilized anti-human DEC-205.mIgG2b fused to Doc (rAb.Doc) or B, anti-human DEC-205.mIgG2b rAb. After washing, the bound AP activity was detected with a chromogenic AP substrate. C, Complexes between immobilized rAb.Doc or mIgG2b (0.25 μg) and a fixed amount (0.1 μg) of Coh.AP or protein G fused to alkaline phosphatase (G.AP) were assembled by incubation for 1 h in microtiter wells. At various times, a 20-fold excess of soluble rAb.Doc or mIgG2b was added and incubation continued. After washing, the bound AP activity was detected with a chromogenic AP substrate. D, Various dilutions of human serum were added to preformed complexes and the incubation was continued for 4 h. After washing, the bound AP activity was detected with a chromogenic AP substrate.
The dockerin domain contains two predicted N-linked glycosylation sites (using NetNGlyc 1.0 Server). We found that rAb.Doc preparations were variably glycosylated within the dockerin domain and glycosylated forms of rAb.Doc did not interact with cohesin fusion proteins (Supplemental Fig. 1A). Therefore, one of these sites was altered by mutagenesis, yielding rAb.Doc products that consistently retained full activity for binding cohesin fusion proteins while reducing product heterogeneity (Supplemental Fig. 1B,C) and possibly extraneous interactions with cell surface lectins. All subsequent experiments used this dockerin variant.
Quantitative parameters of antibody.dockerin interaction with cohesin.antigen
Mixtures of purified rAb.Doc and Coh.antigen fusion proteins were analyzed via gel filtration. Individually, anti-CD40.Doc and an E. coli-expressed cohesin migrated as single peaks (Fig. 3A and Supplemental Fig. 2A). When anti-CD40.Doc was mixed with an excess of cohesin, the anti-CD40 rAb.Doc peak was quantitatively displaced into a faster migrating peak (Fig. 3A). When anti-CD40.Doc was mixed in an equimolar amount with cohesin, the cohesin was quantitatively displaced into a similar faster migrating peak (Supplemental Fig. 2A). In each case, the shifted peaks were relatively homogeneous and migrated consistently with the expected apparent molecular weights of complexes of 1 dimeric rAb.Doc:2 Coh.antigens. Also, complexes between rAb.Doc and Coh.antigen could be purified directly from the supernatant of 293F cells co-transfected with expression vectors directing the synthesis of i) rAb.Doc H chain, ii) rAb L chain, and iii) Coh.antigen (Fig. 3B).
FIGURE 3.
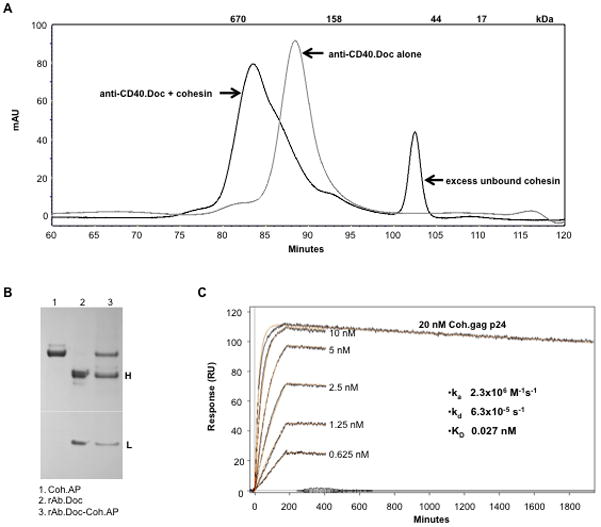
Analysis of antibody.dockerin interaction with cohesin.antigen. A, Analysis by size exclusion chromatography. 40 μg anti-CD40.Doc alone (grey line) and a mixture of anti-CD40.Doc and cohesin (40 μg each, black curve) were used. In the mixture, the cohesin is in 4-fold molar excess relative to anti-CD40.Doc. B, Production of rAb.Doc-Coh.antigen complex within a single fermentation. 293F cells were transiently co-transfected with expression vectors for rAb.Doc H chain, rAb L chain, and Coh.AP. After 2 days, protein A beads were added to the culture supernatant and 2 h later the beads were washed with DPBS + 1 M NaCl and eluted with 20 mM HCl. The eluate was dried, dissolved in SDS sample buffer then analyzed by reduced SDS-PAGE and Coomassie Blue staining. H and L indicate the positions of the IgG4 heavy (~55 kDa) and light (~25 kDa) chains. C, Analysis by surface plasmon resonance of interaction between anti-DC-ASGPR.Doc with Coh.Gag p24. The response curve series are different concentrations of the Coh.Gag p24 flowing over a regenerated protein G surface with freshly immobilized anti-DC-ASGPR.Doc (~233 RU).
In addition, the interaction of Coh.antigen with rAb.Doc immobilized to a surface coated with protein G was investigated with a surface plasmon resonance (SPR) system. The association rate constant was high (ka = 2.3×106 M−1 s−1, Fig. 3C) and the dissociation rate constant was very low (kd = 6.3×10−5 s−1, Fig. 3C). An affinity constant (KD) of 27 pM was determined from the kinetic analysis assuming a 1:1 interaction model and this is in the range of high affinity antibody-antigen interaction (24). In Fig. 3C, 233 RU of protein A-immobilized anti-DC-ASGPR rAb.Doc (162 kDa) bound 111 RU of the Coh.Gag p24 (42 kDa) at ligand saturation (20 nM), indicating that ~1.85 moles of Coh.antigen bound to 1 mole of rAb.Doc, consistent with a dimeric rAb.Doc structure and a 1:1 interaction between the dockerin and cohesin domains. Similar data was found with other Coh.antigen and rAb.Doc interactions, and we did not detect any significant difference in rAb.Doc binding to Coh.antigen secreted from mammalian cells versus expressed as soluble E. coli protein (data not shown).
