Abstract
The cellular mRNA decay machinery plays a major role in regulating the quality and quantity of gene expression in cells. This machinery involves multiple enzymes and pathways that converge to promote the exonucleolytic decay of mRNAs. The transcripts made by RNA viruses are susceptible to degradation by this machinery and, in fact, can be actively targeted. Thus, to maintain gene expression and replication, RNA viruses have evolved a number of strategies to avoid and/or inactivate aspects of the cellular mRNA decay machinery. Recent work uncovering the mechanisms used by RNA viruses to maintain the stability of their transcripts is described below.
Introduction
The cellular mRNA decay machinery plays a major role in influencing gene expression in eukaryotic cells. Differential mRNA stability, for example, is a highly regulated process that accounts for approximately 20–50% of the changes in gene expression levels observed in cells in response to various stimuli (1,2). The quality of RNAs is also constantly monitored by the cellular RNA decay machinery. Transcripts containing premature termination codons, lacking a termination codon, or containing stalled ribosomes are rapidly degraded by the cell (3). Furthermore, unwanted transcripts that arise from intergenic transcription and introns are generally rapidly degraded (4). The transcripts produced by RNA viruses are apt to be placed in this ‘unwanted’ category by the cellular RNA decay machinery for several reasons. These viral transcripts often lack a nuclear experience, thus their messenger ribonucleoprotein (mRNP) organization is likely different than that of a cellular mRNA. Some viral transcripts are uncapped and/or lack a poly(A) tail and thus could be recognized as incomplete or malformed mRNAs. Some viral mRNAs contain multiple open reading frames and thus may be recognized as containing a premature termination codon. How viruses avoid surveillance by the cellular mRNA decay machinery during infection is an understudied area of virus-host interactions. The purpose of this review is to highlight the fundamental pathways and factors of the cellular mRNA decay machinery, discuss recent observations on how the transcripts made by RNA viruses interface with them, and identify a variety of issues for future consideration.
The Cellular RNA Decay Machinery
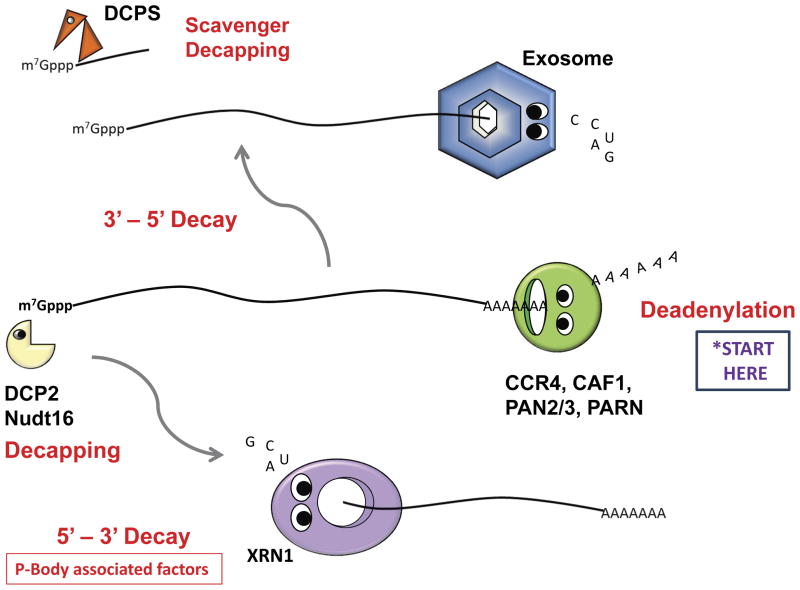
The major pathways of mRNA decay in mammalian cells are diagrammed in Figure 1. The first step in the decay of most mRNAs is the shortening of the poly(A) tail, also known as deadenylation (5). There are multiple deadenylase enzymes in cells, including CCR4, CAF1, PARN and PAN2/3 (6,7). Deadenylation is often the rate limiting step in the decay of many mRNAs. Following deadenylation, the decay of the body of the mRNA is afforded by two exonucleolytic pathways. To be shuttled into the 5′-to-3′ exonucleolytic decay pathway, the 3′ end of deadenylated mRNAs associates with the cytoplasmic LSm1–7 complex and PAT1 initiates the recruitment of factors to remove the m7Gppp cap from the 5′ end (8). There are at least two decapping enzymes in mammalian cells – DCP2 and Nutd16 – which associate with a variety of auxiliary factors (such as DCP1a and HDLS) to effectively remove the 5′ cap from deadenylated transcripts (9). The process of decapping leaves a 5′ monophosphate, creating a substrate for the highly processive 5′-to-3′ exoribonuclease XRN1 (10). XRN1 then degrades the transcript to mononucleotides. Many of the factors in the 5′-to-3′ decay pathway can be found, at least in part, in association with cytoplasmic processing bodies (P-bodies) in cells (11).
