Abstract
Repression of many tumor suppressor genes in cancer is concurrent with aberrantly increased DNA methylation levels at promoter CpG islands (CGIs). About one-fourth of empirically defined human promoters are surrounded by or contain clustered repetitive elements. It was previously observed that a sharp transition of methylation exists between highly methylated repetitive elements and unmethylated promoter-CGIs in normal tissues. The factors that lead to aberrant CGI hypermethylation in cancer remain poorly understood. Here, we established a site-specific integration system with enforced local transcriptional repression in colorectal cancer cells and monitored the occurrence of initial de novo methylation at specific CG sites adjacent to the CGI of the INSL6 promoter, which could be accelerated by binding a KRAB-containing transcriptional factor. Additional repetitive elements from P16 and RIL (PDLIM4), if situated adjacent to the promoter of INSL6, could confer DNA methylation spreading into the CGI particularly in the setting of KRAB-factor binding. However, a repressive chromatin alone was not sufficient to initiate DNA methylation, which required specific DNA sequences and was integration-site (and/or cell-line) specific. Overall, these results demonstrate a requirement for specific DNA sequences to trigger de novo DNA methylation, and repetitive elements as cis-regulatory factors to cooperate with advanced transcriptional repression in promoting methylation spreading.
INTRODUCTION
What determines the pattern of DNA methylation during embryonic or carcinogenic development remains unclear. On the one hand, histone signatures seem to respond faster to upstream signals than DNA methylation as seen from earlier recovery of H3K9me3 and silencing than DNA methylation of the P16 (CDKN2A) promoter in prolonged culture of HCT116 DKO (DNMT1−/−; DNMT3B−/−) cells (1) and from advanced chromatin inactivation of the RASSF1A promoter prior to de novo methylation in epithelial cells (2). Further protein interaction results also suggest the involvement of chromatin configuration system in directing DNA methylation (3–5). Viewed in this way, DNA methylation works as a secondary event to solidify the pre-determined repressive status and sustain epigenetic memory. On the other hand, it was suggested that the DNA methylation machinery is preferentially attracted by certain DNA sequences in the mammalian genome and many genes remain unmethylated in cancers despite a repressed chromatin state (6,7).
A ‘seed and spread model’ has been proposed to explain the distinct patterns of DNA methylation in development (8,9). In this model, ectopic transcriptional silencing of promoters with tumor-suppressive function would arise from adjacent heterochromatin spreading which is normally blocked by barriers and insulators like CTCF, SP1 or USF1. The extension of heterochromatin status is realized by the cooperation of DNA methylation, histone modifications and chromatin remodeling with the help of adaptors like HP1 (10–12). Of note, in mammals, almost 25% of analyzed promoter regions contain repetitive DNA, including many experimentally characterized cis-regulatory elements (13). It has been shown from fungi to mammals that repetitive elements would produce phenotypic variation by subjecting nearby genes to the epigenetic regulation that is targeted to the repetitive elements (14). Therefore, repetitive sequences were hypothesized as methylation centers for original DNA methylation. To assess the possibility, Turker et al. identified two upstream B1 repetitive elements as cis-signals for de novo methylation of the mouse adenine phosphoribosyltransferase (APRT) gene on the X-chromosome, and methylation spreading is resisted by undetermined factors binding to one of the SP1 sites between retrotransposons and the CpG-rich promoter (15–18). Other repetitive elements were also potential targets for methylation triggering, such as human Alus, mouse LINE-1, B2 and IAP.
Experimental validation for the ‘seed and spread’ model is limited, as are the determinants of spreading. Transfected genes generally remain unmethylated, even if introduced into cells where the endogenous ones are methylated. For the glutathione-S-transferase gene (GSTP1), it was shown that both silencing by mutations of transcription factor binding sites and pre-methylation were required for seeing high levels of DNA methylation after spreading (19). One problem in some experiments is the variability associated with insertion site effects. Here, we utilized site-specific integration to assess different aspects of the ‘seed and spread’ model as relevant to tumor-suppressor gene silencing in cancer. We observed that methylation was seeded at specific CG sites and found that the presence of repetitive elements and robust local silencing facilitated methylation spreading into a promoter-CpG island (CGI).
MATERIALS AND METHODS
Generation and characterization of the site-specific integration system
The general guideline to use the Flp-in system (Invitrogen) is available through the commercial instructions. Figure 1A illustrates the steps and constructs revised to fulfill our specific experimental aims. The colorectal cancer cell lines, SW48 and HCT116, are maintained in Leibovitz’s L-15 medium or McCoy’s 5a medium modified with 10% fetal bovine serum. In Step I, after transfection of pFRT/LacZeo (Invitrogen) into HCT116 or SW48, stable selection of single clones was carried out with 50 μg/ml zeocin (Invitrogen). PCR and β-gal staining were performed to verify FLP recognition target (FRT) integration. Tiling primers (sequences available upon request) were used to screen out clones with single integration, and the genomic loci were identified through inverse PCR. The single clones generated in this way were named as Flp-in host cells.
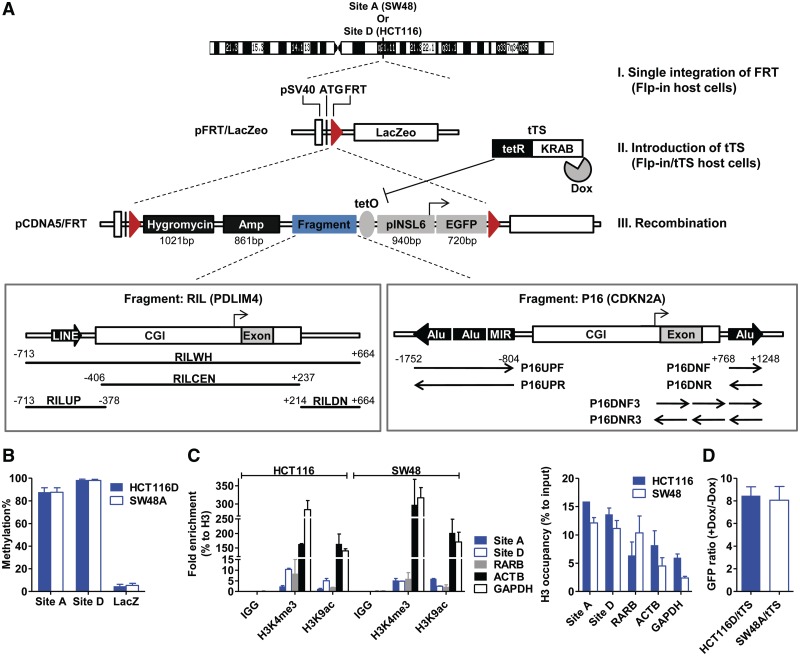
Figure 1.