Anti-DC receptor antibody.dockerin targets cohesin.antigen to DCs in vitro
To assess the capacity of anti-DC receptor rAb.Doc-Coh.antigen complexes to deliver antigens to DCs, we labeled Coh.Flu M1 with biotin and used SA-PE and flow cytometry to detect cell surface-bound complexes. Specific staining was detected on monocyte-derived IFNα-DCs (IFNα-DCs) with > 0.1 μg/ml anti-CD40.Doc but not with isotype control hIgG4.Doc and a maximal signal was observed with ~1.6 μg/ml (Fig. 4A,B). The anti-CD40.Doc-Coh.Flu M1 complex was detected on CD19+ B cells but not on CD3+ T cells (Fig. 4C). On monocyte-derived IL-4-DCs (IL-4-DCs), specific staining was also observed using anti-DC-SIGN/L rAb.Doc (Fig. 4D). Since the SA-PE detecting reagent binds only the biotinylated Coh.Flu M1, these data show that the anti-DC receptor rAb.Doc reagents effectively target the Flu M1 antigen to the cell surface of cells bearing the DC receptor.
FIGURE 4.
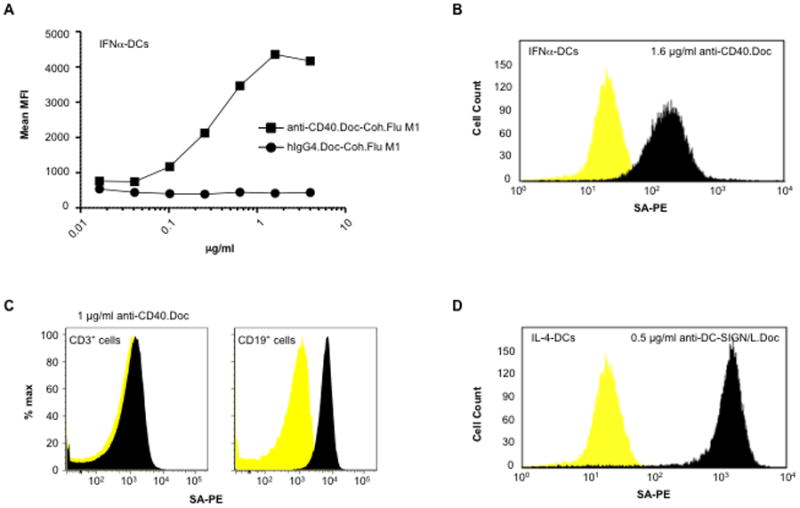
Anti-DC receptor antibody.dockerin mixed with cohesin.Flu M1 fusion protein delivers cohesin.Flu M1 to the surface of human DCs in vitro. A, Various concentrations of anti-CD40.Doc or isotype control hIgG4.Doc and an excess (20 μg/ml) of biotinylated Coh.Flu M1 were pre-incubated and then added to 2–6 × 105 IFNα-DCs After 30 min on ice, cells were washed and incubated for 30 min at R.T. with SA-PE. Cells were then washed and analyzed by flow cytometry. Mean fluorescence intensity values are shown. B, IFNα-DCs stained with 1.6 μg/ml anti-CD40.Doc from panel A. C, CD3+ T cells and CD19+ B cells were stained with 1 μg/ml anti-CD40.Doc and 1 μg/ml Coh.Flu M1. D, IL-4-DCs stained with 0.5 μg/ml anti-DC-SIGN/L.Doc, 1 μg/ml Coh.Flu M1. Dark peaks are staining with rAb.Doc-Coh.Flu M1 complexes, light peaks are staining with isotype control hIgG4.Doc-Coh.Flu M1 complexes.
In dose-ranging studies of the relative affinities for either plate-bound or cell surface CD40, anti-CD40 rAbs with H chain C-terminal dockerin (anti-CD40.Doc) or HA-1 domain fusions (anti-CD40.Flu HA1-1) had only slightly reduced affinities for CD40 compared to recombinant anti-CD40 rAb alone (Supplemental Fig. 3A,B). Similarly, studies with anti-CD40 rAb.Doc-Coh.antigens versus rAb.Doc alone showed that the complexes had no significant impact on interaction with cell surface CD40 (Supplemental Fig. 3C,D).
