Abstract
Drosophila SAYP, a homologue of human PHF10/BAF45a, is a metazoan coactivator associated with Brahma and essential for its recruitment on the promoter. The role of SAYP in DHR3 activator-driven transcription of the ftz-f1 gene, a member of the ecdysone cascade was studied. In the repressed state of ftz-f1 in the presence of DHR3, the Pol II complex is pre-recruited on the promoter; Pol II starts transcription but is paused 1.5 kb downstream of the promoter, with SAYP and Brahma forming a ‘nucleosomal barrier’ (a region of high nucleosome density) ahead of paused Pol II. SAYP depletion leads to the removal of Brahma, thereby eliminating the nucleosomal barrier. During active transcription, Pol II pausing at the same point correlates with Pol II CTD Ser2 phosphorylation. SAYP is essential for Ser2 phosphorylation and transcription elongation. Thus, SAYP as part of the Brahma complex participates in both ‘repressive’ and ‘transient’ Pol II pausing.
INTRODUCTION
Gene activation is a complex process requiring participation of coactivators, a specific group of transcription factors mediating the action of gene-specific activators (1,2). For many years, the recruitment of Pol II and accessory complexes was considered to be the key step in transcription initiation. However, paused Pol II was then detected at the promoters of transcriptionally inactive genes. This phenomenon, initially described for the hsp70 and c-myc genes (3), was subsequently found to be genome-wide (4–6).
Pol II pausing in the repressed state is characterized by several features. Paused Pol II, coactivators and general transcription factors are accumulated on the promoter carrying histone modifications characteristic of active chromatin (7,8). In addition, the CTD of Pol II during initial promoter binding is usually unphosphorylated, whereas that of paused Pol II is phosphorylated at Ser5. This modification is considered to destabilize the interactions between Pol II and promoter-bound factors and mark Pol II ready for promoter escape and early transcription elongation (9).
The Ser5-phosphorylated Pol II molecule leaves the promoter, making the short transcripts, but is usually restrained close (within 100 bp) to the promoter (10,11). This promoter-proximal pausing of Pol II blocks the productive elongation until a certain stimulus appears (12,13). Two factors associated with the elongating Pol II complex, NELF and DSIF, play the essential role in regulation of promoter-proximal pausing (13–15). Thus, promoter-proximal pausing of transcriptionally competent Pol II (also referred to as promoter-proximal stalling) is an important step in the regulation of gene expression.
The promoter-proximal pausing is also observed on actively transcribed genes, where it correlates with the gross rearrangements of the complex of elongating Pol II that lead to transition from early to productive elongation (5,6). This transition is triggered by the recruitment of p-TEFb kinase, which phosphorylates Ser2 residues of Pol II CTD as well as of NELF and DSIF, thereby remodeling the Pol II complex. After transition to productive elongation, this complex can continue transcription without dissociating from the DNA template.
Although the important role of Pol II pausing and promoter-proximal pausing in transcription regulation is evident, many questions concerning its molecular mechanisms remain open. In particular, factors other than NELF and DSIF are also expected to contribute to this process (5,13,15), and the role of histones in pausing the Pol II early elongating complex still remains unclear.
Previously we have described the SAYP coactivator in Drosophila (16–18), which plays an important role in transcription regulation and is indispensable for normal fly development since early embryonic stages. SAYP has homologues in many other metazoans. For example, its human homologue, the PHF10/BAF45a protein, is expressed in different tissues and has been shown to be essential for maintaining the undifferentiated status of neuroblasts (19). All SAYP homologues have an evolutionarily conserved core, which contains the SAY domain involved in transcription activation and two PHD fingers at the C terminus (17).
It has been shown that SAYP/PHF10 interacts with the Brahma complex (PBAP form) in Drosophila (18,20) and mammals (19). More specifically, SAYP in Drosophila interacts with BAP170 subunit of Brahma (18) and unites this chromatin-remodeling complex and the key transcription initiation factor TFIID in a stable, functionally indivisible coactivator supercomplex BTFly (21). SAYP is essential for BTFly interaction with the gene promoter (18), and this mechanism of transcription activation is widespread (20–22).