Establishment of a site-specific integration system. (A) Schematic description of generating Flp-in and Flp-in/tTS host cells. The entire system was established using three steps. I, single integration of FRT; II, expressing tTS in Flp-in cells to generate Flp-in/tTS host cells; III, FRT-mediated homologous recombination of transgenes. Fragments derived from the RIL or P16 promoter are co-existent with the INSL6 promoter (pINSL6). The distance to the transcription start site (TSS) of RIL or P16 is annotated for each subcloned fragment. (B) Methylation levels of the characterized integration sites (Site A for SW48A and SW48A/tTS; Site D for HCT116D and HCT116D/tTS), and the LacZeo in Flp-in host cells (SW48A and HCT116D). Y-axis, the average methylation percentages by pyrosequencing. Error bar, SEM. (C) ChIP-qPCR for the enriched histone marks (H3K4me3 and H3K9ac) and H3 occupancy at the integration sites. ACTB and GAPDH as the positive control regions enriched for active marks, and RARB as the negative control region. Values resulted from biological duplicates. (D) Flow cytometry results to validate the functional tTS in Flp-in/tTS host cells (SW48A/tTS and HCT116D/tTS). Parallel wells of isolated Flp-in/tTS single clones were transiently transfected with a tetO-containing EGFP construct under the presence or absence of doxycycline (Dox) treatment. The percentages of green cells were measured 48 h after transfection and the clones with the highest ratio of Dox (+) to Dox (−) were used as Flp-in/tTS host cells.
In Step II (Figure 1A), to establish transcription silencer (tTS)-containing host cells (Flp-in/tTS), the tetracycline-controlled tTS (Clontech) was transfected into Flp-in host cells and stable single clones were selected with G418 (800 μg/ml). The presence of functional tTS was verified with RT-PCR and transient transfection of an enhanced green fluorescent protein (EGFP) vector carrying a tetO element under conditions of doxycycline(+) (2 μg/ml) or doxycycline(−).
The transgenes were modified from vector pCDNA5/FRT (Invitrogen). In Step III, pCDNA5/FRT constructs and POG44 (for flippase expression) were co-transfected into characterized Flp-in or Flp-in/tTS host cells with 1:9 ratio (w/w) and stably selected with Hygromycin B (150 µg/ml) for 10 days. In the case of Flp-in/tTS cells, 2 μg/ml doxycycline was consistently added in media at the beginning of transfection until 30 days (for HCT116) or 60 days (for SW48) when stable single clones were isolated and split into parallel wells supplemented with or without doxycycline (2 μg/ml). Correct single clones were confirmed by PCR amplification of inserts, zeocin-resistance test (50 μg/ml, 10 days) and β-gal staining. These clones were continuously cultured for 150 days in media and sampled for further examination.
Bisulfite sequencing and bisulfite pyro-sequencing
Genomic DNA was extracted from cell lines or tissues using standard methods, followed by bisulfite conversion with EpiTect Bisulfite Kit (Qiagen). PCR SuperMix High Fidelity (Invitrogen) was used to amplify from genomic DNA and amplicons were subsequently cloned and sequenced in PCR4-TOPO (Invitrogen). Pyrosequencing was performed as previously published (20). Primers for two steps of amplification are listed in Supplementary Table S1, and the locations of the assays are shown in Figure 2B.
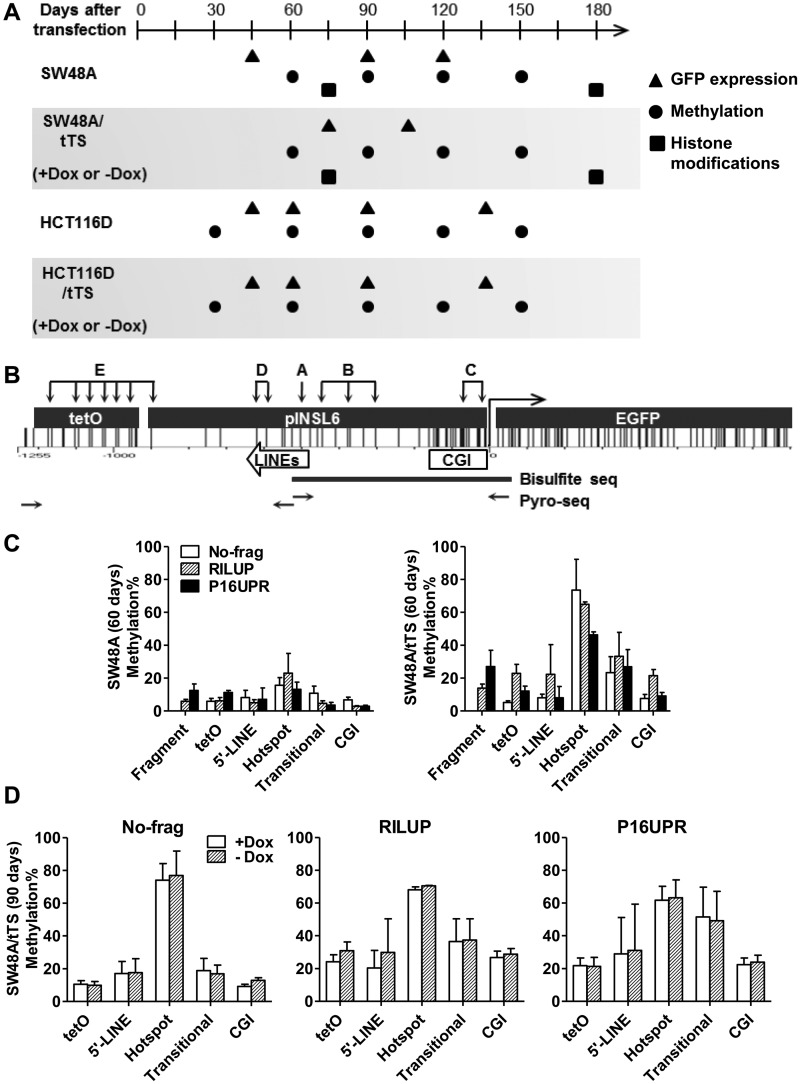
Figure 2.
De novo DNA methylation in the INSL6 promoter. (A) A graph showing the time points to collect long-term cultured cells after stable transfection. The coordinate is the time-scale (days) after constructs were transfected into Flp-in (SW48A and HCT116D) or Flp-in/tTS (SW48A/tTS and HCT116D/tTS) cells. Stable clones were cultured in media supplied with doxycycline (Dox) until 60 days (SW48A/tTS) or 30 days (HCT116D/tTS) before they were split and cultured separately in media with Dox (+Dox) and without Dox (−Dox). Symbols indicate the time to examine GFP expression (triangles), DNA methylation (circles) and histone modifications (squares). (B) Graphical distribution of CG sites in tetO-pINSL6-EGFP. The subcloned 940-bp pINSL6 consists of part of the CGI and two short LINE elements. Primers (horizontal arrows) for the first step of amplification of bisulfite-converted DNA include the tetO sequence or 5′-end of EGFP to distinguish it from the endogenous pINSL6. The capitals and groups of vertical arrows indicate the target sites for pyrosequencing (A, hotspot; B, Transitional; C, CGI; D, 5’-LINE; E, tetO). Assays for fragments are not shown here. Thick line, the amplified region for bisulfite cloning/sequencing. (C) Regional methylation of transgenes (No-frag, RILUP and P16UPR) in SW48A and SW48A/tTS (60 days). (D) Comparison of regional methylation levels of transgenes (No-frag, RILUP and P16UPR) in SW48A/tTS (90 days) under +Dox and −Dox.