Anti-DC receptor antibody.dockerin – cohesin.antigen complexes elicit antigen-specific CD4+ and CD8+ T cell responses in vitro
Our pretext for development of prototype vaccines based on antibody.dockerin – cohesin.antigen complexes was broadening the study of DC-targeting to antigens that could not be expressed as direct rAb.antigen fusion proteins. For example, to demonstrate that Flu M1 delivered to DCs via anti-CD40.Doc is presented to T cells, we incubated IFNα-DCs with preformed anti-CD40.Doc-Coh.Flu M1 complexes, and then co-cultured them with autologous CD8+ T cells for 7 days. Then, HLA-A2 tetramers bearing Flu M1 peptide (58–66) were used to detect Flu M1-specific CD8+ T cell expansion. Antigen doses as low as 10 pM delivered via anti-CD40.Doc-Coh.Flu M1 complexes elicited robust Flu M1-specific CD8+ T cell responses (Fig. 5A). At least 1000-fold higher doses of Coh.Flu M1 alone or isotype control hIgG4.Doc-Coh.Flu M1 complexes were required to elicit detectable antigen-specific CD8+ T cell responses (Fig. 5A). In a second donor, anti-CD40.Doc-Coh.Flu M1 at 1 nM was more potent at inducing expansion of Flu M1-specific CD8+ T cells than the isotype control hIgG4.Doc-Coh.Flu M1 or Coh.FluM1 alone (27% tetramer positive cells vs. 0.35%), and addition of free anti-CD40 rAb did not increase the low potency of hIgG4.Doc-Coh.Flu M1, or Coh.Flu M1 alone (Fig. 5B, upper panel). The potency of targeting via anti-CD40.Doc-Coh.Flu M1 complex was confirmed at 0.1 nM where the hIgG4.Doc-Coh.Flu M1 complex and Coh.Flu M1 were not cross-presented (14.3% of tetramer positive cells vs. 0.36% and 0. 38%, Fig. 5B, lower panel). Thus, this anti-DC rAb.Doc-Coh.antigen DC-targeting reagent can effectively expand in vitro antigen-specific CD8+ T cells against an antigen that could not be configured as a direct rAb.antigen fusion.
FIGURE 5.
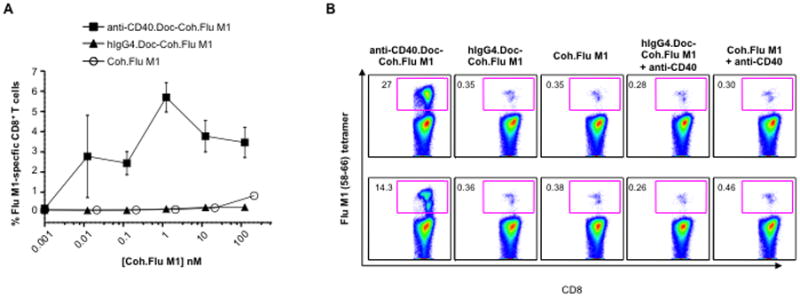
Anti-CD40 rAb.dockerin fusion protein delivers cohesin.Flu M1 fusion protein to human DCs resulting in expansion of Flu M1-specific CD8+ T cells. A, Percentages of Flu M1-specific CD8+ T cells expanded by IFNα-DCs loaded with a dose-range of anti-CD40.Doc or isotype control hIgG4.Doc complexed to Coh.Flu M1 or with Coh.Flu M1 fusion protein alone and analyzed using Flu M1-specific tetramers after 7 days of co-culture. Values are mean of duplicates ±SD. B, IFNα-DCs were incubated with 1 nM or 0.1 nM of the indicated premixed complexes or proteins for 16 h and then co-cultured with autologous CD8+ T cells for 7 days. CD8+ T cells were then analyzed for expansion of Flu M1-specific cells. The inner boxes indicate the percentages of Flu M1 tetramer-specific CD8+ T cells. The background percentage of Flu M1-specific CD8+ T cells without any stimulation was 0.62, and was 0.7 with 1 nM anti-CD40.Doc-Coh.Flu M1 without DCs.
To further study the capacity of antibody.dockerin – cohesin.antigen complexes to direct antigen presentation to CD4+ T cells, we configured Influenza hemagglutinin Flu HA1-1 fused to cohesin (Coh.Flu HA1-1). Stimulation with anti-CD40.Doc-Coh.Flu HA1-1 complexes activated IFNα-DCs in vitro as measured by CD83 or CD86 up-regulation compared to hIgG4.Doc-Coh.Flu HA1-1 or Coh.Flu HA1-1 alone (Fig. 6A). In IFNα-DC/CD4+ T cell co-cultures, anti-CD40.Doc-Coh.Flu HA1-1 complexes expanded memory antigen-specific CD4+ T cells recognizing different epitopes of the Flu HA1-1 protein as measured by IFNγ secretion following peptide stimulation (Fig. 6B). The control hIgG4.Doc-Coh.Flu HA1-1 or Coh.Flu HA1-1 alone resulted in weaker Flu HA1-1 specific responses (Fig. 6B). The responses were dependent on delivery via CD40 since addition of free anti-CD40 rAb did not potentiate the efficacy of control hIgG4.Doc-Coh.Flu HA1-1 complex (Fig. 6B). Flu HA1-1 delivery to surface CD40 via anti- CD40.Doc-Coh.Flu HA1-1 was receptor-dependent, since the incubation of IFNα-DCs with such complexes at 4°C prior to incubation with autologous CD4+ T cells resulted in expansion of Flu HA1-1 peptide-specific CD4+ T cells, while incubation with control hIgG4.Doc-Coh Flu HA1-1 did not (data not shown). To verify that the presence of Doc and Coh sequences in the prototype targeting vaccine was not affecting the breadth of the response, we configured Influenza A hemagglutinin Flu HA1-1 as a direct fusion to the anti-CD40 rAb (anti-CD40.Flu HA1-1), thus permitting direct comparison between the direct fusion and complex. In IFNα-DC/CD4+ T cell co-cultures from a second donor, both reagents were equally effective at expanding autologous CD4+ T cells specific to Flu HA1-1 as determined by the frequency of CFSElow CD4+ T cells producing intracellular IFNγ in response to the same specific Flu HA1-1 peptides (Fig. 6C). These data show that this anti-DC rAb.Doc-Coh.antigen reagent is able to elicit DC-directed antigen-specific MHC class II-restricted CD4+ T cell responses. In addition, the presence of cohesin and dockerin sequences in the prototype targeting vaccine did not appear to affect the magnitude and the breath of the antigen-specific CD4+ T cells responses in vitro.