Figure 1. The major enzymes and pathways of cellular mRNA decay.
As indicated by the ‘Start Here’ sign, the majority of mRNA degradation in eukaryotic cells is initiated by poly(A) shortening. The four best characterized deadenylase enzymes (CCR4, CAF1, PAN2/3 and PARN) are shown. Following deadenylation, the body of the mRNA is then degraded by one of two exonuclease pathways (or both acting in concert). The exosome complex degrades mRNAs in a 3′-5′ direction (top panel). Exosome-mediated decay leaves a short RNA fragment with a 5′ cap that gets removed by the scavenger decapping activity DCPS. In the 5′-3′ decay pathway (bottom panel), the mRNA is first decapped by DCP2 or Nudt16 and then the body of the mRNA is degraded by the XRN1 exoribonuclease. Many of the components of the 5′-3′ decay pathway can often be found associated in a cytoplasmic granule referred to as the P-body.
For 3′-to-5′ decay, the deadenylated transcript is acted on by the cytoplasmic exosome, a ~400 kDa multi-protein complex that contains a subunit (hDIS3/RRP44) which possesses both RNase II-like hydrolytic exonucleolytic and a PIN domain-mediated endonucleolytic activities (although in humans the cytoplasmic form of hDIS3 (hDIS3L) does not retain an active endonuclease) (12). The activity of the exosome is influenced by the SKI complex (SKI2, 3 and 8) which contains helicase and other activities (13). Following processive decay of the majority of the body of the mRNA by the exosome, the 5′ cap is removed from the small fragment by a scavenger decapping activity (DCPS) (14).
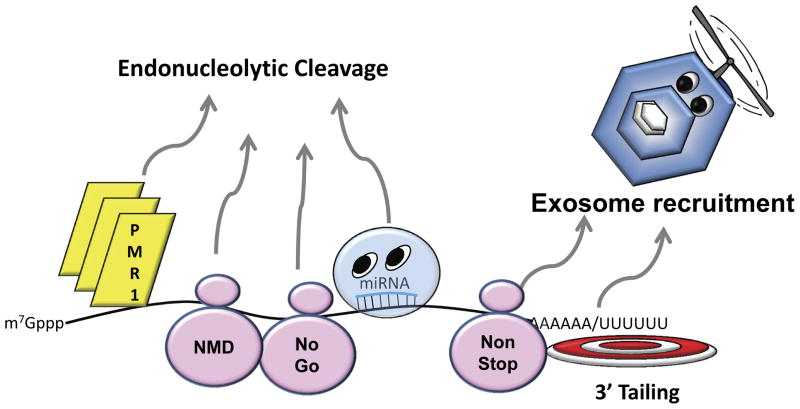
Specialized RNA decay/surveillance pathways also exist in cells (Fig. 2). mRNA decay may be initiated by an endonucleolytic cleavage event through endonucleases such as PMR1, IRE1, G3BP, SMG6, APE1 and Zc3h12a/MCPIP (15). The inducible RNase L protein is also an endonuclease whose activity has been shown to increase upon viral infection (16). The RNA interference-associated decay pathway is also initiated by an endonucleolytic cleavage event mediated by an argonaute protein in the RISC complex (17). Nonsense mediated decay shuttles targeted mRNAs into one or more decay pathways, and involves a series of auxiliary factors, in particular UPF1–3 and a series of SMG proteins (18). The turnover of mRNAs that lack a termination codon, referred to as nonstop mRNA decay, is mediated through the SKI complex outlined above (19). The decay of mRNAs with stalled ribosomes (no-go decay), as well as the decay of nonfunctional 18S rRNA, is mediated by the DOM34-Hbs1 complex (20). The RNA deamination enzymes APOBEC3G and 3F have also been shown to localize to P-bodies (21), suggesting that this form of RNA editing is also associated with RNA degradation in some fashion. Finally, the decay of structured RNAs can be initiated by the attachment of a short poly(A) or poly(U) stretch on the 3′ end by a non-canonical poly(A/U) polymerase to provide a landing pad for the exosome (22).