ChIP
The procedures of chromatin immuno-precipitation (ChIP) assays were adapted from the online protocol (http://myers.hudsonalpha.org/documents/Myers Lab ChIP-seq Protocol v041610.pdf, date last accessed 7 May 2012). About 1 × 106 cells were used for each immuno-precipitation. Antibodies (10 µg) used are IgG (ab6709, Abcam), H3 (ab1791, Abcam), H3K4me3 (07-473, Millipore), H3K9ac (07-352, Millipore), H3K27me3 (07-449, Millipore) and H3K9me3 (ab8898, Abcam). Following immuno-precipitation, qPCR was performed in 7500 Real-Time PCR System (Applied Biosystems) to get Ct values. All the fold enrichment of histone marks was normalized to H3 (percent of H3) and the nucleosome density measured by H3 occupancy (percent of input) was calculated against the 1/50 input control. Primers and probes are listed in Supplementary Table S2.
Drug treatment and detection of GFP expression
For reversion of DNA methylation and reactivation of green fluorescent protein (GFP), we used 200 nM 5-aza-2′-deoxycytidine (DAC, Sigma) and/or 800 nM trichostatin A (TSA, MP Biomedicals). Cells were split 24 h before each experiment, and given one of the following treatments. (i) DAC was given every day for 96 h, and media were replaced every day, (ii) TSA was added at the last 24 h and (iii) Combined treatment of the above DAC and TSA. Flow cytometry (FACSCalibur, BD Biosciences) was performed to detect GFP expression as instructed by the manufacturer.
RESULTS
Establishing a site-specific integration system with local repressive status
Our aim was to evaluate the possibility of de novo DNA methylation in cancer cell lines using exogenous sequences without in vitro methylation. Hence, an Flp/FRT-mediated integration system was utilized to make each transgene integrated into the same single genomic locus due to our concern for position effects arising from multiple or inconsistent chromatin environment. First, a vector with an FRT site (pFRT/LacZeo) was introduced into two colorectal cancer cell lines, SW48 and HCT116 (Figure 1A), because of their dense methylation background as shown before (21,22). Using inverse PCR, we identified clones with a single insert in each Flp-in host cells (SiteA on Chr7q21.11 in SW48A and SiteD on Chr3q13.31 in HCT116D, Table 1). Both loci were intragenic and outside CGIs. Epigenetic analyses suggested that the endogenous loci are inactive with high levels of CG methylation (SiteA in SW48, 87.6% ± 4.0%; SiteD in HCT116, 98.4% ± 0.8%, mean ± SEM, Figure 1B), and absence of active histone marks (H3K4me3 and H3K9ac) (Figure 1C). However, CGs of the inserted LacZ lacked methylation (SW48A, 5.4% ± 1.8%; HCT116D, 4.7% ± 1.7%, Figure 1B), excluding the possibility of methylation recruitment caused by endogenous sequences nearby or sequences from the first construct.
Table 1.
Characterization of the single integration sites in Flp-in host cells
| Site (Single clone) | Location | Position(hg18) | Nearby genes | Orientation | Elements |
|---|---|---|---|---|---|
| SiteD (HCT116D) | Chr3q13.31 | 115,911,000–115,913,000 | ZBTB20 | + | Intron |
| SiteA (SW48A) | Chr7q21.11 | 80,138,271 | CD36 | − | Exon |
The frequent association of abnormal promoter silencing with gain of DNA methylation in tumorigenesis (23) reminds us that the integrated sequences comprise potential promoters (e.g. pSV40) which may interfere with methylation recruitment. One strategy to avoid it could be to impose robust transcriptional silencing on the transgenes once they enter characterized Flp-in cells. As shown in Figure 1A, the tetracycline-controlled tTS (a tetR DNA-binding domain fused with a KRAB domain) was expressed to generate a parallel set of Flp-in/tTS host cells (SW48A/tTS and HCT116D/tTS), where tTS is capable of binding tetO and subsequently inhibiting the transgenes. We confirmed the stable expression and repressive capability of tTS by comparing the expression of a tetO-containing EGFP construct in the presence and absence of doxycycline (Figure 1D). Usage of Flp-in and Flp-in/tTS host cells would construct two conditions for each transgene as to the extent of silencing.
As a second way to avoid interference from active promoters, all transgenes were driven by the insulin-like 6 (INSL6) promoter, a CGI-promoter which is frequently found methylated in somatic cells including many cancer cell lines (24). After characterization of the host cells, transgenes were integrated into the FRT site through flippase-mediated homologous recombination. The constructs were composed of a reporter (EGFP, 720 bp, 69 CGs), the INSL6 promoter (pINSL6, TSS-918 to + 21, 940 bp, 42 CGs), a tetO element (287 bp, 17 CGs) and fragments of interest (variable sizes) (Figure 1A). The INSL6 promoter has two short LINE elements (L2, 89 bp, 4 CGs; L4, 52 bp, 1 CG; in-between, 13 bp, 1 CG) upstream of its CGI. TetO is the response element for binding tTS in the absence of doxycycline, and here was located upstream of pINSL6. In order to determine the elements that would facilitate DNA methylation, we chose the RIL (PDLIM4) and P16 (CDKN2A) promoter regions to generate variable fragments. Both promoters contain consistently methylated repetitive elements (one upstream LINE for RIL; three upstream SINEs and one downstream SINE for P16) surrounding their CGIs (25), and were found to become highly methylated in many types of cancers (25,26). The transgenes are named after the fragments of interest (Figure 1A and Supplementary Table S3), which include the entire RIL promoter region (RILWH), or isolated fragments containing the upstream (RILUP), central (RILCEN) or downstream (RILDN) regions. In the case of P16, fragments are derived from three upstream SINEs (P16UPF and P16UPR) and one downstream Alu (P16DNF and P16DNR; P16DNF3 and P16DNR3). Isolated stable clones were selected by Hygromycin B for 10 days and continually cultured without selection up to 180 days, as our unpublished observation indicated that long-term treatment with Hygromycin B would confer a selection priority on clones that remain unmethylated. Figure 2A shows the time points to test the expression, DNA methylation and chromatin modifications of transgenes in Flp-in or Flp-in/tTS cells.
Robust transcriptional repression of the exogenous INSL6 promoter by tTS
Integration of transgenes gradually led to repression for all 10 fragments as well as for the control (No-frag) which does not comprise any sequences from the RIL or P16 promoter. pINSL6 was the common promoter used to drive all transgenes (Figure 2B) which were expressed in both SW48A (Supplementary Figure S1) and HCT116D (Supplementary Figure S2B) cells at Day 45 after transfection and fully silenced by Day 120 in SW48 or Day 135 in HCT116D (Supplementary Figure S2A and S2B). Therefore, the exogenous INSL6 promoter underwent progressive silencing, mimicking the endogenous promoter as shown before (24).
By comparison, when tTS was expressed, GFP was undetectable at the earliest sampling time (75 days for SW48A/tTS; 45 days for HCT116D/tTS) if doxycycline was not supplied in the media after splitting (Supplementary Figures S1, S2A and S2B). Thus, tTS could induce robust gene silencing, which emerged at an earlier time point than cells without tTS. Of note, persistent addition of doxycycline resulted in intermediate GFP expression as observed in every HCT116D/tTS (+Dox) clone (Supplementary Figure S2A and S2B), presumably due to incomplete repression of tTS. Because of this suboptimal effect of doxycycline, we chose to compare Flp-in cells with Flp-in/tTS cells to demonstrate different effects of gradual silencing versus rapid silencing on DNA methylation.