Recently, SAYP has been shown to interact with the DHR3 (Hr46) activator, an orphan nuclear receptor (NR) that is an essential member of the ecdysone cascade, and to be necessary for the high level of DHR3-dependent activation (23). The DHR3 gene is directly induced by a pulse of 20 hydroxyecdysone (below, referred to simply as ecdysone) at the onset of metamorphosis (24), and the main known target for DHR3 is the ftz-f1 gene encoding FTZ-F1 NR (25–27). This gene is activated by DHR3 only after the decay of the first ecdysone pulse and is repressed by the next ecdysone pulse, its expression during development being restricted to a narrow time window (28). The ftz-f1 expression is important for subsequent events in metamorphosis (29) and is apparently controlled by several mechanisms ensuring precision in its activation.
Paused Pol II was detected on the DHR3-dependent ftz-f1 gene in inactive state. Taking into account the role of SAYP in the recruitment of TFIID and Brahma and its interaction with DHR3, we supposed that SAYP may have a role in the ftz-f1 transcription and, in particular, in Pol II pausing. To reveal this role, we performed experiments with S2 cells in which the transcription of DHR3 and ftz-f1 was induced by the addition and subsequent removal of ecdysone, respectively. We analyzed Pol II pausing on the ftz-f1 gene in the repressed state (at a high ecdysone titer) and at different time points after repression cessation (30 min and several hours after ecdysone removal).
MATERIALS AND METHODS
Antibodies
Experiments were performed with antibodies against SAYP (17,18), TAF1 and TBP (18), Brahma subunits (30,31), Pol II (32), phosphoS5 Pol II Ab5131 (Abcam), phosphoS2 Pol II Ab5095 (Abcam), and acetylated and total histone H3 (clones 4G12 Upstate and Ab1791 Abcam). Antibodies against DHR3 (112–487 aa fragment) were raised in rabbits immunized with the corresponding His6-tagged fragment in our laboratory. An antibody against β-tubulin, obtained by M. Klymkowsky (University of Colorado, Boulder), was from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained at the University of Iowa, Department of Biological Sciences.
Northern blotting, RT–PCR and 5′-RACE analysis
Northern blot analysis of poly(A)+ RNA (flies at different stages of development) or total RNA (S2 cells) was performed as described (17). For measuring gene expression, RNA was extracted with Trizol (Ambion) and treated with DNase I. Reverse transcription (RT) was performed from random hexanucleotide primers and measured by quantitative PCR (qPCR). Measurements at each point were made in at least three replications, and the mean value was calculated. Samples for RT contained equal amounts of RNA, and ras transcription was measured in all experiments to evaluate the total RNA level that remained unchanged under activation or knockdown conditions. To estimate the level of ftz-f1 transcription at different points along the gene (Figure 2D), the content of RNA was normalized with respect to the level of the corresponding DNA fragment (in genomic DNA) to estimate the efficiency of probe amplification.
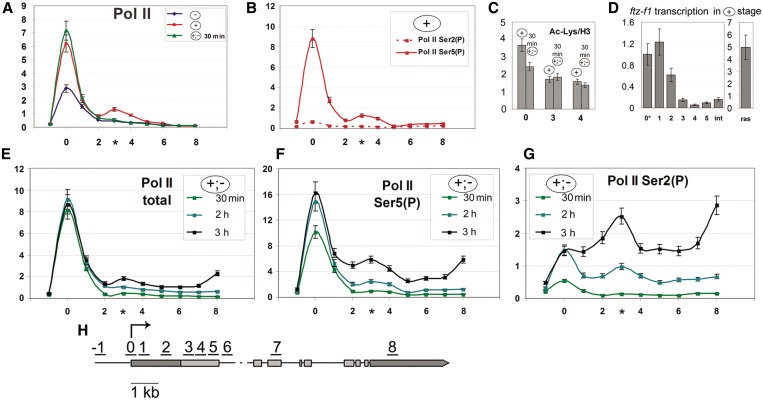
Figure 2.
