Summary
Explorations of human microbiota have provided substantial insight into microbial community composition; however, little is known about interactions between various microbial components in human ecosystems. In response to the powerful impact of viral predation, bacteria have acquired potent defenses, including an adaptive immune response based on the CRISPR/Cas system. To improve our understanding of the interactions between bacteria and their viruses in humans, we analyzed 13,977 streptococcal CRISPR sequences and compared them with 2,588,172 virome reads in the saliva of 4 human subjects over 17 months. We found a diverse array of viruses and CRISPR spacers, many of which were specific to each subject and time point. There were numerous viral sequences matching CRISPR spacers; these matches were highly specific for salivary viruses. We determined that spacers and viruses coexist at the same time, which suggests that streptococcal CRISPR/Cas systems are under constant pressure from salivary viruses. CRISPRs in some subjects were just as likely to match viral sequences from other subjects as they were to match viruses from the same subject. Because interactions between bacteria and viruses help to determine the structure of bacterial communities, CRISPR-virus analyses are likely to provide insight into the forces shaping the human microbiome.
Keywords: CRISPRs, Saliva, Virus, Virome, Microbiome
Introduction
The human body consists of many different habitats colonized by various microbes. As greater attention is given to the microbial inhabitants of each human environment, we gain broader understanding of the role these organisms may serve in the physiology of the host. There now are numerous studies characterizing the interplay between diet and bacterial community members in the human gastrointestinal tract [1-3] that highlight the important contribution of bacteria to human physiology, and conversely, diet on community composition. Others have suggested that human indigenous microbial communities are involved in homeostasis [4], and that community perturbations may have consequences for human health [5]. Following antibiotic perturbation, microbial community composition may slowly return to a profile similar to that which pre-dated the antibiotic administration [5, 6]. The indigenous microbiota is known to be altered in chronic periodontal disease, implicating microbial communities as potential etiological agents [7-9].
Much of the study of human microbial communities has focused on the bacterial component. However, there now is substantial evidence that viral communities coexist in these human habitats [10-15]. In human saliva, viral communities vary substantially between subjects, and carry numerous virulence factors potentially involved in adaptation of bacterial hosts to the human oral environment [11]. Similar to the findings of research focused on human feces [13], lysogenic viruses are known to be common inhabitants of human saliva [11]. Despite recent data characterizing these human viral communities, relatively little is known about their interactions with their bacterial hosts.
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) loci are part of the CRISPR/Cas system in bacteria and archaea, and are known to be involved in acquired immunity against viruses and plasmids [16, 17]. CRISPR loci evolve through the acquisition of new spacers at sites between palindromic repeats. These spacer sequences contain short fragments of DNA from viruses or plasmids, and confer resistance to matching viruses and plasmids through nucleic acid interference [18-20]. CRISPRs have previously been shown to reflect adaptation of host organisms to local virus populations in acid mine drainage systems [21]. In human saliva, streptococcal CRISPRs represent unique characteristics of individual subjects at each time point studied [11]. We studied two separate groups of streptococcal CRISPRs in human saliva and compared their sequences with those of salivary viruses to determine if streptococci in the human oral cavity develop resistance to the specific viruses they encounter in these ecosystems.
Results
Identification of salivary viruses
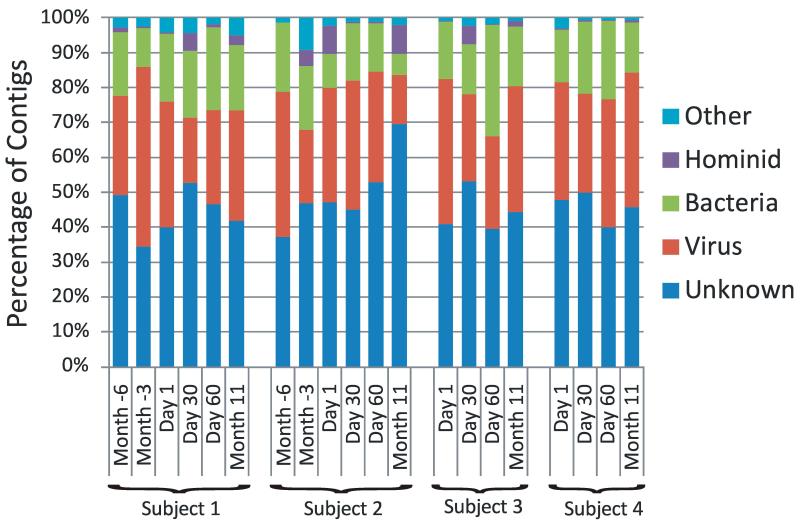
We recruited 4 human subjects in good overall oral and periodontal health and collected saliva from January 2008 to July 2009 (Supplemental Table 1). All subjects donated saliva on Day 1, Day 30, Day 60, and Month 11. Two additional samples were collected from two of the subjects on Month -3 and Month -6. No specific intervention took place at any time during the study. To isolate salivary viruses, the saliva samples were sequentially filtered, subjected to cesium chloride density gradient purification, and the resulting DNA subjected to pyrosequencing using FLX titanium technology as previously described [11, 22]. Viromes were sequenced for each subject at all time points for a total of 2,588,172 reads, including subjects #1 and #2, which were sequenced as part of a prior study [11]. With a goal to characterize the most abundant viruses present, we obtained approximately 129,000 reads per time point and assembled each into larger contigs (Table 1). Each contig was subjected to blastX analysis based on the NCBI non-redundant (NR) database, and contigs with homologues to known viral elements were identified. The majority of the contigs in each subject across each time point had no identifiable homologues in the NR database; however, many contigs did have homologues to known viral elements (Figure 1). While there were homologues to sequences from known bacteria, they may represent bacterial elements in viral genomes or prophage in bacterial genomes. The presence of homologues to hominid and other sequences mostly reflects similarity to redundant eukaryote DNA; however, compared to other studies there was a far lower percentage of sequences homologous to eukaryote DNA (Supplemental Table 2).