FIGURE 6.
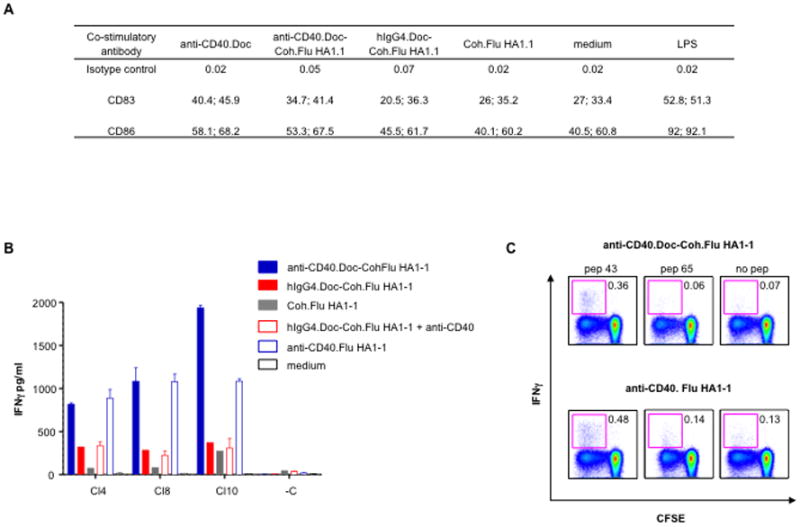
Anti-CD40 rAb.dockerin fusion protein mixed with cohesin.Flu HA1-1 fusion protein targets human DCs as efficiently as a direct anti-CD40.Flu HA1-1 fusion protein. A, Cell surface expression analysis of CD83 and CD86 on IFNα-DCs. IFNα-DCs were incubated with 1.8 nM of premixed Coh.Flu HA1-1 complexes, single proteins, LPS or medium alone for 16 h. Values represent the percentage of the positive cell population for 2 independent healthy donors. B, IFNα-DCs were incubated with 0.01 nM of premixed Coh.Flu HA1-1 complexes, single proteins or medium alone for 16 h and then co-cultured with purified CD4+ T cells for 10 days. Samples were then restimulated for 48 h with or without Flu HA1-1 peptide pools. C, IFNα-DCs were incubated with 0.22 nM of premixed Coh.Flu HA1-1 complexes or 0.22 nM anti-CD40.Flu HA1-1 fusion protein for 16 h and then co-cultured with CFSE-labeled CD4+ T cells for 10 days. Proliferated CD4+ T cells were then restimulated with 5 μM of 17-mer Flu HA1-1 peptides (pep 43 LEPGDTIIFEANGNLIA and pep 65 DQKSTQNAINGITNKVN) for 6 h in the presence of brefeldin A and analyzed for IFNγ production. The inner boxes indicate the percentages of intracellular IFNγ produced by the CFSElow CD4+ T cells.
Anti-Langerin antibody.dockerin – cohesin.antigen complexes elicit immune responses in vivo
We further tested the efficacy of anti-DC receptor rAb.Doc-Coh.antigen complexes in vivo. We used transgenic mice expressing human Langerin (huLangerin-DTR) specifically on Langerhans cells (LCs) and an anti-human Langerin rAb (anti-hLang rAb) previously characterized to bind to LCs and to induce antigen-specific immune responses via LC-targeting (7). In this study anti-hLang mAb was administered intraperitoneally (i.p.) and LCs isolated from the epidermis efficiently acquired the antibody, while keratinocytes, dendritic epidermal T cells, and LCs from littermate control mice (i.e., without human Langerin) did not acquire the antibody (7). huLangerin-DTR mice were injected i.p. with 1 μg of anti-hLang.Doc-Coh.Flu HA1-1 complex (Fig. 7A), and a control group were injected with a 1 μg mixture of anti-hLang rAb (i.e., without dockerin) and Coh.Flu HA1-1 (Fig. 7A). In the huLangerin-DTR group injected with anti-hLang.Doc-Coh.Flu HA1-1 complexes, Flu HA1-1-specific antibodies were detected in serum sampled 2 weeks post vaccination and this response was strongly boosted with a second injection (Supplemental Fig. 4A). In the non-complexed control group, only a very weak anti-Flu HA1-1 response could be detected (Fig. 7A and Supplemental Fig. 4A) and then in only one of 3 mice after the boost (Fig. 7A and Supplemental Fig. 4B), indicating that the targeting antibody is required to be complexed to the antigen through the dockerin-cohesin domains interaction for efficacy. The mice from both groups also raised antibodies against the antibody vehicle itself, which had human IgG4 constant regions (Supplemental Fig. 4C,D). In the huLangerin-DTR mice, the anti-hIgG titers were generally higher and developed with a more rapid kinetic when immunized with the dockerin-cohesin assembled complex rather than the rAb alone mixed with Coh.antigen (Supplemental Fig. 4C,D), and these complex-elicited antibodies were predominantly of the IgG1 isotype (data not shown). Additional controls were non-transgenic littermate mice injected with the identical mixtures. Littermate control groups immunized with the same reagents gave no significant anti-Flu HA1-1 or anti-hIgG antibody responses (Fig. 7A and Supplemental Fig. 4E,F), indicating that the prototype vaccine was acting specifically through hLangerin expressed on LCs.
FIGURE 7.