Figure 2. Alternative and specialized pathways of mRNA decay.
Two of the major routes of alternative mRNA decay are highlighted. First, decay can be initiated by endonucleolytic cleavage through the direct recruitment of endonucleases. These enzymes can interact directly with their target RNAs (e.g PMR1, RNase L), be recruited as part of the nonsense-mediated decay pathway (NMD) at a premature termination codon (Smg6 endonuclease), by stalled ribosomes in the ‘no-go’ decay pathway by recruitment of the Dom34-Hbs1 complex, or by miRNAs directing Ago ‘slicer’ proteins of the RISC complex to the transcript. Alternatively, the exosome can be recruited to the 3′ end of malformed RNAs such as those lacking a translation termination codon (non-stop decay) or onto transcripts with structured 3′ ends by poly(A) or poly(U) tailing.
The process of mRNA decay is highly regulated (Fig. 3). Numerous mRNA binding proteins have been identified that destabilize mRNAs. Some of the best characterized mRNA instability factors include TTP, AUF1, and KSRP (23–25). Small RNA regulators such as miRNAs can also regulate the stability of targeted transcripts (26). Major factors that stabilize mRNAs include HuR and PCBP2 proteins (27, 28). Combinatorial association of these factors with the targeted mRNA, in association in some fashion with the translation machinery and subcellular localization, likely prescribe the fate of the transcript.
Figure 3. Regulation of mRNA stability.
The decay of mRNAs is a highly regulated process. It can be promoted by the interaction of destabilizing factors such as the TTP, AUF1 and KSRP proteins, or by miRNAs. These destabilizing factors can serve to attract deadenylases as shown in the figure, or by alternative mechanisms such as endonucleolytic cleavage. Alternatively, mRNAs can be selectively stabilized by the recruitment of specific proteins such as HuR or PCBP2 to their 3′ UTRs.
Strategies of RNA Viruses to Avoid Deadenylation
Given that poly(A) shortening is often the first and rate limiting step in mRNA decay, RNA viruses likely have developed ways to repress it or avoid it altogether. Several families of RNA viruses, including flaviviruses, bunyaviruses and arenaviruses, have evolved 3′ terminal stem loop structures reminiscent perhaps of those found on non-polyadenylated histone mRNAs (29) that maintain the stability of the transcript while still affording translatability. For RNA viruses that possess a 3′ poly(A) tail on their mRNAs, two strategies to evade deadenylation have been uncovered to date. Poliovirus targets the deadenylase PAN3, which is postulated to initiate deadenylation of many cellular mRNAs before the transcript is handed over to more processive deadenylases, for rapid degradation during infection (30). Sindbis virus recruits the cellular HuR protein to the 3′ untranslated region (UTR) of its transcripts which stabilizes the ~60 base poly(A) tail of these alphaviruses (31,32). Deletion of the high affinity HuR binding site in the 3′UTR of Sindbis virus results in very unstable viral transcripts that become effective substrates for cellular deadenylases (32). Other mechanisms could be used to stabilize the poly(A) tail, including forming structures between the poly(A) and internal sequences of viral transcripts as has been shown for the abundant non-coding PAN mRNA made by Kaposi’s sarcoma associated herpesvirus (KSHV) (33). However, this and other mechanisms of repressing deadenylation have not been demonstrated to date for an RNA virus.