Table 1.
Virome sequencing reads
| Reads | Contigs | ||||
|---|---|---|---|---|---|
|
|
|||||
| Number | Lengtha | Singletonsb | Number | Lengtha | |
| Subject 1 | |||||
| Month -6 | 95,701 | 408bp | 34,104 (36%) | 806 | 1,065bp |
| Month -3 | 358,287 | 307bp | 13,993 (4%) | 2,010 | 2,015bp |
| Day 1 | 180,164 | 369bp | 9,028 (5%) | 1,930 | 1,750bp |
| Day 30 | 105,605 | 196bp | 25,904 (25%) | 956 | 566bp |
| Day 60 | 152,858 | 393bp | 38,521 (25%) | 4,039 | 595bp |
| Month 11 | 178,097 | 428bp | 32,422 (18%) | 3,506 | 937bp |
|
| |||||
| Subject 2 | |||||
| Month -6 | 76,960 | 410bp | 9,876 (13%) | 1,873 | 883bp |
| Month -3 | 234,219 | 272bp | 31,657 (14%) | 2,183 | 925bp |
| Day 1 | 117,081 | 347bp | 6,318 (5%) | 1,985 | 1,218bp |
| Day 30 | 208,243 | 392bp | 17,225 (8%) | 4,365 | 966bp |
| Day 60 | 68,061 | 416bp | 4,535 (7%) | 2,650 | 755bp |
| Month 11 | 90,557 | 410bp | 65,337 (72%) | 3,302 | 421bp |
|
| |||||
| Subject 3 | |||||
| Day 1 | 126,644 | 421bp | 5,649 (4%) | 4,615 | 961bp |
| Day 30 | 121,005 | 411bp | 18,569 (15%) | 3,915 | 699bp |
| Day 60 | 79,465 | 403bp | 10,073 (13%) | 1,497 | 682bp |
| Month 11 | 80,511 | 418bp | 20,301 (25%) | 2,047 | 754bp |
|
| |||||
| Subject 4 | |||||
| Day 1 | 73,639 | 400bp | 4,830 (7%) | 1,360 | 816bp |
| Day 30 | 73,282 | 372bp | 12,098 (17%) | 8,843 | 407bp |
| Day 60 | 111,935 | 397bp | 22,699 (20%) | 14,757 | 571bp |
| Month 11 | 55,858 | 418bp | 7,829 (14%) | 2,124 | 712bp |
Average length
Number of reads that do not assemble to contigs
Figure 1.
Putative biological assignments for contigs from human salivary viromes. Contigs were assigned to biological groups based on significant blastX E-scores based on the NCBI non-redundant database. The percentage of contigs assigned to each biological group is demonstrated for each subject at all time points. Subjects #1 and #2 were sampled at additional the additional time points of Month -6 and Month -3, however, no intervention took place during the study.
Analysis of Streptococcus Group II (SGII) CRISPR spacers
We amplified SGII CRISPR spacers from each specimen, as previously described [12]. We chose a metagenomic approach to evaluating CRISPRs based on their repeat sequences known to be present in the genomes of Streptococcus gordonii and S. thermophilus (Supplemental Table 3), as we had previously analyzed Streptococcus Group I (SGI) CRISPR spacers present in S. mutans, S. pyogenes, S. mitis, S. salivarius, S. anginosus, S. sanguinis, and S. agalactieae [12]. The benefit of this metagenomic approach is that numerous different CRISPR loci harboring these repeat motifs can be evaluated simultaneously; however, the technique is limited in that we cannot assign individual CRISPR spacers to a single species or strain. From all subjects combined, we characterized 21,118 spacers, of which 3,473 (16%) were unique (Table 2). As was previously seen for SGI spacers [12], the estimated SGII spacer richness varies over time and between subjects (Supplemental Figure 1), while Good’s coverage estimate was >71 for all individual time points and >88 for all subjects when all time points are combined (Table 2).
Table 2.
SGII and SGI CRISPR Spacers
| SGII | SGI | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Number of Spacers |
Number of Singletons |
New Spacersa |
Good's Estimateb |
Number of Spacers |