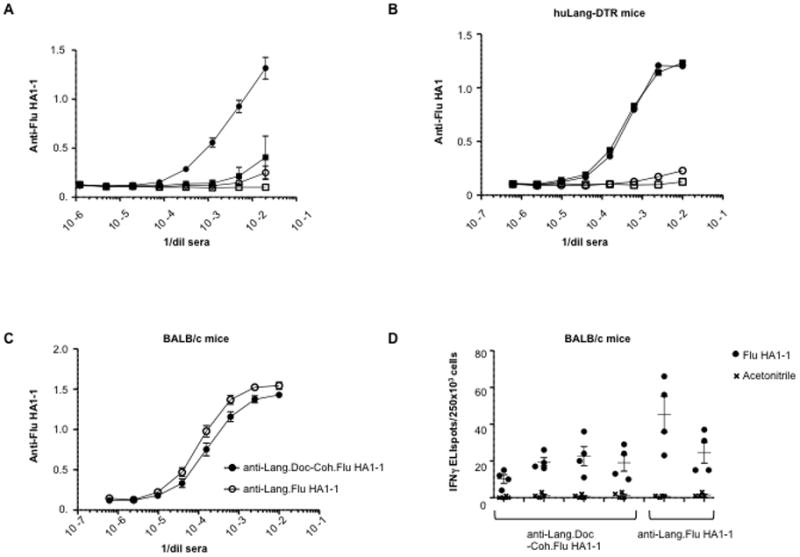
Targeting Flu HA1-1 in vivo to LCs in mice via an anti-hLang.Doc-Coh.Flu HA1-1 complex enhances Flu HA1-1-specific responses. A, At day 0, huLangerin-DTR mice were primed by injection with 1 μg of either anti-hLang.Doc-Coh.Flu HA1-1 conjugate (●) or anti-hLang rAb mixed with Coh.Flu HA1-1 (■). Littermate mice were primed by injection with 1 μg of either anti-hLang.Doc-Coh.Flu HA1-1 conjugate (○) or anti-hLang rAb mixed with Coh.Flu HA1-1 (□). At day 14, mice were boosted with the same reagents respectively. Serum samples were collected at day 21. Serum anti-Flu HA1-1 titers were measured by ELISA. Symbols represent average of 3 individual mice per group +/− SEM. O.D. at 450 nm is plotted vs. serum dilution. B, At day 0, huLangerin-DTR mice were primed by injection with 10 μg of either anti-hLang.Doc-Coh.Flu HA1-1 conjugate (●, ○) or anti-hLang.Flu HA1-1 (■, □). At days 14 and 21, mice were boosted with the same reagents. Serum samples were collected at day 28. Serum anti-Flu HA1 (PR8, filled symbols or CAL04, open symbols) titers were measured by ELISA. Symbols represent an average of 4 individual mice per group +/− SEM. C, At day 0, BALB/c mice were primed by injection with 10 μg of either anti-mLang.Doc-Coh.Flu HA1-1 conjugate (●) or anti-mLang.Flu HA1-1 (○). At days 14 and 21, mice were boosted with the same reagents. Serum samples were collected at day 28. Serum anti-Flu HA1-1 titers were measured by ELISA. Symbols represent average of 3 to 4 individual mice per group +/− SEM. D, As in C, but the mice were boosted one more time at day 30 with the indicated reagents respectively and then 4 days later, splenocytes were taken and restimulated in vitro with Flu HA1-1 peptide pools (●) or peptide solvent as negative control (x). IFNγ was evaluated by ELISPOT. 2 to 4 mice per group were analyzed in quadruplicate, and there was no significant difference between the two groups (Student’s t test).
To address possible differences in immunity against Flu HA1-1 delivered via cohesin-dockerin interaction versus direct fusion and their potential for development of neutralizing anti-Flu HA1-1 antibodies, two additional groups of huLangerin-DTR mice were vaccinated as above, but with 10 μg of complex versus 10 μg of direct fusion. The sera were tested one week after a third injection. We did not observe any differences in anti-Flu HA1-1 responses between delivering Flu HA1-1 with anti-hLangerin.Doc-Coh.Flu HA1-1 versus anti-hLangerin.Flu HA1-1 (Fig. 7B). The resulting Flu HA1 specific antibodies had only a minimal cross-reactivity against a different Influenza A hemagglutin strain (H1N1, Flu HA1-Cal04). Using the same protocol, we also vaccinated two groups of BALB/c mice with an anti-mouse Langerin rAb.Doc complexed to Coh.Flu HA1-1 complex or with an anti-Lang.Flu HA1-1 direct fusion protein. Both the cohesin-dockerin complex and the direct fusion protein-vaccinated groups mounted anti-Flu HA1-1-specific antibody responses (Fig. 7C). The anti-Flu HA1-1 positive sera shown in Fig. 7A did not neutralize the homologous Influenza A PR8 virus (hemagglutination inhibition HAI <1:1). However, the increased anti-Flu HA1-1 titers obtained in the 2 high-dose studies (Fig. 7B,C) did yield potentially neutralizing antibodies (HAI range: 1:1-1:2) (data not shown) and there were no observed significant differences between the efficacy of the cohesin-dockerin interaction versus the direct fusion nor between the huLangerin-DTR and normal mice (Fig. 7B,C).