Pol II pausing occurs at ∼1.5 kb downstream of the transcription start site, whether the ftz-f1 gene is in the repressed or transcriptionally active state. Here and in Figures 3–5, numerals on the x-axis correspond to those in the scheme (H) indicating the positions of probes used in RT–PCR (ChIP) or qPCR experiments; the asterisk indicates the position of second Pol II peak. Each distribution profile represents the average data of at least five experiments. (A) Distribution profile of Pol II along the ftz-f1 gene at different stages of induction—without ecdysone (–) shown in blue, at a high ecdysone titer (+) shown in red, and 30 min after ecdysone removal (+;–) shown in light green—as determined by ChIP with antibodies against total Pol II recognizing both its modified and unmodified forms. (B) Distribution profile of CTD Ser5- or Ser2-phosphorylated Pol II along the ftz-f1 gene in the repressed (+) state as determined by ChIP. (C) The levels of nucleosome acetylation at the promoter and downstream regions (indicated on the x-axis) of the ftz-f1 gene at the (+) and (+;–) 30-min stages as determined by ChIP. The level of acetylated histone is shown relative to the total H3 level. (D) The abortive transcription of ftz-f1 in the repressed state. The transcription level was measured at different points along the gene by quantitative RT–PCR and normalized with respect to the level of the corresponding DNA fragment (in genomic DNA) to estimate the efficiency of probe amplification. (E) Distribution profile of Pol II along the ftz-f1 gene at different time points [30 min (light green), 2 h (cyan) and 3 h (black)] after ecdysone removal as determined by ChIP with antibodies against total Pol II. (F) Distribution profile of Pol II Ser5-(P) along the ftz-f1 gene at different time points [30 min (light green), 2 h (cyan) and 3 h (black)] after ecdysone removal as determined by ChIP with antibodies against CTD Ser5-phosphorylated Pol II. (G) Distribution profile of Pol II Ser2-(P) along the ftz-f1 gene at different time points [30 min (light green), 2 h (cyan) and 3 h (black)] after ecdysone removal as determined by ChIP with antibodies against CTD Ser2-phosphorylated Pol II.
The effect of SAYP mutation on ftz-f1 expression during fly development was analyzed using a Drosophila line carrying the mutant allele e(y)3u1 of the SAYP-encoding e(y)3 gene. Flies of the control line carried the wild-type allele of this gene. The late third instar larvae of the mutant and control lines were collected at equal time points to isolate RNA, which was purified with the Trizol reagent (Ambion).
Primers used to synthesize probes and measure gene expression are given next in the Supplement in Supplementary Data. The 5′-RACE analysis of the pupal ftz-f1 transcript was performed with the primer located at point 1 (see Supplement in Supplementary Data) and RNA isolated from S2 cells at the (+;–) stage, using a Mint cDNA synthesis kit (Evrogen).
Run-on assay
Nuclear purification and in vitro transcription were performed as described (3). Newly synthesized RNA was detected by qPCR instead of radioactive labeling. RNA from the reaction mixture was extracted with the Trizol reagent (Ambion). The level of transcription induction upon adding sarcosyl (0.6% in 80 mM KCl) or high salt buffer (800 mM KCl) was calculated relative to the level of control reaction in 80 mM KCl buffer. To analyze the ftz-f1 gene at the (+) activation stage, cells were preliminary treated with 1 µM 20-hydroxyecdysone (Fluka) overnight. The run-on assay was performed in the presence of 1 µM ecdysone to exclude that activation of ftz-f1 transcription was caused by a drop in ecdysone titer.
Experiments with S2 cell culture
Drosophila Schneider line 2 (S2) cells were maintained at 25°C in charcoal-treated ecdysone-free Schneider’s insect medium (Sigma) containing 10% FBS (HyClone). Conditions optimal for ecdysone cascade initiation were determined experimentally. The cells were treated with 1 µM 20-hydroxyecdysone (Fluka) overnight and analyzed for Pol II pausing on ftz-f1. For subsequent ftz-f1 activation, ecdysone was removed by triple washing, and the cells were incubated in the ecdysone-free medium for additional 30 min and 3 h (for ChIP analysis) or 5 h (for measuring gene induction). Probes for measuring the kinetic of ftz-f1 transcription induction were collected every hour after ecdysone removal. RNAi knockdown experiments were performed as described (18); in sham experiments, the cells were treated with dsRNA corresponding to a fragment of pBluescript II (pSK) vector. SAYP knockdown was performed with two different dsRNAs, #1 and #2 (see Supplement in Supplementary Data). As in the previous study [18], SAYP RNAi did not affect the levels of test factors in the cells, except for PB: its level decreased upon SAYP knockdown.