De novo DNA methylation center in the INSL6 promoter
DNA methylation patterns were mapped by bisulfite pyrosequencing of several regions shown in Figure 2B. We found three CG sites (Region A) located in the proximal LINE (L2) of the INSL6 promoter which harbored higher sensitivity to DNA methylation. Figure 2C illustrates the fact that Region A had the highest methylation level of all regions studied in both SW48A and SW48A/tTS. Thereby it is designated as a ‘hotspot’ for its quicker captivation of methyl groups. By contrast, the CGI (Region C), 5′-LINE (L4, Region D) and the tetO cassette (Region E) achieved low levels of methylation, while the CG sites of Region B, located between the CGI and hotspot, obtained an intermediate methylation level, suggesting that it may be a ‘transitional’ region in methylation spreading. This was the most apparent in SW48A/tTS where the hotspot had on average ∼61.8% methylation, the transitional region ∼27.8% and the others ∼12.7% (CGI), ∼19.2% (5′-LINE) and ∼13.4% (tetO). The fragments of both RILUP and P16UPR were repetitive elements (LINE and SINEs, respectively), but their methylation levels (RILUP, 13.9%; P16UPR, 27.0%) did not reach those of the hotspot even in SW48A/tTS. Altogether, these data imply that the CG sites of the hotspot (Region A) were rapidly ‘seeded’ after transfection and thus could function as a methylation center.
Notably, neither of the fragments of RILUP or P16UPR located adjacent to the INSL6 promoter significantly changed DNA methylation of the hotspot (t-test: RILUP, P = 0.70; P16UPR, P = 0.41) in SW48A/tTS (Figure 2C) or other host cells (SW48A, HCT116D and HCT116D/tTS). Thus, the sensitivity of the hotspot to DNA methylation could be an inherent characteristic of these CG sites. Earlier, we have shown that the addition of doxycycline could not fully maintain GFP expression, probably due to incomplete release of tTS from the tetO element. Consistent with this, the presence or absence of doxycycline did not make a difference to DNA methylation (Figure 2D).
The effect of repetitive elements on DNA methylation spreading
In order to view the methylation status of every CG site of the examined INSL6 promoter, bisulfite sequencing was performed for all 11 transgenes in SW48A/tTS (60 days, Figure 3A). All the peaks of regional methylation (41.6%–75.0%, mean) overlapped with the location of the hotspot examined by pyrosequencing; moreover, none of the 10 transgenes affected methylation at the hotspot significantly (t-test: P > 0.05). The transitional methylation between the hotspot and the CGI is consistent with the ‘seed and spread’ model. According to this model, methylation spreading leads to CGI methylation once the protective boundaries are disrupted (8,9). However, inherent cis-signals for initiating DNA methylation within CGIs independent of a hotspot might be an alternative mechanism that does not require spreading. To address this possibility, we separately constructed a truncated INSL6 promoter (tr-pINSL6) by removing all the sequences upstream of the CGI including the hotspot and 5′-LINE, but still retaining the tetO cassette (Figure 3B). In SW48A/tTS, truncation caused 1.8- to 2.9-fold decrease at 60 days and 2.4- to 5-fold decrease at 90 days in the methylation of the INSL6 CGI region (Figure 3B), while GFP was silenced to the same level. We conclude that CGI methylation of pINSL6 arose more easily from spreading from a hotspot. Nevertheless, the CGI itself was not absolutely methylation-free in the absence of the hotspot, suggesting that DNMTs may target randomly independent of methylation centers although in a less efficient way.
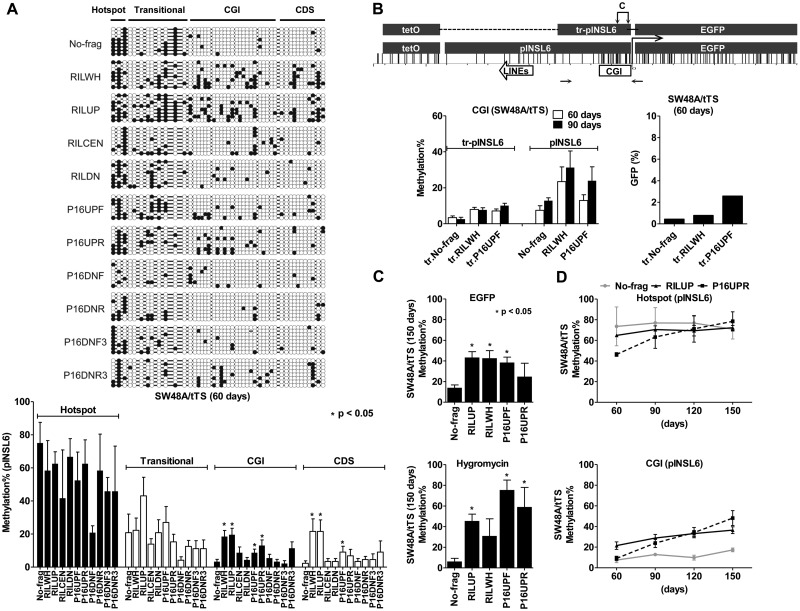
Figure 3.
Methylation spreading from the seeding sites to the adjacent regions. (A) Methylation patterns of the INSL6 promoter in 11 transgenes (SW48A/tTS, 60 days). Bisulfite-cloning/sequenced CGs are displayed in circles (closed, methylated; open, unmethylated); the average levels, calculated in respect to four regions (hotspot, transitional, CGI and CDS), are shown in the bar graph (mean ± SEM). *Statistically significant difference in comparison to No-frag (t-test, P < 0.05). (B) Removal of the seeding sites impaired CGI methylation in SW48A/tTS. The graph shows the distribution of CG sites in the truncated INSL6 promoter (tr-pINSL6), which retains the CGI and the tetO cassette. Horizontal arrows, the amplicon used for pyrosequencing (C, assays for CGI methylation). Pyrosequencing was performed for transgenes (No-frag, RILWH and P16UPF) in their truncated and complete forms (60 and 90 days). The repression of the truncated transgenes at 60 days was measured by GFP expression (flow cytometry). (C) Methylation extended to regions of EGFP and hygromycin sequences in SW48A/tTS (150 days). Pyrosequencing was used to examine 5 transgenes (No-frag, RILUP, RILWH, P16UPF and P16UPR). *Significantly methylated transgenes in comparison to No-frag (P < 0.05). (D) Gradual accumulation of transgene methylation in SW48A/tTS over time (60, 90, 120 and 150 days). Please refer to Supplementary Figure 5B for the cases of HCT116D and HCT116D/tTS.