To compare the efficacy of anti-Lang.Doc-Coh.Flu HA1-1 versus anti-Lang.Flu HA1-1 for eliciting T cell responses, we again boosted the BALB/c mice and analyzed splenic Flu HA1-1-specific T cell responses 4 days after the final boost. IFNγ-secreting Flu HA1-1-specific T cells detected by ELISPOT were induced without adjuvant and the levels of responses were similar for the complex versus the direct fusion protein (Fig. 7D). However, no such responses that cross-reacted to Flu HA1-Cal04 peptides were observed (data not shown).
Collectively, these data in mice show that anti-hLang.Doc-Coh.Flu HA1-1 complex can effectively target the antigen to Langerin in vivo, resulting in specific antibody and T cell responses.
Discussion
DC-targeting vaccines, typically recombinant anti-DC receptor antibodies fused directly to antigens via the heavy chain C-terminus, offer a very promising new tool for enhancing and controlling humoral and cellular immune responses. Here we demonstrate that many desirable antigens cannot be produced in this direct antibody fusion configuration. To circumvent this problem, recombinant antibodies directly linked to a dockerin domain can be assembled into fully functional DC-targeting agents via non-covalent interaction with separately produced cohesin.antigen fusion proteins. We considered assembling such antibody-antigen complexes based on modifications of existing technologies for making heterodimeric antibodies or proteins, e.g., amphiphilic Fos-Jun leucine zippers (25, 26), CH3 domain ‘knob-in-hole’ pairs (27), or E-coil-K-coil peptides (28, 29). However, we were intrigued with the possibilities offered by the well characterized dockerin-cohesin interaction that is a hallmark of the cellulosome. These very high affinity interacting dockerin and cohesin modules were designed by nature to function when fused to other domains. The cellulosome proteins are secreted and glycosylated bacterial products, but their dockerin and cohesin domains are also functional when expressed as intracellular E. coli products (30).
The results presented here establish the utility of dockerin and cohesin modules for making very specific and stable anti-DC receptor antibody-antigen complexes, which retain full DC-targeting function in vitro and in vivo. These bacterial domains were well expressed and functional, either as antibody.dockerin secreted by mammalian cells, or as cohesin.antigen fusion proteins produced in E. coli or mammalian cells. Recombinant anti-DC receptor antibody-antigen complexes assembled through the dockerin-cohesin interaction are remarkably stable, especially in the presence of human serum, and retain native antibody binding properties to the DC receptor. Quantitative analysis of the interaction between antibody.Doc and Coh.antigen confirmed a high affinity interaction between the 2 domains, within the range of high affinity antibody-antigen interaction (24).
Anti-DC receptor antibody.dockerin – cohesin.antigen complexes are potentially very useful tools for in vitro and in vivo antigen-targeting studies. They effectively deliver antigen to the DC surface, permitting expansion in vitro of both CD4+ and CD8+ antigen-specific T cells in a DC receptor-dependent fashion. This was exemplified via targeting two key Influenza antigens, M1 and HA1-1 to DCs through CD40. Infection with Influenza virus generates a broad range of CD4+ and CD8+ T cells that are reactive against most of the viral proteins (31), and many of these T cell epitopes are conserved across the various Influenza virus strains, making them attractive as potential antigens for invoking cellular immunity. Targeting the whole Flu M1 antigen to human DCs with anti-CD40.Doc-Coh.Flu M1 complex could efficiently expand Flu M1-specific memory CD8+ T cells. Also, targeting Flu HA1-1 to human DCs via either, a direct anti-CD40.Flu HA1-1 fusion protein, or via a dockerin-cohesin mediated complex, expanded anti-Flu HA1-1-specific memory CD4+ T cells in vitro with equal efficiency. The Influenza HA1 domain is one of the relevant antigens for raising protective humoral responses (32), and augmenting anti-Flu HA1 antibody responses via DC-targeting is an attractive option for novel Influenza vaccine development. Targeting HA1-1 in vivo to mouse LCs via an anti-Lang.Doc-Coh.Flu HA1-1 complex evoked strong anti-Flu HA1-1 antibody responses at very low doses and those antigen-specific antibody responses were dependent on both the dockerin-cohesin interaction and the targeted receptor.
Targeting Flu HA1-1 in vivo to mouse Langerin via an anti-Lang.Doc-Coh.Flu HA1-1 complex versus a direct fusion protein yielded identical neutralizing serum anti-Flu HA1-1 antibody responses, as well as Flu HA1-1-specific T cell responses. These data indicate that this form of delivery does not affect the presentation of crucial epitopes for neutralization. These responses had minimal cross-reactivity to a different Influenza A strain hemagglutinin (H1N1, Flu HA1-Cal04), but these studies were without adjuvant which should increase the magnitude of responses and thus the potential for cross-reactivity. An obvious difference between injection with direct fusion rAb versus a complex is the development of responses against the dockerin and cohesin components, which are neo-antigens with no major cross-reactivity to human proteins. In this respect, the similar efficacy of anti-Lang.Doc-Coh.Flu HA1-1 versus anti-Lang.Flu HA1-1 for both humoral and cellular responses suggests that anti-Doc and anti-Coh antibodies are not a hindrance in such a ‘prime-boost’ setting. However, much like viral vector-based vaccines, such anti-vector immunity could impede subsequent vaccinations. However, a wide selection of very distantly related Doc-Coh pairs are available that could be adapted to circumvent this possibility, or dominant epitopes could be dulled by protein engineering (33).