ChIP and qPCR analysis
ChIP was performed as described (18), with DNA sheared to ∼500 bp. Approximately 3 × 106 cells and 10 mg of an antibody were taken for one experiment. The primers used for analysis are given in the Supplement in Supplementary Data. After ChIP, the recovered DNA was analyzed by qPCR with MiniOpticon (BioRad). Each point was measured in at least five experiments, and the mean value was calculated. For baseline (control) measurements, ChIP with preimmune IgG was used. In all experiments, this control did not exceed 0.1% of the input. A fragment of 28S rDNA was used as a negative control in each experiment. Its values did not exceed 0.2% of the input with all affinity-purified antibodies, increasing to 0.3% with unpurified α-Moira antisera.
RESULTS
Induction of DHR3-driven transcription in S2 cells: SAYP is present on the DHR3-driven ftz-f1 promoter
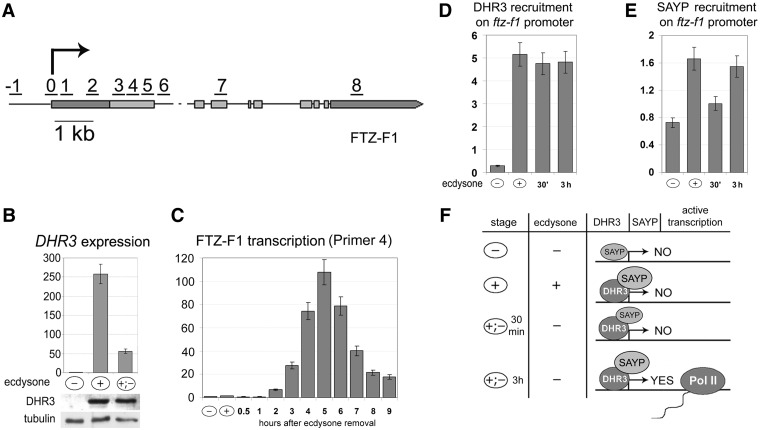
Paused Pol II was found on the inactive ftz-f1 promoter at stages preceding its activation, which was confirmed by the modENCODE database search (33). Therefore, we were interested to find out whether SAYP participates in ftz-f1 regulation and, in particular, in Pol II pausing. To this end, a system for ftz-f1 activation in Schneider (S2) cells was constructed. S2 cells were initially grown in the ecdysone-free medium (–), then incubated overnight in the ecdysone-containing medium (+), and finally washed of ecdysone and again transferred to the ecdysone-free medium (+;–) (for details, see ‘Materials and methods’ section). As expected, the DHR3 expression was near the baseline level before ecdysone treatment (–) but increased significantly in the (+) state, which was confirmed at both mRNA and protein levels (Figure 1B). After ecdysone was washed off (+;–), the level of DHR3 mRNA dropped, but the level of the protein remained unchanged for ∼5 h.
Figure 1.
Induction of DHR3-driven transcription in S2 cells. (A) Scheme of the DHR3-dependent ftz-f1 transcript. Numerals indicate positions of probes used in qPCR to study ftz-f1 transcription and in ChIP experiments. The arrow indicates the transcription start site. (B) Levels of DHR3 transcription in cells grown without ecdysone (–), in the presence of 1 µM ecdysone (+), and after its removal by washing (+;–) as estimated by qPCR. Here and in (C), the data are shown relative to the transcription level in uninduced S2 cell (taken as 1). The level of the DHR3 protein was measured by western blotting. (C) Levels of DHR3-inducible ftz-f1 transcription in cells grown without ecdysone (–), in the presence of 1 µM ecdysone (+), and at different time points after ecdysone removal (+;–) as estimated by qPCR. The ftz-f1 transcription was detected with a pair of primers corresponding to position 4 in Figure 1A. The x-axis shows time (hours) after ecdysone removal. (D) ChIP analysis of DHR3 recruitment on the ftz-f1 promoter (probe 0 in the scheme) at different stages of ftz-f1 activation. Here and in Figures 2–5, ChIP data are presented as a percentage of the input. (E) ChIP analysis of SAYP recruitment on the ftz-f1 promoter (probe 0 in the scheme) at different stages of ftz-f1 activation. (F) Schematic representation of different stages of ftz-f1 activation in S2 cells. Cell treatment with ecdysone induces the expression of the DHR3 activator, which binds to the ftz-f1 promoter and remains on it at all subsequent stages. Its presence strongly facilitates SAYP binding to the promoter. The ftz-f1 transcription is at about the background level in untreated S2 cells and in the presence of ecdysone, increasing within ∼1 h after ecdysone removal. In contrast to DHR3, the level of SAYP on the promoter drops during the first 30 min after ecdysone removal but is restored as the level of transcription increases.