Number of Singletons |
New Spacersa |
Good’s Estimateb |
||
| Subject 1 | |||||||||
| Month -6 | 969 | 53 | 162 | 94.5 | 1038 | 240 | 279 | 76.9 | |
| Month -3 | 1429 | 42 | 54 | 97.1 | 1288 | 88 | 80 | 93.2 | |
| Day 1 | 1254 | 60 | 57 | 95.2 | 1242 | 145 | 111 | 88.3 | |
| Day 30 | 1217 | 51 | 42 | 95.8 | 1512 | 60 | 61 | 96.0 | |
| Day 60 | 1080 | 93 | 66 | 91.4 | 1218 | 228 | 83 | 81.3 | |
| Month 11 | 1057 | 67 | 53 | 93.4 | 1323 | 226 | 220 | 82.9 | |
| Total c | 7006 | 173 | 434 | 97.5 | 7621 | 529 | 834 | 93.1 | |
|
| |||||||||
| Subject 2 | |||||||||
| Month -6 | 1030 | 286 | 479 | 72.2 | 1207 | 132 | 228 | 89.1 | |
| Month -3 | 1067 | 197 | 211 | 81.5 | 1132 | 161 | 275 | 85.8 | |
| Day 1 | 1020 | 217 | 168 | 78.7 | 916 | 261 | 188 | 71.5 | |
| Day 30 | 998 | 234 | 123 | 76.6 | 1266 | 224 | 78 | 82.3 | |
| Day 60 | 937 | 257 | 120 | 72.6 | 1044 | 129 | 61 | 87.6 | |
| Month 11 | 1094 | 208 | 219 | 81.0 | 1488 | 136 | 88 | 90.9 | |
| Total c | 6146 | 511 | 1320 | 91.7 | 7053 | 525 | 918 | 92.6 | |
|
| |||||||||
| Subject 3 | |||||||||
| Day 1 | 839 | 172 | 330 | 79.5 | 1339 | 235 | 491 | 82.4 | |
| Day 30 | 1036 | 208 | 307 | 79.9 | 1237 | 157 | 142 | 87.3 | |
| Day 60 | 841 | 212 | 166 | 74.8 | 1191 | 293 | 290 | 75.4 | |
| Month 11 | 950 | 251 | 217 | 73.6 | 1357 | 158 | 103 | 88.4 | |
| Total c | 3666 | 411 | 1020 | 88.8 | 5124 | 425 | 1026 | 91.7 | |
|
| |||||||||
| Subject 4 | |||||||||
| Day 1 | 997 | 114 | 286 | 88.6 | 1169 | 71 | 186 | 93.9 | |
| Day 30 | 1086 | 146 | 150 | 86.6 | 1028 | 84 | 86 | 91.8 | |
| Day 60 | 1210 | 149 | 116 | 87.7 | 962 | 73 | 70 | 92.4 | |
| Month 11 | 1023 | 194 | 291 | 81.0 | 1308 | 114 | 220 | 91.3 | |
| Total c | 4316 | 353 | 843 | 91.8 | 4467 | 260 | 562 | 94.2 | |
Number of unique spacer sequences that were not identified at prior time points
Estimate of Good’s coverage
Based on all time points combined prior to analysis of spacer composition
Newly identified spacers
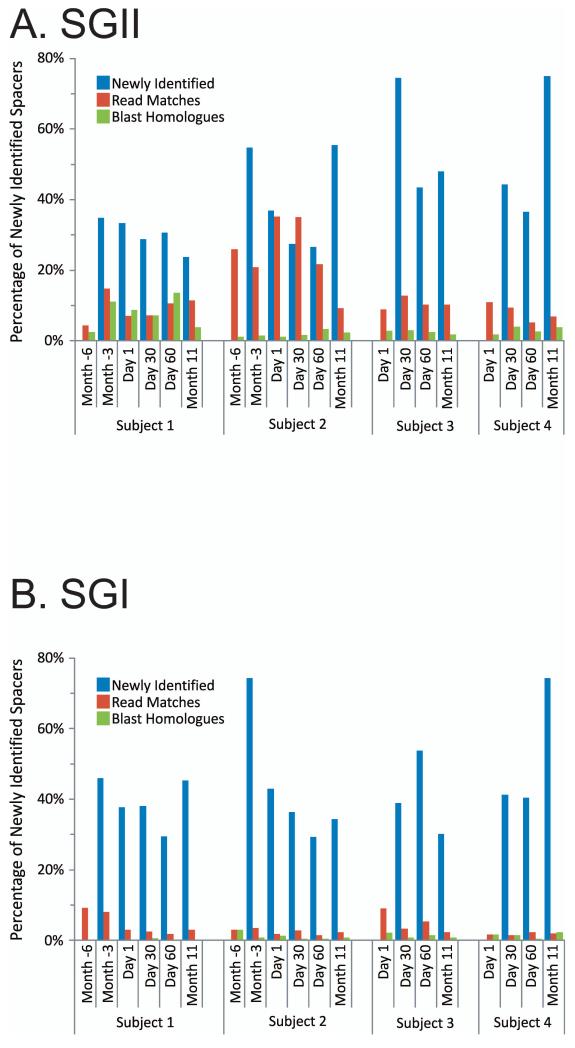
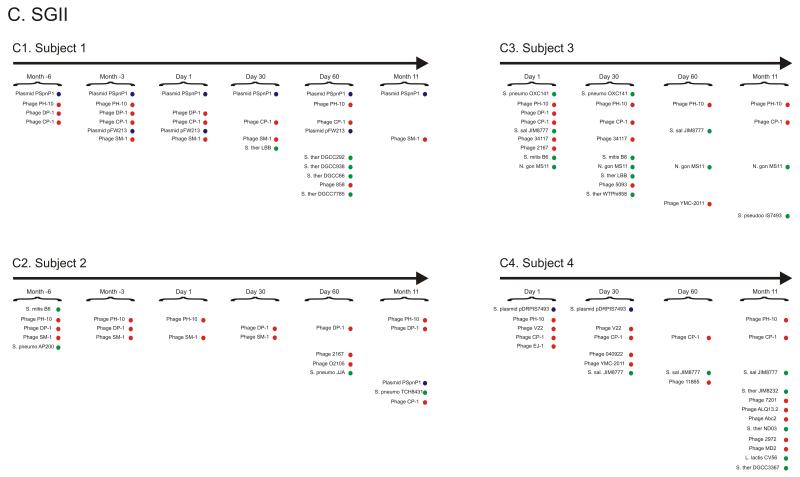
Unique SGII and SGI spacers were identified for each subject at each time point (Figure 2). While many SGII spacers were shared for all time points in each subject (18% for subject #1, 3% for subject #2, 4% for subject #3, and 3% for subject #4) (Supplemental Figure 2), there were newly identified spacers at each time point in all subjects (ranging from 25% to 75%). Because mutations occur frequently in viral genomes, we measured CRISPR spacers that match virome matches (also referred to as protospacers) with an allowance for a single mismatch in any nucleotide along the length of the sequence [23]. Many of the newly identified SGII and SGI spacers at each time point have matches to virome reads (Figure 2 Panels A and B), suggesting that the streptococcal community may be adapting to local viruses. A much smaller proportion of the newly identified CRISPR spacers have homologues to various known bacteriophage, plasmids, and streptococcal genomes (Supplemental Table 4). Through examination of the homologues to newly identified spacers at each time point, we find that many are homologous to the same bacteriophage and plasmids as spacers from prior time points (Figure 2 Panel C and Supplemental Figure 3), suggesting that many bacteriophage and plasmids are repeatedly targeted by an evolving oral CRISPR repertoire.