We next examined the effect of adjacent sequences on DNA methylation spreading by comparing CGI methylation between the 10 transgenes and No-frag control. Statistical analysis (t-test) of the bisulfite sequencing results in Figure 3A showed significantly higher CGI methylation of pINSL6 for both RILWH (18.5%, P = 0.0004) and RILUP (19.6%, P = 0.0003), comprising the upstream LINE element (L2) of the RIL promoter; whereas transgenes with the CGI of the RIL promoter (RILCEN, 8.9%), the downstream portion (RILDN, 4.4%) and No-frag (3.3%) displayed lower levels of methylation. Likewise, the presence of three upstream SINE elements of the P16 promoter made the CGI of pINSL6 attract more methyl groups (P16UPR, 13.0%, P = 0.01) than No-frag, while the downstream SINE (P16DNF, P16DNR, P16DNF3 and P16DNR3) did not lead to significant differences compared to No-frag (P > 0.05). The rest of the bisulfite-sequenced region (CDS) which included the 5′-end of EGFP also showed the same effects (Figure 3A). Assays of bisulfite pyrosequencing and bisulfite cloning/sequencing revealed a good correlation (hotspot, r = 0.78; transitional, r = 0.82; CGI, r = 0.88) and similar results that repetitive elements adjacent to the INSL6 promoter could significantly enhance CGI methylation. For convenience, therefore, we used bisulfite pyrosequencing for subsequent analyses.
Next, in order to understand how far DNA methylation could spread in transgenes, we measured methylation levels of CG sites inside EGFP (∼750 bp distant to the hotspot) and Hygromycin (∼5.7 kb to ∼6.7 kb distant to the hotspot) in SW48A/tTS. At 150 days, 24.7%–43.4% of the examined CG sites in EGFP and 31.0%–71.5% in Hygromycin became methylated for transgenes containing repetitive elements (RILUP, RILWH, P16UPF and P16UPR), as opposed to No-frag (EGFP, 14.0%; Hygromycin, 6.2%) (Figure 3C). Thus, both the LINE (from RIL) and SINEs (from P16) enhanced adjacent CGI methylation, as well as methylation spreading 6–7 kb away from the seeding sites.
We also investigated the accumulation of methyl groups at CG sites by monitoring the changes over time. In Figure 3D, DNA methylation of three representative transgenes (No-frag, RILUP and P16UPR) in SW48A/tTS are plotted at four time points (60, 90, 120 and 150 days). In the hotspot, between 60 and 150 days, methylation increased by 6.0% for No-frag, 7.3% for RILUP and 32.1% for P16UPR; in the CGI, the increases were 10.8% for No-frag, 15.2% for RILUP and 39.0% for P16UPR. Together with data not shown, almost all CG sites tended to recruit methyl groups under long-term culture; some CGs rapidly established high levels of methylation at an early stage (e.g. in the hotspot, 73.6% for No-frag and 64.9% for RILUP at 60 days), while others increased slowly (e.g. CGI methylation only increased to 17.3% for No-frag at 150 days).
Enforced transcriptional repression promoted both methylation seeding and spreading
When the Flp/FRT system was established, two comparable host cells, Flp-in and Flp-in/tTs were also generated in parallel in order to evaluate the impact of repression on methylation seeding and spreading. Compared with SW48A, SW48A/tTS cells shortened time to complete transgene silencing by at least 45 days (Supplementary Figure S2A); pyrosequencing showed that, at 60 days, SW48A/tTS cells had 1.3- to 4.7-fold higher methylation of the hotspot (Supplementary Figure S3), and this difference persisted to 150 days with 1.2- to 3.1-fold higher levels (Figure 4A). A validation experiment was carried out by removing tetO [tetO(−)-pINSL6] from two transgenes (No-frag and RILUP) to make tTS incapable of binding pINLS6. As anticipated, even in SW48A/tTS, tetO(−) transgenes were still actively expressing GFP at 60 days (flow cytometry: avg. 74% for No-frag and 65% for RILUP, Figure 4B); loss of rapid repression resulted in 60.3% (No-frag) and 42.4% (RILUP) reduction of methylated CGs of the hotspot, and the same pattern was maintained at 90 days (58.9% reduction for No-frag and 46.7% for RILUP, Figure 4B). This experiment also ruled out possible interference with methylation seeding caused by cell engineering, such as non-specific effects due to tTS insertion in the course of generating SW48A/tTS host cells.
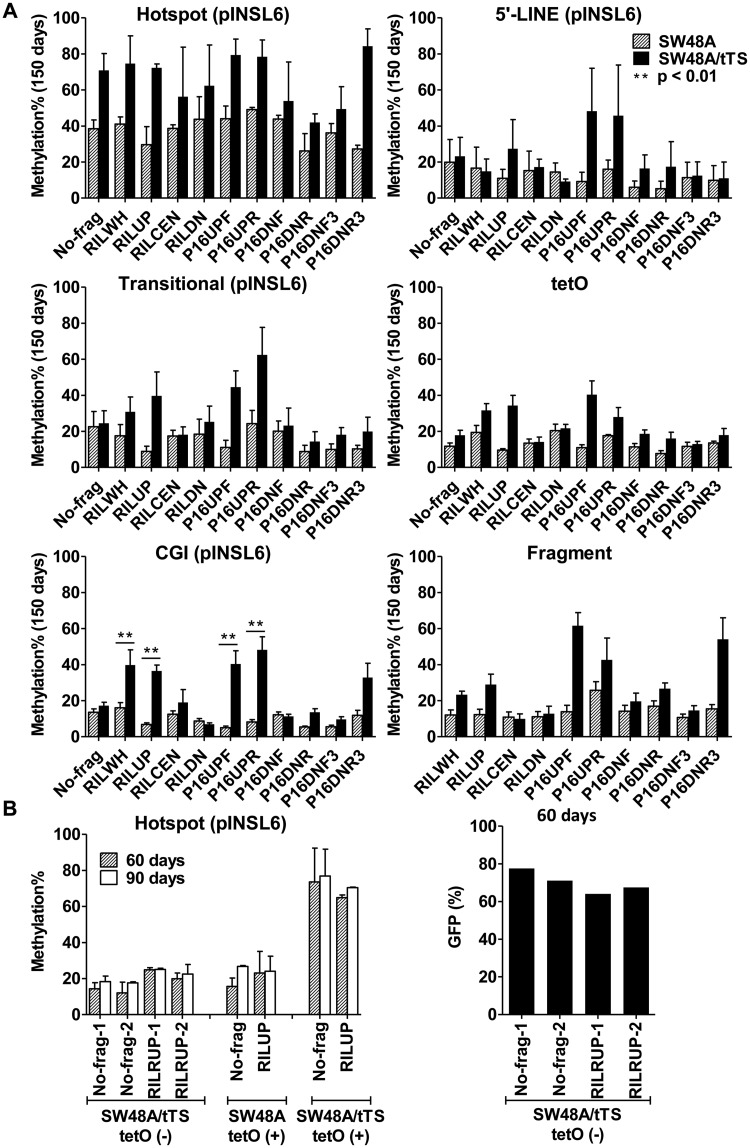
Figure 4.
The effects of enforced silencing by tTS on methylation seeding and spreading. (A) Comparing the methylation profiles of pINSL6 in SW48A/tTS and SW48A cells (150 days). **Significantly different methylation of each transgene between host cells (P < 0.01). (B) Comparison of methylation and GFP expression between tet(−)-pINSL6 in SW48A/tTS, tetO(+)-pINSL6 in SW48A and tetO(+)-pINSL6 in SW48A/tTS. Pyrosequencing was performed for transgenes (No-frag and RILUP) at 60 and 90 days; flow cytometry was used to detect GFP expression at 60 days.