Strategies of RNA Viruses to Avoid the Enzymes of the 5′-to-3′ mRNA Decay Pathway
A variety of evidence suggests that RNA viruses are indeed subject to degradation by the 5′-3′ decay pathway. Overexpression of isoforms of the XRN1 exoribonuclease in mammalian or plant systems, for example, has been shown to inhibit Hepatitis C Virus (HCV) or Tomato Bushy Stunt Virus (TBSV) (34, 35). The XRN1 exonuclease is also required to generate a small subgenomic RNA (sfRNA) as a decay intermediate that is observed during infections with most, if not all, insect-borne flaviviruses (36,37). Overexpression of auxiliary factors associated with the 5′-to-3′ decay pathway have also been associated with the inhibition of viral growth – for example the MOV10 P-body-associated helicase and Human Immunodeficiency Virus (HIV) inhibition (38). Thus the need for viruses to avoid the 5′-to-3′ mRNA decay pathway is starting to move from theoretical considerations to a well-documented reality.
Four strategies that can be associated with evasion of the 5′-to-3′ decay pathway have been identified to date. First, poliovirus infection is associated with degradation of the XRN1 exoribonuclease as well as the auxiliary decapping factor DCP1a (30). This should severely limit the action of this decay pathway during poliovirus infection. Second, many viruses usurp 5′-to-3′ decay pathway factors and disrupt the formation of P-bodies during infection. Brome mosaic virus has been known for years to use the LSm1–7 complex as well as the auxiliary decay factor PAT1 to promote its replication (39). More recently, HCV has been also shown to hijack these same P-body components along with the RCK/p54 helicase, to promote its translation and replication (40). P-body disruption during infection has clearly been documented in a number of RNA virus infections, including flavivirus and picornaviruses (41–43). Third, sequences and structures in the 5′ UTR of viral mRNAs may have evolved under pressure from the 5′-to-3′ decay pathway to provide some resistance. TBSV passaged under the pressure of overexpressed XRN4p is, for example, associated with the emergence of 5′ UTR variants and knock out of Xrn1 leads to viral RNA recombination in a yeast model (34,44). Since the activity of cellular decapping enzymes varies depending on the sequence context of the cap structure, such 5′UTR variations can have a large potential impact on viral resistance to this decay pathway. Finally, HCV usurps the cellular factors miR-122 and Ago2 to stabilize its RNAs via interactions near the 5′ end (45,46).
Strategies of RNA Viruses to Avoid the Exosome and the 3′-to-5′ mRNA Decay Pathway
The association of aspects of the 3′-to-5′ decay pathway with viral infections allowed the initial discovery (and naming) of the SKI complex of proteins as ‘suppressors of yeast killer virus’ (13). This observation, along with the observation that non-polyadenylated RNA viruses all possess large structured elements directly at their 3′ ends that can be inferred to protect viral transcripts from the exosome (47), clearly imply that exosome-mediated decay can have an impact on RNA virus infections. However additional insights into virus escape from exosome-based surveillance await future experimentation.
Strategies of RNA Viruses to Avoid and Interface with Other RNA Decay Pathways and Regulatory Factors
There are several studies which clearly demonstrate that RNA viruses have also taken steps to avoid more specialized RNA decay pathways as well as usurp regulatory factors that normally target mRNAs for decay. Poliovirus, for example, contains an RNA element that interacts with and inactivates the RNase L endonuclease (48). Pseudoknot structures present at the 5′ border of the 3′ UTR of insect-borne flaviviruses stall the XRN1 enzyme (36,37). Rous sarcoma virus contains an RNA element that insulates unspliced viral mRNAs from the nonsense-mediated decay pathway (49). As outlined above, several RNA viruses utilize cellular RNA binding proteins that regulate decay to either stabilize their RNAs or for other aspects of viral replication/gene expression. Interestingly, rabies virus appears to utilize the cellular RNA decay machinery through the PCBP2 regulatory protein to fine tune its gene expression via differential stability of its glycoprotein mRNA (50). Thus some negative-sense RNA viruses that encode multiple independent mRNAs may use the RNA decay machinery in a manner similar to cells to fine tune overall gene expression. Finally, some RNA viruses through cap snatching (51) or viral-encoded nucleases (52) may simply attempt to dysregulate the entire process of mRNA decay in infected cells in order to re-model host gene expression and make the cell less able to respond to various aspects of the infection.