Figure 2.
Percentage of spacers (Panels A and B) and diagram of SGII CRISPR spacer homologous to various bacteria, phage, and plasmids (Panel C). The percentage of spacers in each subject and time point that were not identified at prior time points is demonstrated in blue. The percentage of those spacers that match virome reads is demonstrated in red, and the percentage of those spacers that have homologues in the NCBI NR database are shown in green. Panel A - SGII spacers and Panel B - SGI spacers. For the initial time point for each subject, homologues to CRISPR spacer complements are demonstrated in Panel C. At each subsequent time point, only homologues to newly identified spacers that were not present in prior time points are shown. For example, in subject #3 (Panel C3), spacers homologous to streptococcal phage PH-10 are identified on Day 1, while on Day 30 newly identified spacers that were not present on Day 1 also have homology to phage PH-10. Panel C1-Subject #1, Panel C2 - Subject #2, Panel C3 - Subject #3, and Panel C4 - Subject #4. Homologues to phage are shown in red, plasmids in blue, and bacteria in green.
While most SGII spacers were unique to each subject, the greatest degree of inter-individual spacer sharing (approximately 8%) is shared between subjects #1 and #2 (Supplemental Figure 4). That these subjects reside in the same household, suggests that their shared spacers result from common environmental factors or direct contact. Principal coordinates analysis demonstrates that the variation present in SGII CRISPR spacers was unique to each subject (Supplemental Figure 5), similar to the variation previously found for SGI CRISPR spacers [11]. No SGII spacers were identical to any SGI spacers, indicating that SGI and SGII CRISPRs likely reflect exposures to different virus populations.
CRISPR spacer matches in human salivary and other viromes
We tested whether SGI and SGII CRISPR spacers are specific to salivary viruses by comparing them with virome reads from each subject. For all subjects, there were 3422 (0.13% of the virome reads) unique salivary virome reads with SGI CRISPR spacer matches and 3964 (0.15% of the virome reads) unique reads with SGII CRISPR spacer matches (Supplemental Figure 6). There were some oropharyngeal virome reads that had SGI and SGII spacer matches (0.02% and 0.04% of the virome reads for SGI and SGII spacers, respectively) from a different group of subjects [15]. No marine virome reads [24] or human fecal virome reads [13] had CRISPR spacer matches, and only 3 reads from a pooled human respiratory tract virome [14] had CRISPR spacer matches (<0.00% of the virome reads), suggesting that SGI and SGII spacers are the result of relatively specific exposures to human oral viruses.
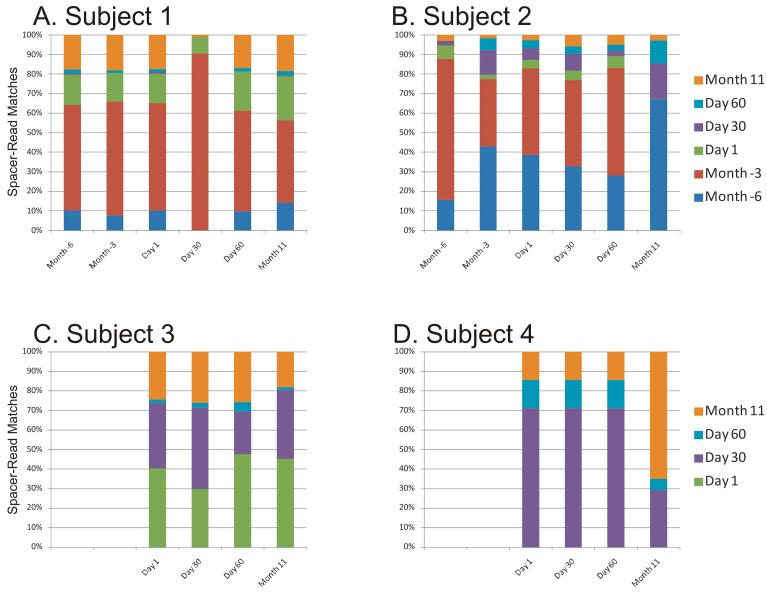
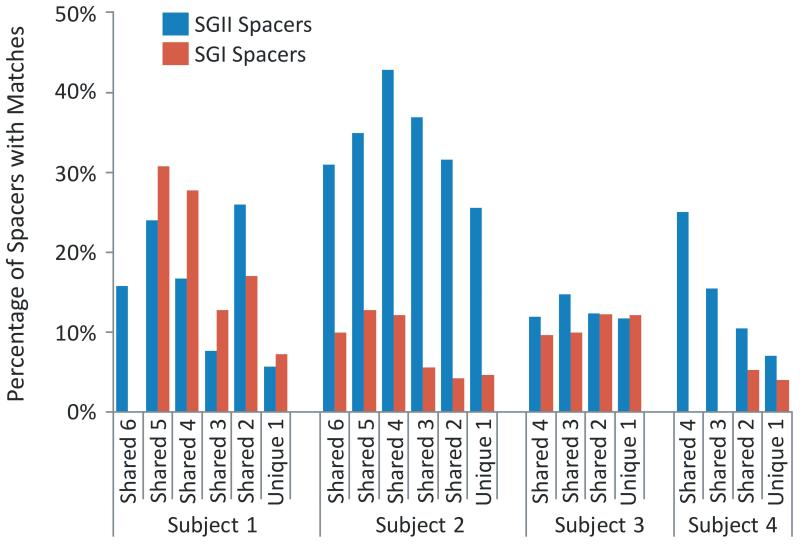
When comparing spacers with virome reads from that same subject, we found numerous reads matching SGI and SGII spacers (Supplemental Figure 7), with the majority containing a single nucleotide mismatch (Supplemental Figure 8). Within individual subjects, the SGI and SGII spacers present on any particular day also had corresponding viral protospacers from that day as well as most other days studied (Figure 3 and Supplemental Figure 9). These findings indicate that CRISPR spacers coexist with corresponding protospacers and that certain CRISPR spacers persist in the community. In subjects #1 (p<0.0005), #2 (p<0.005), and #4 (p<0.005), SGII spacers present at multiple time points have a much higher proportion of matching virome reads than spacers that are unique to single time points (Figure 4); for SGI spacers, only subject #1 (p<0.0005) had a significantly higher proportion of spacers shared between multiple time points matching virome reads.