Besides the seeding sites, robust silencing by tTS extended its impact on DNA methylation to the adjacent areas. All analyzed sites (5′-LINE, tetO cassette, fragments, transitional region and CGI) showed higher methylation levels in SW48A/tTS than SW48A when examined at 60 days (Supplementary Figure S3) and 150 days (Figure 4A). Remarkably, the presence of tTS enabled more prominent enhancement of CGI methylation by transgenes containing repetitive sequences from RIL or P16. At 60 days, compared with SW48A, RILWH and RILUP achieved 14.4% (P = 0.11) and 18.7% (P = 0.002) higher methylation in SW4A/tTS cells, while P16UPF and P16UPR achieved 8.7% (P = 0.02) and 6.4% (P = 0.007) higher levels; the differences for the other fragments were much less or not significant (Supplementary Figure S3). At 150 days, tTS-induced silencing demonstrated more apparent effects with levels raised by 23.7% (P = 0.0061), 29.7% (P = 0.0001), 35.4% (P = 0.0018) and 40.0% (P = 0.0008), respectively, for RILWH, RILUP, P16UPF and P16UPR (Figure 4A). These results suggest that the impact of repetitive elements on methylation spreading could be limited by the extent of local repression, and both repetitive elements and gene silencing would have to cooperate for a CGI to gain high levels of methylation.
Repressive chromatin signatures of transgenes
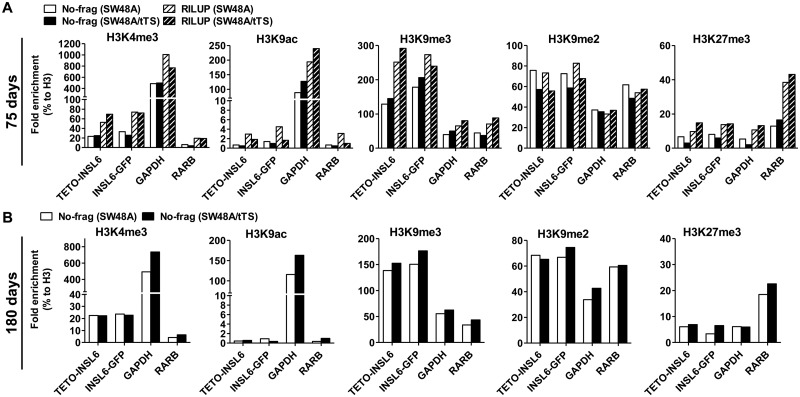
ChIP analysis was performed to examine histone modifications in the transgenes. Active marks (H3K4me3 and H3K9ac) and inactive marks (H3K9me3, H3K9me2 and H3K27me3) were analyzed in the cells sampled from 75 and 180 days of culture. Compared with the control regions (GAPDH and RARB), pINSL6 of the transgenes was highly enriched for H3K9me3 (2- to 3-fold more than GAPDH and RARB), moderately enriched for H3K9me2 (0.5- to 1.5-fold more than GAPDH) and devoid of H3K4me3 and H3K9ac, indicating a local repressive environment (Figure 5A and B); by contrast, H3K27me3 was low in pINSL6 (Figure 5A and B). However, no differences in the inactive histone marks (H3K9me3, H3K9me2 and H3K27me3) were observed between SW48A/tTS and SW48A, nor were there differences between the repetitive-elements-containing transgene (RILUP) and pINSL6 only (No-frag). Thus, we speculate that the enrichment for repressive marks in the exogenous pINSL6 had already reached a stable level at the earliest time points examined (75 days).
Figure 5.
ChIP-qPCR to analyze the enrichment for histone marks in pINSL6. Antibodies against active marks (H3K4me3 and H3K9ac) and repressive marks (H3K9me3, H3K9me2 and H3K27me3) were used to pull down sonicated chromatin. All the values of fold enrichment were normalized to H3. GAPDH as the control region for active marks; RARB as the positive control for H3K27me3. TETO-INSL6 and INSL6-GFP are targets designed at the 5′-end and 3′-end of pINSL6 in order to distinguish it from the endogenous one. (A) Two transgenes (No-frag and RILUP) examined in SW48A and SW48A/tTS at 75 days. (B) The transgene (No-frag) examined in SW48A and SW48A/tTS at 180 days.
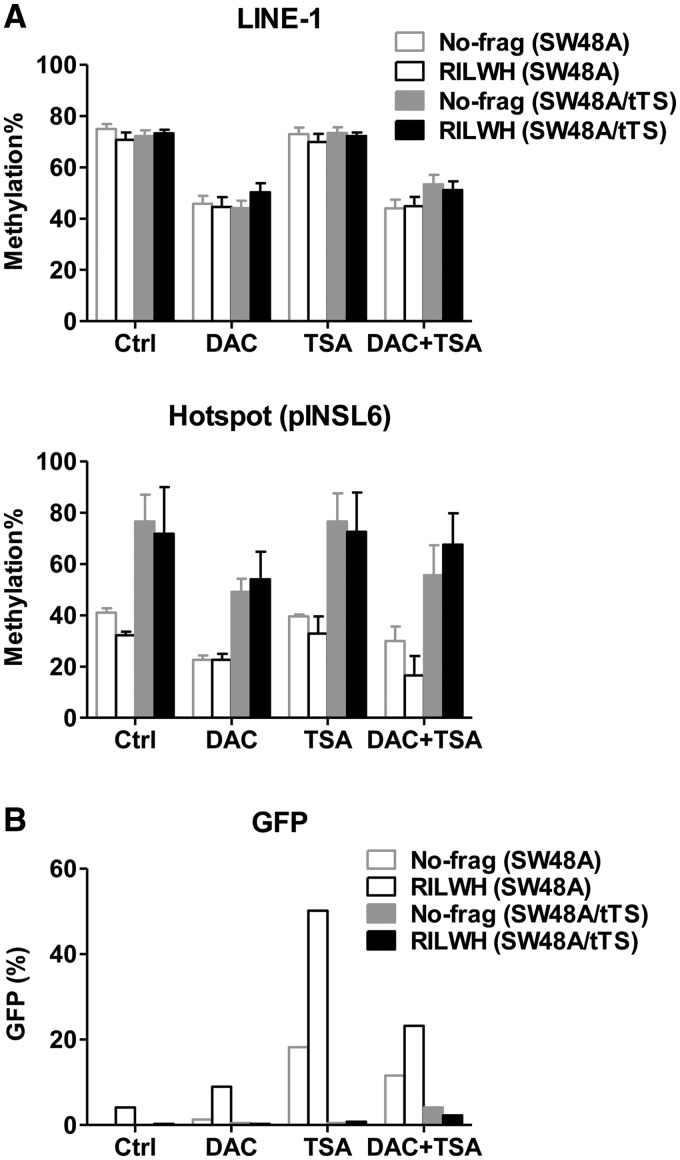
We also tested whether robust silencing and methylation seeding induced by tTS would be reverted through treating 120-day-clones (No-frag and RILWH) of SW48A and SW48A/tTS with epigenetic activating drugs. Compared with mock treatment (Ctrl), the DNA methylation inhibitor 5-aza-2′-deoxycytidine (DAC) induced global LINE-1 de-methylation by 20%–30% as well as local ‘hotspot’ de-methylation by 5%–18% (Figure 6A), but was not able to reactivate GFP expression (Figure 6B). The HDAC inhibitor TSA could not de-methylate the seeding sites (hotspot) (Figure 6A), but GFP expression experienced recovery of 18.2% (No-frag) and 46.0% (RILWH) for SW48A clones, whereas clones of SW48A/tTS showed little response to TSA (0.5% increase for No-frag and RILWH, Figure 6B). Therefore, this confirmed that persistent tTS binding conferred a robust repression which may not be entirely dependent on histone deacetylation.