In summary, our work demonstrates that non-covalent assembly of prototype DC-targeting vaccines is practical and greatly expands options for development of such vaccines delivering a multitude of antigens that cannot be efficiently expressed as direct fusions of antibody to antigen. Importantly, a single antibody-dockerin vaccine preparation can be efficiently linked to different cohesin.antigen fusion proteins, e.g., different seasonal Influenza HA antigens, or a single cohesin.antigen preparation can be linked to different anti-DC receptor antibodies, bringing the flexibility of combinatorial approaches to DC-targeting vaccine design and optimization. Prototype vaccines based on this technology enabled in vitro studies of targeting Flu M1 to different DC subsets, including human blood DCs and skin DCs for expanding antigen-specific CD8+ T cell responses (16), and cross-priming of the neo-antigen MART-1 via targeting human DCs (16, 18). Our new data provides the rationale, characterization, and validation of this technology for further use in both in vitro and in vivo studies.
Supplementary Material
Acknowledgments
This work was funded by grants from the Baylor Health Care System Foundation and the National Institutes of Health (RO-1 CA78846 and U-19 AI-57234).
We thank Drs. Michael Ramsay and William Duncan for their support, and Keiko Akagawa, Julien Blond, Brandon Hahm, Guocheng He for some vector constructs and protein purifications, Aaron Martin for help with SPR, and Yaming Xue for HAI assays. We thank Drs. Ralph Steinman and Christine Trumpfheller for kindly providing vectors for anti-human DEC-205, mIgG2b rAb and isotype control mIgG2b and the HIV Gag p24 component of our rAb.Gag p24 construct; and the Adolfo Garcia-Sastre Laboratory for providing Influenza cDNAs.
Abbreviations used in this paper
- Coh
cohesin
- Doc
dockerin
- rAb.antigen
recombinant antibody-antigen fusion protein
- Coh.antigen
cohesin-antigen fusion protein
- H chain
heavy chain
- DCs
dendritic cells
Footnotes
Disclosures
A.L.F., E.K., and G.Z. are inventors on an issued patent concerning the dockerin-cohesin technologies described in this study. The application is held by the Baylor Research Institute, a nonprofit research arm of the Baylor Heath Care System. J.Q. is employed by ICx Nomadics, which develops and markets SPR instrumentation.
References
- 1.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 3.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 4.Delneste Y, Magistrelli G, Gauchat J, Haeuw J, Aubry J, Nakamura K, Kawakami-Honda N, Goetsch L, Sawamura T, Bonnefoy J, Jeannin P. Involvement of LOX-1 in dendritic cell-mediated antigen cross-presentation. Immunity. 2002;17:353–362. doi: 10.1016/s1074-7613(02)00388-6. [DOI] [PubMed] [Google Scholar]
- 5.Trumpfheller C, Finke JS, Lopez CB, Moran TM, Moltedo B, Soares H, Huang Y, Schlesinger SJ, Park CG, Nussenzweig MC, Granelli-Piperno A, Steinman RM. Intensified and protective CD4+ T cell immunity in mice with anti-dendritic cell HIV gag fusion antibody vaccine. J Exp Med. 2006;203:607–617. doi: 10.1084/jem.20052005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caminschi I, Proietto AI, Ahmet F, Kitsoulis S, ShinTeh J, Lo JC, Rizzitelli A, Wu L, Vremec D, van Dommelen SL, Campbell IK, Maraskovsky E, Braley H, Davey GM, Mottram P, van de Velde N, Jensen K, Lew AM, Wright MD, Heath WR, Shortman K, Lahoud MH. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 2008;112:3264–3273. doi: 10.1182/blood-2008-05-155176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Igyarto BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, Edelson BT, Zurawski SM, Malissen B, Zurawski G, Berman J, Kaplan DH. Skin-Resident Murine Dendritic Cell Subsets Promote Distinct and Opposing Antigen-Specific T Helper Cell Responses. Immunity. 2011 doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, Steinman RM, Nussenzweig MC. Differential antigen processing by dendritic cell subsets in vivo. Science (New York, NY. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 9.Tacken PJ, I, de Vries J, Gijzen K, Joosten B, Wu D, Rother RP, Faas SJ, Punt CJ, Torensma R, Adema GJ, Figdor CG. Effective induction of naive and recall T-cell responses by targeting antigen to human dendritic cells via a humanized anti-DC-SIGN antibody. Blood. 2005;106:1278–1285. doi: 10.1182/blood-2005-01-0318. [DOI] [PubMed] [Google Scholar]
- 10.Ramakrishna V, Treml JF, Vitale L, Connolly JE, O’Neill T, Smith PA, Jones CL, He LZ, Goldstein J, Wallace PK, Keler T, Endres MJ. Mannose receptor targeting of tumor antigen pmel17 to human dendritic cells directs anti-melanoma T cell responses via multiple HLA molecules. J Immunol. 2004;172:2845–2852. doi: 10.4049/jimmunol.172.5.2845. [DOI] [PubMed] [Google Scholar]