Figure 3.
Fraction of virome reads with SGI CRISPR spacer matches over time within each subject. The virome reads with CRISPR spacer matches are normalized by virome size. Each column represents the SGI CRISPR spacer repertoire characterized at the individual labeled time point, and the Y axis represents the normalized percentages of unique reads with matches to CRISPR spacers recovered from each of the time points. Blue represents spacer-read matches from Month -6, red represents Month -3, green represents Day 1, purple represents Day 30, cyan represents Day 60, and orange represents Month 11.
Figure 4.
Percentage of spacers with matches to virome reads in each subject. The percentage of spacers that are unique to individual time points and those that are shared between multiple time points are shown. The percentage of spacers with matches to reads in each virome is demonstrated on the Y axis.
CRISPR spacers match virome reads among different subjects
We tested whether CRISPR spacers from each subject were found in the virome reads from other subjects to determine whether streptococci from individual subjects might have the potential to counteract viruses from others. Many SGI and SGII CRISPR spacers were identified from individual subjects that had matches in the viromes of others (Figure 5). We also tested whether each subject was more likely to have CRISPR spacers matching their own viruses than viruses in other subjects. At each time point, the fraction of CRISPR spacers with matches to each of the subject-associated viromes was compared (Supplemental Figure 10). For SGI, each subject was no more likely to have spacers with matches in their own virome than in the viromes of other subjects. For SGII spacers, both subjects #1 and #2 were more likely across nearly all time points to have spacers that matched their own viruses than viruses from other subjects (p = 0.00024 and p = 0.0046, respectively).
Figure 5.
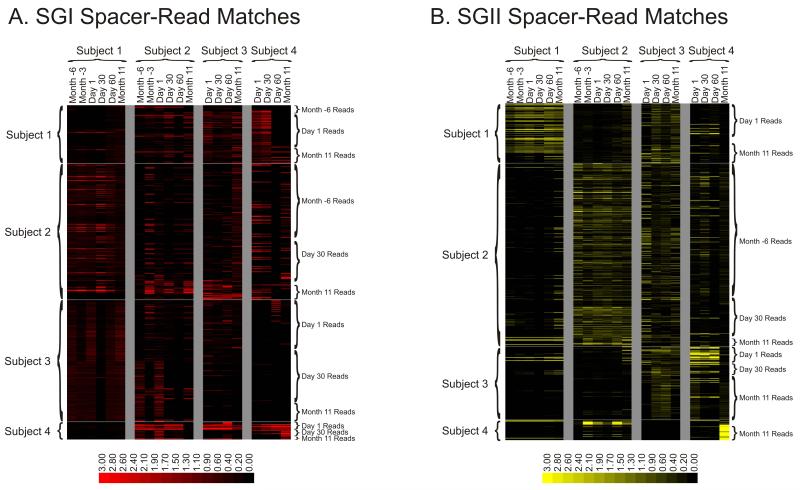
Heatmap of virome reads with CRISPR spacer matches for SGI and SGII CRISPR spacers. Each row represents a unique virome read, and each column represents the CRISPR spacer repertoire found on that day for that subject. Reads from viromes of each subject are identified on the left of each diagram, and the day from which each read was recovered is shown on the right of each diagram. The intensity scale bar is shown below each panel, and its values correspond to the percentage of CRISPR spacer matches present in each subject and time point. Panel A – SGI CRISPR spacer-read matches, Panel B – SGII CRISPR spacer-read matches.
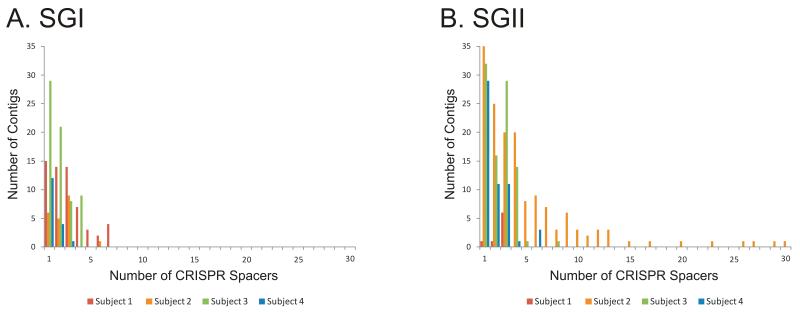
We analyzed the virome contigs from each subject at each time point, and found numerous viral contigs that were matched by CRISPR spacers from each subject and time point (Supplemental Figure 11). The relative proportion of contigs that had CRISPR spacer matches was no different for contigs with known viral homologues than for those without (0.7% vs. 0.6%, respectively), suggesting that many of the contigs without viral homologues likely also are viral in origin. To determine if many of these virome contigs have been repeatedly sampled by streptococci, we analyzed the number of CRISPR spacers that matched each individual virome contig. We found that for SGI and SGII CRISPR spacers, there were numerous CRISPR spacers that matched virome contigs (ranging from 1 to 30 different spacers per virome contig) (Figure 6). In general, there were more SGI CRISPR spacers that matched virome contigs for each subject than SGII CRISPR spacer matches. Some viral contigs with matching CRISPR spacers have homologues to known streptococcal viruses (Supplemental Figure 12). There were no SGI or SGII spacers that targeted the same virome read or contig, suggesting that different viruses are targeted by each CRISPR system.