Figure 6.
The reaction of SW48A and SW48A/tTS clones to epigenetic drug treatment. The 120-day-clones (No-frag and RILWH) were treated with DAC (200 nM) for four days and/or TSA (800 nM) during the last day and controls (mock treatment) were cultured in regular media. (A) Global DNA methylation (LINE-1) and local methylation of the seeding sites (hotspot) in pINSL6 were measured and compared with the controls. (B) GFP expression was detected by flow cytometry. All the values were averaged from biological duplicates.
The effect of cell lines or integration sites on DNA methylation recruitment
In addition to the host cells from SW48, we utilized a different cell line HCT116 with a different integration site (HCT116D) to assess methylation seeding and spreading. In both HCT116D and HCT116D/tTS, the three CGs of the hotspot were also the seeding sites for rapid recruitment of methyl groups (Supplementary Figure S5A), although at lower levels compared with SW48A and SW48A/tTS. Methylation of most regions gradually increased after 150 days of culture (Supplementary Figure S5B). However, methylation spreading progressed slowly in these cell lines. It was not distinctly promoted by the upstream repetitive elements and the presence of tTS in HCT116D/tTS did not effectively lead to spreading to the CGI (Supplementary Figure S5A). Thus, DNA methylation of the hotspot appeared to be an intrinsic property of the CG sites, whereas methylation spreading was greatly influenced by cell line context and/or integration sites.
DISCUSSION
Our investigation of de novo methylation and spreading in cancer cells was realized through a site-specific integration system with enforced local transcriptional repression. By studying expression, DNA methylation and histone modifications of transgenes at a single integration site, we were able to distinguish several aspects involved in DNA methylation of promoter-CGIs. We find that (i) DNA methylation originates from very specific CG sites, in this case, within a LINE element, (ii) methylation spreading into a promoter-CGI is facilitated by enforced transcriptional repression, presence of additional repetitive elements and is site-specific and (iii) transcriptional repression is required but not sufficient to promote DNA methylation.
Repetitive elements comprise ∼45% of human genome, most of which are derived from the activity of transposable elements (27). They are thought to influence global DNA methylation in normal somatic cells, while they become hypomethylated in cancers and increase the risk of genomic instability (28). In mammals, almost 25% of the analyzed promoter regions contain repetitive DNA (13), some of which still maintain methylation in cancer cells, such as SINE sequences located upstream of the P16/CDKN2A promoter (29) and the LINE element upstream of the RIL promoter (26). We evaluated the roles of repetitive elements in DNA methylation recruitment and spreading using site-specific integration of a single transgene, which could overcome problems associated with position effects and multi-copy gene effects. The first observation of our experiments is ‘seeding’ of DNA methylation in transgenes. The exogenous 940 bp-INSL6 promoter consists of two short LINEs (L2, divergence 27.3%, RepeatMasker; L4, 20.0%) upstream of a CGI. There are six CGs across the repeat region, but only the proximal two CGs of the L2 with an adjacent non-LINE CG site achieved distinct methylation levels at the earliest time point in almost all eleven transgenes examined. Methylation seeding was induced independently of cell lines (HCT116 and SW48), genomic loci or the strength of transcriptional repression (by tTS). But the extent of methylation was elevated by the presence of tTS and affected by cell line and/or loci used. When these three CG sites were deleted, DNA methylation of the CGI was markedly diminished. Therefore, certain sequences do serve as seeding targets for de novo DNA methylation in cancer cells. Nonetheless, not all CGs with LINE homology can serve this function, but it is unknown why some do and some do not. It is also interesting to note that methylation accumulated much slower in these colorectal cancer cells than what was expected from experiments in ES cells where the DNA methylation machinery is a lot more active (30), so culture time has to be extended in order to observe appreciable levels of methylation.
The hypothesis of ‘methylation centers’ was proposed on the basis of studies of the APRT gene which possesses B1 repetitive elements, signaling de novo methylation when transfected into embryonic carcinoma cells (8,16,17). Moreover, de novo methylation was initiated at discrete sites of the mouse Oct-4 regulatory region (30). However, since very few promoters were analyzed in this way, it is not yet possible to do meaningful sequence alignments. This may be elucidated by genome-wide analysis. While they could be direct and specific targets for de novo DNMTs, ‘methylation centers’ could also be due to other regulatory mechanisms, such as transcription factor binding sites and/or histone modifications. A recent paper described that small methylation-determining regions proximal to some promoters could be necessary and sufficient to mediate de novo DNA methylation in cis (31). Also, the ‘seeding’ event may be determined by dynamic nucleosome deposition as suggested by de novo methylation of the P16/CDKN2A CGI in post-selection primary human mammary epithelial cells (HMECs) (32).
The second effect of repetitive elements located further upstream is to cis-regulate methylation spreading into adjacent regions from de novo sites, especially into CGIs. The LINE element (L2, divergence 26.9%) from the RIL promoter and three concatenated upstream SINEs (MIR, divergence 24.1%; Alu, 8.7%; Alu, 9.0%) from the P16 promoted striking methylation of the CGI in SW48A/tTS. In another host cell (SW48A), spreading was not significant. So these repetitive elements we studied here may be unable to overcome the protective machinery independently of transcriptional repression. Alternatively, repetitive elements may have to cooperate with stronger inactivation in order to render methylation spreading. Importantly, not all repetitive elements contribute to methylation spreading equally. For example, the downstream Alu (divergence 11.9%) of P16 did not raise the adjacent methylation to the same level as the above repetitive elements did. The functions, if any, of repetitive elements in biological processes have been mysterious. Previously, LINE (L1) elements on the X-chromosome were proposed as candidates for X-inactivation spreading over ∼160 Mb range (33). Moreover, some repetitive elements were empirically defined as cis-regulatory elements (13) and genome-wide analyses have shown that some human and mouse promoters are derived from specific repetitive elements (34). Thus, the LINE of RIL or SINEs of P16 may work as cis-signals to recruit either stronger transcription repressors or chromatin remodeling factors, thereby facilitating the access of DNMTs to the CGI. On the other hand, repetitive elements may also play roles in protecting genomic regions from silencing. It was reported in studies of the murine growth hormone (GH) gene locus that tissue-specific transcription of a SINE B2 element serves as a boundary to compartmentalize local chromatin so as to regulate gene activation during organogenesis (35). Genome-wide computation from mouse and human cancer methylomes discovered the association of a lower frequency of retrotransposons (SINEs and LINEs) and methylation-prone genes, which could facilitate prediction of methylation states from the proximal features of a promoter (36). Therefore, repetitive DNA could supply signals for diverse epigenetic behaviors (e.g. DNA methylation or protection), and the actual outcomes may arise from the activity or structural features of every individual repetitive element (14).