Conclusions
Work to date likely has only scratched the surface on how RNA viruses interface with aspects of the cellular RNA decay machinery. The question has simply not yet been addressed for many virus families. Therefore future work in this area will likely yield interesting strategies of viral RNA stabilization that may have a significant impact on viral replication and provide new insights into factors involved in cellular mRNA stability. The overall impact of viral-mediated disruption of cellular RNA decay pathways on host cell gene expression is also an understudied area for most viruses. This disruption may dysregulate the expression of numerous cellular mRNAs – particularly those transcripts with short, highly regulated half-lives. Since many of these short-lived cellular transcripts include cell cycle genes and factors implicated in innate and adaptive immunity (53), investigations into this area may shed important new light on the underlying molecular mechanisms of aspects of viral replication and pathogenesis. Finally, disarming viral defense mechanisms against the cellular mRNA decay machinery may afford a novel avenue for therapeutic intervention to ameliorate the effects of viral infection. This is particularly attractive since viral stability mechanisms described to date appear to be well-conserved throughout individual virus families (32), allowing the possibility of broad spectrum drugs against specific virus groups.
Highlights.
Numerous factors and pathways are used by the cellular RNA decay machinery to regulate transcript stability.
The cellular RNA decay machinery has a significant influence on viral gene expression in infected cells.
Strategies used by RNA viruses to avoid the cellular RNA decay machinery include degrading/inactivating key components, recruiting cellular stability factors and evolution of nuclease resistant RNA structures.
Instead of simply avoiding it, RNA viruses may also use the cellular RNA decay machinery to increase or fine tune their gene expression.
Acknowledgments
We wish to thank members of the Wilusz laboratories for critical comments. Research in the area of viral RNA stability in the Wilusz lab is supported by funds associated with the Rocky Mountain Regional Center for Excellence in Biodefense awarded to J.W. (AI065357).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1*.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;472:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3*.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012 Mar 9; doi: 10.1101/gr.131037.111. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 7.Collart MA, Panasenko OO. The Ccr4--not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury A, Raju KK, Kalurupalle S, Tharun S. Both Sm-domain and C-terminal extension of Lsm1 are important for the RNA-binding activity of the Lsm1–7-Pat1 complex. RNA. 2012 Mar 26; doi: 10.1261/rna.029876.111. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Li Y, Song M, Kiledjian M. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA. 2011;17:419–428. doi: 10.1261/rna.2439811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Chang JH, Xiang S, Xiang K, Manley JL, Tong L. Structural and biochemical studies of the 5′→3′ exoribonuclease Xrn1. Nat Struct Mol Biol. 2011;18:270–276. doi: 10.1038/nsmb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng D, Chen CY, Shyu AB. Unraveling regulation and new components of human P-bodies through a protein interaction framework and experimental validation. RNA. 2011;17:1619–1634. doi: 10.1261/rna.2789611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 2011;8:61–66. doi: 10.4161/rna.8.1.14237. [DOI] [PubMed] [Google Scholar]
- 13*.Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–134. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu SW, Rajagopal V, Patel SS, Kiledjian M. Mechanistic and kinetic analysis of the DcpS scavenger decapping enzyme. J Biol Chem. 2008;283:16427–16436. doi: 10.1074/jbc.M800341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenberg DR. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2011;2:582–600. doi: 10.1002/wrna.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc Natl Acad Sci U S A. 2011;108:2366–2371. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 21*.Martin KL, Johnson M, D’Aquila RT. APOBEC3G complexes decrease human immunodeficiency virus type 1 production. J Virol. 2011;85:9314–9326. doi: 10.1128/JVI.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- 23*.Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, Hartweger H, Vielnascher R, Karaghiosoff M, Rülicke T, Müller M, Hofacker I, Lang R, Kovarik P. Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol Syst Biol. 2011;7:560. doi: 10.1038/msb.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gherzi R, Chen CY, Trabucchi M, Ramos A, Briata P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip Rev RNA. 2010;1:230–239. doi: 10.1002/wrna.2. [DOI] [PubMed] [Google Scholar]