Figure 6.
Number of virome contigs with multiple SGI and SGII CRISPR spacer matches. The number of contigs is represented on the Y-axis, the number of CRISPR spacer matches for each contig is represented on the X-axis, and the different colors represent the subject from which each CRISPR spacer was derived.
Protospacer-associated motifs
Previous studies have suggested that the CRISPR/Cas system is only capable of sampling viral sequences that are flanked by motifs called protospacer-associated motifs (PAMs), which generally are located within a few nucleotides of the 3′ end of the protospacer [25]. Our analysis of SGI CRISPRs demonstrated that there are specific 3′ motifs (Supplemental Figure 12), consistent with previously identified PAMs for these specific repeat motifs [25]. Because we cannot identify with certainty the positive strand of each virus, we analyzed the 3′ ends of the protospacers from both strands, likely resulting in an underestimation of the presence of PAMs by 50%. For SGII spacers, no previously identified PAMs were recognized; however, there were some motifs present at conserved positions (Supplemental Figure 13). The presence of these conserved motifs supports the hypothesis that SGI and SGII CRISPRs are specific for salivary viruses.
Discussion
While previous studies have examined CRISPR-virus interactions in various ecosystems [21, 26, 27], we provide the first such analysis in humans. By sampling our subjects at various time points over a 17-month period, we were able to assess whether potential interactions between streptococcal CRISPRs and viruses persist over time. We analyzed CRISPR spacers from two distinct repeat sequence families in order to examine dynamics in different groups of streptococci. That no SGI or SGII CRISPR spacers were identical in any subject, nor any individual virome contigs targeted by both SGI and SGII CRISPRs, supports the assumption that SGI and SGII CRISPRs tend to be harbored by different streptococci. As has previously been noted for SGI CRISPRs [12], SGII spacer richness varies considerably between different subjects and generally is greater than that of SGI spacers (Table 2 and Supplemental Figure 1).
The abundance of SGI and SGII CRISPR protospacers in the salivary viromes lends support to the concept that oral streptococci adapt to salivary viruses and that the history of host-viral encounters can be deciphered from CRISPR sequences [12, 21]. Our finding of multiple streptococcal protospacers in many viral contig sequences provides further support to this contention. The finding of numerous protospacers in virome contigs (Supplemental Figure 11) and the newly identified spacers at each time point matching viruses that also were targeted at earlier time points (Figure 2) suggests that these viruses have been repeatedly targeted by the CRISPR/Cas system. While there are limited viromes available for comparison, the presence of streptococcal protospacers in pooled oropharyngeal viromes from unrelated subjects suggests that there are viruses common to saliva and the oropharynx. Human sputum is commonly known to be contaminated with saliva, which likely explains our finding a few protospacers present in a pooled human respiratory virome. Interestingly, for both SGI and SGI CRISPR spacers, there are certain time points that when compared to others are highly enriched for viruses with matching protospacers (Figure 3 and Supplemental Figure 9). These matching protospacers are independent of virome sampling depth, and we believe reflect a true biological phenomenon such as a higher proportion of CRISPR-targeted lysogenic phage present in the community at these time points. A much finer sampling effort than the one presented here would be necessary to decipher how virome characteristics may vary over short time periods.
Our analysis clearly demonstrates that CRISPR spacers and viruses with corresponding protospacers coexist at the same time in a given subject; however, our amplification methods preclude assessment of CRISPR locus evolution because our analysis suggests that the repeat based priming leads to altered spacer order in CRISPR loci [12]. The coexistence of viruses and matching CRISPR spacers presented here is similar to those previously reported for acid mine drainage systems [21]. There also were many instances of CRISPR spacers with matches in viromes from other subjects. While this could potentially reflect a history of shared streptococci harboring these CRISPRs in different subjects, the vast majority of the CRISPR spacers were subject specific (Supplemental Figure 4), and thus strain ancestry cannot account for most of the CRISPR spacers that match virome reads from different subjects. One potential explanation for viral protospacers that match CRISPR spacers from different subjects may relate to the fact that PAMs determine which portions of salivary viral genomes can be sampled. As such, relatively conserved portions of virus genomes adjacent to PAMs may be sampled, resulting in CRISPR spacer repertoires more highly adapted to conserved portions of salivary viruses.
While there are many aspects of the human microbiome that have yet to be thoroughly examined, the analysis presented here substantially broadens our understanding of the interplay between bacteria and their viruses in the human oral cavity. From the identification of salivary viruses, to the analysis of CRISPR groups in various streptococci, we now know that these prominent members of the oral microbiome possess a substantial immune repertoire to counteract oral viruses. Through analysis of the most abundant viruses present, we also have demonstrated that there are robust oral viral communities present in the saliva of each subject. Many of these viromes have similar features including an abundance of identifiable bacteriophage, high estimated diversity, and numerous factors putatively involved in virulence functions similar to previous analysis of salivary viromes [11]. That each subject studied has CRISPR spacers that match viruses from all other subjects studied, suggests that streptococcal CRISPR/Cas systems may have the ability to respond to viruses with matching protospacers they have yet to encounter regardless of whether their matching spacers are newly acquired at the 5′ end of the locus or the entire locus is inherited. As we continue to explore the human microbiome, the interactions between viruses and their host bacteria are critical to an understanding of the processes that shape the human indigenous microbiota.
Experimental Procedures
Subject enrollment
Recruitment of each subject and enrollment in the current study was approved by the Stanford University Administrative Panel on Human Subjects in Medical Research. Each subject donated saliva samples over an 11 to 17 month time period from January 2008 to July 2009. Each subject received a baseline periodontal examination including measurements of probing depths, clinical attachment loss, Gingival Index, Plaque Index, and gingival irritation [28], and were all found to be periodontally healthy with an overall average clinical attachment loss of <1mm. In addition, all subjects had normal oral mucous membranes and were free from non-restored carious lesions. Exclusion criteria included antibiotic administration during the 12 months prior to the beginning of the study and preexisting medical conditions that could result in immunosuppression.