One strategy of our experiments was to control the local repression strength by using the tetracycline-controlled tTS. The tTS is usually used in inducible expression systems, and here we are employing its role in sequential recruitment of the H3K9-specific histone methyltransferase (e.g. SETDB1), HP1, and the histone deacetylase (HDAC)-containing complex via KRAB–KAP1 interaction (37,38). Hence, even over a range of euchromatin, a highly compact heterochromatin patch can be generated and maintained for quite a few generations. On the other hand, without tTS, pINSL6 was still gradually silenced and the promoter was enriched for repressive histones (H3K9me3) probably through adopting the endogenous regulators targeting pINSL6. Therefore, pINSL6 could set up a repressive background, and usage of tTS accelerated the silencing process and sustained it stringently. The earlier the localized repressive heterochromatin was established, the faster de novo methylation occurred and the more methylated the CGI could become.
The variation of position effect was another trans-regulatory aspect taken into account in our experiments. It is interesting that in HCT116D/tTS with another integration site, tTS was not able to lead to methylation spreading in spite of initial methylation seeding. The mechanisms involved here are not clear, although we found the integration site was under inactive status marked by cytosine methylation and lacked active histone modifications (H3K4me3 and H3K9ac). It is likely that transgene methylation is subjected to effects of a large domain centered over the position, which may construct a non-permissive environment for DNA methylation. The role of long-range domains in epigenetic regulation in cancers needs detailed investigation in the future, but recent data in long-range epigenetic silencing in cancer support this concept (39).
Finally, our data demonstrated that the connection between repressive histone modifications and DNA methylation in CGI-promoter silencing may not be as tight as previously considered. The first one was insignificant methylation spreading in several host cells (except SW48A/tTS) even though reporter expression was gradually suppressed and the promoter was highly enriched for H3K9me3 in all cases. Second, methylation was not efficiently recruited into the CGI of the truncated pINSL6 devoid of the LINE element, even though transgenes experienced accelerated silencing in SW48A/tTS. Moreover, treatment with DAC only was not able to recover GFP expression along with DNA demethylation in SW48A/tTs under robust binding of tTS, nor was TSA treatment. Thus, it seems likely that factors other than (or in addition to) HDACs caused by tTS binding are required for sustained silencing and enhanced DNA methylation. Among the set of factors recruited by tTS binding, HP1 and the H3K9-specific HMT (e.g. SETDB1) have been shown to interplay with DNMTs (37,38), and could contribute to DNA methylation. These data suggest that DNA methylation is not an inevitable consequence of transcriptional silencing, but a gradual event that requires relatively strong and sustained local repression. The loose connection of histone modifications and DNA methylation, therefore, could afford the flexibility to dynamically modulate gene transcription via epigenetic machinery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–5.
FUNDING
National Institutes of Health [CA098006 and CA100632 to J.P.I.]; American Cancer Society and F. M. Kirby Foundation (to J.P.I.); National Institutes of Health (Core Grant; CA16672). Funding for open access charge: Fels Institute, Temple University.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 2.Strunnikova M, Schagdarsurengin U, Kehlen A, Garbe JC, Stampfer MR, Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol. Cell. Biol. 2005;25:3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AHFM. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, Li E, Muegge K. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De La Fuente R, Baumann C, Fan T, Schmidtmann A, Dobrinski I, Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat. Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 6.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turker MS. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene. 2002;21:5388–5393. doi: 10.1038/sj.onc.1205599. [DOI] [PubMed] [Google Scholar]
- 9.Toyota M, Issa J-PJ. Epigenetic changes in solid and hematopoietic tumors. Semin. Oncol. 2005;32:521–530. doi: 10.1053/j.seminoncol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 11.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat. Rev. Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 12.Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 14.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 15.Turker MS. The establishment and maintenance of DNA methylation patterns in mouse somatic cells. Semin. Cancer Biol. 1999;9:329–337. doi: 10.1006/scbi.1999.0133. [DOI] [PubMed] [Google Scholar]
- 16.Yates PA, Burman RW, Mummaneni P, Krussel S, Turker MS. Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J. Biol. Chem. 1999;274:36357–36361. doi: 10.1074/jbc.274.51.36357. [DOI] [PubMed] [Google Scholar]
- 17.Mummaneni P, Bishop PL, Turker MS. A cis-acting element accounts for a conserved methylation pattern upstream of the mouse adenine phosphoribosyltransferase gene. J. Biol. Chem. 1993;268:552–558. [PubMed] [Google Scholar]
- 18.Yates PA, Burman R, Simpson J, Ponomoreva ON, Thayer MJ, Turker MS. Silencing of mouse aprt is a gradual process in differentiated cells. Mol. Cell. Biol. 2003;23:4461–4470. doi: 10.1128/MCB.23.13.4461-4470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song JZ, Stirzaker C, Harrison J, Melki JR, Clark SJ. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–1061. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- 20.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 21.Estecio MRH, Gharibyan V, Shen L, Ibrahim AEK, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang THM, et al. LINE-1 Hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards KL, Zhang B, Baggerly KA, Colella S, Lang JC, Schuller DE, Krahe R. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS One. 2009;4:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 24.Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa J-PJ. Genome-wide profiling of DNA methylation reveals a class of normally methylated cpg island promoters. PLoS Genet. 2007;3:e181. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 26.Boumber YA, Kondo Y, Chen X, Shen L, Gharibyan V, Konishi K, Estey E, Kantarjian H, Garcia-Manero G, Issa J-PJ. RIL, a LIM gene on 5q31, is silenced by methylation in cancer and sensitizes cancer cells to apoptosis. Cancer Res. 2007;67:1997–2005. doi: 10.1158/0008-5472.CAN-06-3093. [DOI] [PubMed] [Google Scholar]
- 27.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 28.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 29.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang GN, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 2004;2:62–72. [PubMed] [Google Scholar]
- 30.Athanasiadou R, de Sousa D, Myant K, Merusi C, Stancheva I, Bird A. Targeting of de novo DNA methylation throughout the Oct-4 gene regulatory region in differentiating embryonic stem cells. PLoS One. 2010;5:e9937. doi: 10.1371/journal.pone.0009937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 32.Hinshelwood RA, Melki JR, Huschtscha LI, Paul C, Song JZ, Stirzaker C, Reddel RR, Clark SJ. Aberrant de novo methylation of the p16INK4A CpG island is initiated post gene silencing in association with chromatin remodelling and mimics nucleosome positioning. Hum. Mol. Genet. 2009;18:3098–3109. doi: 10.1093/hmg/ddp251. [DOI] [PubMed] [Google Scholar]
- 33.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 34.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lunyak VV, Prefontaine GG, Nez E, Cramer T, Ju B-G, Ohgi KA, Hutt K, Roy R, García-Díaz A, Zhu X, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 36.Estécio MRH, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, Oki Y, Kondo Y, Jelinek J, Shen L, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010;20:1369–1382. doi: 10.1101/gr.107318.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fahrner JA, Baylin SB. Heterochromatin: stable and unstable invasions at home and abroad. Genes Dev. 2003;17:1805–1812. doi: 10.1101/gad.1123303. [DOI] [PubMed] [Google Scholar]
- 39.Coolen MW, Stirzaker C, Song JZ, Statham AL, Kassir Z, Moreno CS, Young AN, Varma V, Speed TP, Cowley M, et al. Consolidation of the cancer genome into domains of repressive chromatin by long-range epigenetic silencing (LRES) reduces transcriptional plasticity. Nat. Cell Biol. 2010;12:235–246. doi: 10.1038/ncb2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.