- 11.Bozzacco L, Trumpfheller C, Siegal FP, Mehandru S, Markowitz M, Carrington M, Nussenzweig MC, Piperno AG, Steinman RM. DEC-205 receptor on dendritic cells mediates presentation of HIV gag protein to CD8+ T cells in a spectrum of human MHC I haplotypes. Proc Natl Acad Sci U S A. 2007;104:1289–1294. doi: 10.1073/pnas.0610383104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho AL, Dias FM, Prates JA, Nagy T, Gilbert HJ, Davies GJ, Ferreira LM, Romao MJ, Fontes CM. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc Natl Acad Sci U S A. 2003;100:13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annual review of microbiology. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 14.Bayer EA, Morag E, Lamed R. The cellulosome--a treasure-trove for biotechnology. Trends in biotechnology. 1994;12:379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 15.Reddy MP, Kinney CA, Chaikin MA, Payne A, Fishman-Lobell J, Tsui P, Dal Monte PR, Doyle ML, Brigham-Burke MR, Anderson D, Reff M, Newman R, Hanna N, Sweet RW, Truneh A. Elimination of Fc receptor-dependent effector functions of a modified IgG4 monoclonal antibody to human CD4. J Immunol. 2000;164:1925–1933. doi: 10.4049/jimmunol.164.4.1925. [DOI] [PubMed] [Google Scholar]
- 16.Klechevsky E, Flamar AL, Cao Y, Blanck JP, Liu M, O’Bar A, Agouna-Deciat O, Klucar P, Thompson-Snipes L, Zurawski S, Reiter Y, Palucka AK, Zurawski G, Banchereau J. Cross-priming CD8+ T cells by targeting antigens to human dendritic cells through DCIR. Blood. 2010;116:1685–1697. doi: 10.1182/blood-2010-01-264960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bates EE, Fournier N, Garcia E, Valladeau J, Durand I, Pin JJ, Zurawski SM, Patel S, Abrams JS, Lebecque S, Garrone P, Saeland S. APCs express DCIR, a novel C-type lectin surface receptor containing an immunoreceptor tyrosine-based inhibitory motif. J Immunol. 1999;163:1973–1983. [PubMed] [Google Scholar]
- 18.Ni L, Gayet I, Zurawski S, Duluc D, Flamar AL, Li XH, O’Bar A, Clayton S, Palucka AK, Zurawski G, Banchereau J, Oh S. Concomitant activation and antigen uptake via human dectin-1 results in potent antigen-specific CD8+ T cell responses. J Immunol. 2010;185:3504–3513. doi: 10.4049/jimmunol.1000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bobr A, Olvera-Gomez I, Igyarto BZ, Haley KM, Hogquist KA, Kaplan DH. Acute ablation of Langerhans cells enhances skin immune responses. J Immunol. 2010;185:4724–4728. doi: 10.4049/jimmunol.1001802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Q, V, Zarnitsyn G, Ye L, Wen Z, Gao Y, Pan L, Skountzou I, Gill HS, Prausnitz MR, Yang C, Compans RW. Immunization by vaccine-coated microneedle arrays protects against lethal influenza virus challenge. Proc Natl Acad Sci U S A. 2009;106:7968–7973. doi: 10.1073/pnas.0812652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawiger D, Inaba K, Dorsett Y, Guo M, Mahnke K, Rivera M, Ravetch JV, Steinman RM, Nussenzweig MC. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J Exp Med. 2001;194:769–779. doi: 10.1084/jem.194.6.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, Lu Y, Meng YG, Ng C, Yang J, Lee CC, Duenas E, Gorrell J, Katta V, Kim A, McDorman K, Flagella K, Venook R, Ross S, Spencer SD, Lee Wong W, Lowman HB, Vandlen R, Sliwkowski MX, Scheller RH, Polakis P, Mallet W. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nature biotechnology. 2008;26:925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 23.Achari A, Hale SP, Howard AJ, Clore GM, Gronenborn AM, Hardman KD, Whitlow M. 1.67-A X-ray structure of the B2 immunoglobulin-binding domain of streptococcal protein G and comparison to the NMR structure of the B1 domain. Biochemistry. 1992;31:10449–10457. doi: 10.1021/bi00158a006. [DOI] [PubMed] [Google Scholar]
- 24.Delves Peter J, Seamus IMR, Martin J, Burton Dennis. Roitt’s Essential Immunology. Blackwell Publishing; 2006. [Google Scholar]
- 25.Kostelny SA, Cole MS, Tso JY. Formation of a bispecific antibody by the use of leucine zippers. J Immunol. 1992;148:1547–1553. [PubMed] [Google Scholar]
- 26.de Kruif J, Logtenberg T. Leucine zipper dimerized bivalent and bispecific scFv antibodies from a semi-synthetic antibody phage display library. J Biol Chem. 1996;271:7630–7634. doi: 10.1074/jbc.271.13.7630. [DOI] [PubMed] [Google Scholar]
- 27.Ridgway JB, Presta LG, Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein engineering. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 28.Lian Q, Szarka SJ, Ng KK, Wong SL. Engineering of a staphylokinase-based fibrinolytic agent with antithrombotic activity and targeting capability toward thrombin-rich fibrin and plasma clots. J Biol Chem. 2003;278:26677–26686. doi: 10.1074/jbc.M303241200. [DOI] [PubMed] [Google Scholar]
- 29.Pack P, Pluckthun A. Miniantibodies: use of amphipathic helices to produce functional, flexibly linked dimeric FV fragments with high avidity in Escherichia coli. Biochemistry. 1992;31:1579–1584. doi: 10.1021/bi00121a001. [DOI] [PubMed] [Google Scholar]
- 30.Fierobe HP, Bayer EA, Tardif C, Czjzek M, Mechaly A, Belaich A, Lamed R, Shoham Y, Belaich JP. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277:49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 31.Doherty PC, Turner SJ, Webby RG, Thomas PG. Influenza and the challenge for immunology. Nature immunology. 2006;7:449–455. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 32.Ahmed R, Oldstone MB, Palese P. Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nature immunology. 2007;8:1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones TD, Crompton LJ, Carr FJ, Baker MP. Deimmunization of monoclonal antibodies. Methods Mol Biol. 2009;525:405–423. xiv. doi: 10.1007/978-1-59745-554-1_21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