Isolation and analysis of viruses
Saliva from human subjects was filtered sequentially through 0.45μ and 0.2μ filters to remove cellular debris, and the remaining fraction purified on a cesium chloride gradient as previously described [11]. Only the fraction at the density of most known viruses [29] was retained; it was then further purified on Amicon YM-100 protein purification columns (Millipore, Inc., Bellerica, MA), and treated with DNASE I, followed by lysis and DNA purification using Qiagen UltraSens virus kit (Qiagen, Valencia, CA). Resulting DNA was amplified using Qiagen RepliG MDA amplification (Qiagen, Valencia, CA), fragmented and bar-coded libraries created, followed by sequencing using primer A on a 454 Life Sciences Genome Sequencer FLX instrument using Titanium chemistry (Roche Applied Science, Indianapolis, IN). Reads from each subject at each time point were subjected to quality control to remove low quality reads, defined as short reads (reads <100 nucleotides), reads with >10 homopolymer tracts, and reads with ambiguous characters. Remaining reads were analyzed using CLC Genomics workbench 3.65 (CLC bio USA, Cambridge, MA) to construct assemblies based on 98% identity with a minimum of 20% read overlap, consistent with criteria developed to discriminate between highly related viruses [30]. Because the shortest reads were 100 nucleotides, the minimum tolerated overlap was 20 nucleotides, and the average overlap ≥20 nucleotides (range 20-43 nucleotides) depending on the characteristics of each virome. Contigs were assigned to categories (virus, bacteria, hominid, unknown, and other) based on the presence of known homologues using blastX analysis of the NCBI non-redundant database with an E-score cutoff value of 10−3. Reads were assigned to the category ‘Hominid’ if they were assigned to Homo sapiens or Pan troglodytes. Reads were assigned to the category ‘Other’ if they did not meet the criteria for the other outlined categories, and usually represented homologues to redundant eukaryote DNA. For subject #1 on Day 60, subject #3 on Day 60, and subject #4 on Day 1, there likely was clonal Ralstonia eutropha contamination (a significant proportion of the virome reads map to different areas of the Ralstonia eutropha genome) that represented less than 15% of the reads in each of these respective viromes. Contigs with significant homology (blastN E-score ≤10−5) to Ralstonia were removed from subsequent analysis. No reads or contigs with significant homology to Ralstonia had protospacers matches to SGI or SGII CRISPR spacers. Viral contigs were analyzed using FGenesV (Softberry Inc, Mount Kisco, NY) for ORF prediction, and individual ORFs analyzed using blastX analysis against the NCBI non-redundant database. If the best hit were to a hypothetical gene or to a gene with no known putative function, lower level hits with known putative function and an E-score of at least 10−3 were used for the annotation (Supplemental Figure 10). Specific viral homologues were determined by parsing blastX results for known viral genes including replication, structural, transposition, restriction/modification, hypothetical, and other genes previously found in viruses for which the E-score was at least 10−3 [11]. Results then were manually analyzed to ensure accuracy.
Amplification of streptococcal CRISPR spacers
From each subject, genomic DNA was prepared from saliva using Qiagen QIAamp DNA MINI kit (Qiagen, Valencia, CA). Primers SGRPF-1 (CAGTTACTTAAATCTTGAGAG) and SGRPR-1 (AGATTTAAGTAACTGTACAAC) were designed based on their specificity to the CRISPR repeat motifs present in S. gordonii str. Challis substr. CH1, S. thermophilus LMD-9, S. thermophilus LMG-18311, and S. thermophilus CNRZ-1066, and were used to amplify CRISPRs from salivary DNA by PCR. Reaction conditions included 5μl 10X PCR buffer (Applied Biosystems, Foster City, CA), 3μl MgCl2 (25mM), 1μl of each of the forward and reverse primer (20pmol each), 0.5μl AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA), 5μl salivary DNA template, and 34.5μl H2O. The cycling parameters were 3 minutes initial denaturation at 95°C, followed by 30 cycles of denaturation (60 seconds at 95°C), annealing (60 seconds at 45°C), and extension (5 minutes at 72°C), followed by a final extension (10 minutes at 72°C). CRISPR amplicons were purified using Qiagen QIAquick PCR Purification kit (Qiagen, Valencia, CA), and purified amplicon mixtures were cloned into the pCR4 vector using Invitrogen TOPO TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA). For each subject at each time point, 384 clones were picked, and subjected to Sanger sequencing using standard M13 primers.
Analysis of streptococcal spacers and repeats
CRISPR sequences were analyzed using Sequencher 4.9 (Gene Codes Corporation, Ann Arbor, MI). Primer sequences were removed and only those sequences with a length of >80 nucleotides and a quality score >80% were chosen for further analysis. CRISPR repeats were identified based on an algorithm that searches for the first 5 nucleotides of the CRISPR repeat motif (GTTTT) followed by the last 5 nucleotides (ACAAC) of the motif, with allowances for single nucleotide polymorphisms at any nucleotide position. The repeats were defined as any set of nucleotides approximately 36 nucleotides long (range 30-42 nucleotides, median 36 nucleotides, average 36 nucleotides) that begins and ends with the aforementioned nucleotides. In addition, for all subjects at all time points the sequences were manually examined to ensure that no repeat motifs went undetected and that no errors occurred in the classification of repeat motifs. Spacers were defined as any nucleotide sequence (length ≥ 20) located between repeat motifs. Only clone sequences containing at least 2 full repeat motifs and 1 spacer were retained, and all others were removed from the analysis. Spacers were grouped according to 3 rules: 1) spacers that were identical, 2) spacers that were identical, with the exception of a single nucleotide polymorphism, and 3) spacers that differed in length, but were identical over the length of the shorter spacer. For each subject at each time point, a database of spacers and repeats was generated, and databases were compared to determine shared spacers and repeats, and to create heat maps using Java Treeview [31]. Good’s coverage was determined as the estimation of the number of singletons in the population (n), compared to the total number of sequences (N), using the equation [1-(n/N)] x 100 [32]. Rarefaction analysis was performed based on species richness estimates of 10,000 iterations using EcoSim [33]. Beta Diversity was determined using Sorensen’s similarity, which also was used as the input for principal coordinates analysis. Spacers from each subject were subjected to blastN analysis based on the NCBI non-redundant database. Hits were considered significant based on bit scores ≥50, which roughly correlates to 2 nucleotide differences over the 30 nucleotide average length of the spacers.