- 26.Palanisamy V, Jakymiw A, Van Tubergen EA, D’Silva NJ, Kirkwood KL. Control of cytokine mRNA expression by RNA-binding proteins and microRNAs. J Dent Res. 2012 Feb 1; doi: 10.1177/0022034512437372. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin Z, Han W, Zhao Z, Xia Q, Yin B, Yuan J, Peng X. PCBP2 enhances the antiviral activity of IFN-α against HCV by stabilizing the mRNA of STAT1 and STAT2. PLoS One. 2011;6:e25419. doi: 10.1371/journal.pone.0025419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Schmidt MJ, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Dougherty JD, White JP, Lloyd RE. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol. 2011;85:64–75. doi: 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J. The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and mammalian cells. J Virol. 2008;82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe. 2010;8:196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng CP, Jaag HM, Jonczyk M, Serviene E, Nagy PD. Expression of the Arabidopsis Xrn4p 5′-3′ exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology. 2007;368:238–248. doi: 10.1016/j.virol.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 35*.Jones DM, Domingues P, Targett-Adams P, McLauchlan J. Comparison of U2OS and Huh-7 cells for identifying host factors that affect hepatitis C virus RNA replication. J Gen Virol. 2010;91:2238–2248. doi: 10.1099/vir.0.022210-0. [DOI] [PubMed] [Google Scholar]
- 36**.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, Delviks-Frankenberry KA, Hu WS, Pathak VK. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J Virol. 2010;84:10241–10253. doi: 10.1128/JVI.00585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Galão RP, Chari A, Alves-Rodrigues I, Lobão D, Mas A, Kambach C, Fischer U, Díez J. LSm1–7 complexes bind to specific sites in viral RNA genomes and regulate their translation and replication. RNA. 2010;16:817–827. doi: 10.1261/rna.1712910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol. 2011;85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward AM, Bidet K, Yinglin A, Ler SG, Hogue K, Blackstock W, Gunaratne J, Garcia-Blanco MA. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2–3′ UTR structures. RNA Biol. 2011;8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng CP, Serviene E, Nagy PD. Suppression of viral RNA recombination by a host exoribonuclease. J Virol. 2006;80:2631–2640. doi: 10.1128/JVI.80.6.2631-2640.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson JA, Zhang C, Huys A, Richardson CD. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J Virol. 2011;85:2342–2350. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ford LP, Wilusz J. 3′-Terminal RNA structures and poly(U) tracts inhibit initiation by a 3′-->5′ exonuclease in vitro. Nucleic Acids Res. 1999;27:1159–1167. doi: 10.1093/nar/27.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA. 2008;14:1026–1036. doi: 10.1261/rna.958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Withers JB, Beemon KL. The structure and function of the Rous sarcoma virus RNA stability element. J Cell Biochem. 2011;112:3085–3092. doi: 10.1002/jcb.23272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Palusa S, Ndaluka C, Bowen RA, Wilusz CJ, Wilusz J. The 3′ untranslated region of the rabies virus glycoprotein mRNA specifically interacts with cellular PCBP2 protein and promotes transcript stability. PLoS One. 2012;7:e33561. doi: 10.1371/journal.pone.0033561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujimura T, Esteban R. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc Natl Acad Sci U S A. 2011;108:17667–17671. doi: 10.1073/pnas.1111900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schott J, Stoecklin G. Networks controlling mRNA decay in the immune system. Wiley Interdiscip Rev RNA. 2010;B1:432–456. doi: 10.1002/wrna.13. [DOI] [PubMed] [Google Scholar]