Analysis of protospacers
All CRISPR spacers from each subject were placed into a database and used to search virome reads and contigs. Protospacers were defined as virome sequences that were identical or had a single nucleotide mismatch when compared to the CRISPR spacers sequences. Protospacers could be present on either the sequenced strand for each virome sequence or its reverse complement. Control databases included a virome from the Sargasso Sea [24], human respiratory tract viromes [14], human oropharyngeal viromes [15], and viromes from human feces [13]. Numerous other aquatic and environmental viromes had no protospacer matches to the streptococcal CRISPR spacers, but were not reported here. CRISPR spacers for each subject and time point were used to search all of the virome reads and contigs for protospacer matches, and the number of spacer matches per virome read or contig were used to create heatmaps. These heatmaps were normalized by the total number of protospacers per virome read or contig for each time point, and were generated using Java Treeview [31]. For the analysis of CRISPR-spacer read matches (Figure 4 and Supplemental Figure 7), the number of reads that had matching spacers was normalized by the number of reads present for each time point. PAMs (Protospacer Associated Motifs) were determined by taking the 8 nucleotides at either end of protospacers found in viral sequences. All of these 3′ and 5′ sequences were added to a database and analyzed using Weblogo [34]. Because we were unable to determine whether we were examining the forward or reverse DNA strand for each virome read with a protospacer, we examined the 3′ and 5′ ends for both DNA strands, which likely resulted in a 50% reduction in the frequency of the PAMs presented (Supplemental Figure 13).
Statistical analysis
A rank test was used to assess whether a subjects’ virome reads were more likely to match CRISPR spacers from that individual compared to the CRISPR spacers from other individuals in the study. The approach was used to eliminate bias that might be introduced by the fact that the number of spacers observed for each subject and time point is variable. For each subject and time point, all subjects were ranked according to the fraction of virome reads from that subject with spacer matches in that subject and time point. Thus, a rank was produced for each observation of each subject. For example, subject #1 was observed at 7 time points, producing a total of 7 rankings of 4 numbers. For each (subject i, time j) point, the data were reduced to binomial counts by counting a ’1′ if among the fractions of virome reads from all subjects, the fraction of virome reads from subject i had the most spacers that matched in that subject, time pair. The R programming language was used to perform an exact binomial test of p=1/4. Fisher’s exact test was used to determine whether the proportion of shared and unique CRISPR spacers have equal distributions of virome read matches.
Supplementary Material
Acknowledgements
Supported by the Robert Wood Johnson Foundation, the Stanford University Institute of Infection, Transplantation and Immunity, the Burroughs Wellcome Fund, and NIH 1K08AI085028 to DTP, and the National Institutes of Health Director’s Pioneer Award DP1OD000964 to DAR. DAR is supported by the Thomas C. and Joan M. Merigan Endowment at Stanford University. We thank Christine Sun, Jill Banfield, Peter Loomer, and Gary Armitage for their contribution to this work.
Footnotes
Authors Contributions Conceived and designed experiments: DTP and DAR. Performed the experiments DTP. Analyzed the data: DTP and JS. Wrote the manuscript: DTP and DAR.
References
- 1.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449(7164):804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dethlefsen L, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77(6):2367–75. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledder RG, et al. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007;73(2):516–23. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13(12):589–95. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto M, et al. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J Med Microbiol. 2004;53(Pt 6):563–71. doi: 10.1099/jmm.0.45576-0. [DOI] [PubMed] [Google Scholar]
- 10.Breitbart M, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159(5):367–73. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2011 doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pride DT, et al. Analysis of streptococcal CRISPRs from human saliva reveals substantial sequence diversity within and between subjects over time. Genome Res. 2011;21(1):126–36. doi: 10.1101/gr.111732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozytotic twins and their mothers. Nature. 2010:466. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willner D, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4(10):e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willner D, et al. Microbes and Health Sackler Colloquium: Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 17.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marraffini LA, Sontheimer EJ. Invasive DNA, chopped and in the CRISPR. Structure. 2009;17(6):786–8. doi: 10.1016/j.str.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320(5879):1047–50. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 22.Thurber RV, et al. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4(4):470–83. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 23.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190(4):1390–400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angly FE, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4(11):e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mojica FJ, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 26.Heidelberg JF, et al. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One. 2009;4(1):e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10(1):200–7. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 28.Loe H. The Gingival Index, the Plaque Index and the Retention Index Systems. J Periodontol. 1967;38(6):610–6. doi: 10.1902/jop.1967.38.6.610. Suppl. [DOI] [PubMed] [Google Scholar]
- 29.Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD. Virus Taxonomy: Sizth Report of the International Committee on Taxonomy of Viruses. Supplement 10. Springer-Verlag; New York: 1995. [Google Scholar]
- 30.Breitbart M, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci U S A. 2002;99(22):14250–5. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 32.Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 33.Lee SG, Kim CM, Hwang KS. Development of a software tool for in silico simulation of Escherichia coli using a visual programming environment. J Biotechnol. 2005;119(1):87–92. doi: 10.1016/j.jbiotec.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 34.Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.