The ftz-f1 gene has two promoters (for details, see Supplement in Supplementary Data and Supplementary Figure S1), one of them being activated by DHR3 (25). In this study, we focused on the inducible transcript and the corresponding promoter with its downstream region (Figure 1A). The level of the inducible ftz-f1 transcript in S2 cells was low either without ecdysone (–) or in its presence (+), remaining so for 1 h after ecdysone removal; thereafter, it gradually increased >100-fold, reaching a peak within 5 h (Figure 1C). This result correlates with in vivo data (34). In contrast, the level of non-inducible transcript remained unchanged after ecdysone addition or removal (Supplementary Figure S1).
To clarify the molecular mechanisms underlying ftz-f1 induction, we checked the kinetics of DHR3 and SAYP recruitment onto the ftz-f1 promoter by ChIP (Figure 1D and E). It can be seen that a high level of DHR3 was detected on the promoter as soon as its expression at the (+) stage started, before the onset of transcription, and remained approximately the same either 30 min or 3 h after ecdysone removal. Therefore, the DHR3 activator is already present on the promoter when it is inactive. This indicates that ftz-f1 is activated in at least two steps; with the (+) stage being a prerequisite for subsequent gene activation.
SAYP was detected on the ftz-f1 promoter at all stages (Figure 1D). Its level significantly increased at the (+) stage (in parallel with DHR3 binding), dropped within 30 min after ecdysone washing (at the onset of transcription), but was restored after 3 h. Thus, SAYP is also present on the ftz-f1 promoter and is likely a component of ftz-f1 induction.
Thus, we succeeded in inducing the DHR3–ftz-f1 fragment of the ecdysone cascade in S2 cells. Our experimental scheme provided the possibility to study the consecutive stages of ftz-f1 gene induction (Figure 1F): in the absence of DHR3 (–); at the stage preliminary to the onset of transcription, when the DHR3 activator is expressed but transcription is blocked (+); at the onset of transcription, after ecdysone was removed but transcription level has not yet increased [(+;–) 30 min], and at the stage of active transcription, 3 h after ecdysone removal [(+;–) 3 h].
Repressive Pol II pausing occurs at about 1.5 kb downstream of the ftz-f1 transcription start
We then studied the occupancy of the inducible promoter and downstream region by Pol II at all stages. At the (–) stage, a certain amount of Pol II was detected on the ftz-f1 promoter; at the (+) stage, the Pol II level strongly increased and remained unchanged after the removal of repression at the (+;–) stage (Figure 2A). The observed accumulation of Pol II on the promoter in the absence of active transcription is a distinctive feature of Pol II pausing. Next, the phosphorylation status of Pol II at the (+) stage was analyzed by ChIP with antibodies against Ser5- or Ser2-phosphorylated Pol II CTD domain [for brevity, these Pol II modifications are referred to below as Ser5-(P) or Ser2-(P), respectively]. The specificity of antibodies was confirmed in experiments with the hsp70 gene (Supplementary Figure S2) (9). The high level of Ser5-(P) characteristic of genes with paused Pol II was revealed on the promoter (Figure 2B). Moreover, another characteristic feature—an elevated level of Ac H3, the mark of transcriptionally active chromatin—was detected on the promoter and in the proximal region (Figure 2C). In contrast, the level of Ser2-(P) was very low (Figure 2B), indicating the absence of productive transcription elongation. Thus, we found indications of promoter activity at the stage when transcription was blocked. The observed distribution of Pol II modifications was similar to that on repressed hsp70 (Supplementary Figure S2).
Unlike in experiments with the hsp70 gene, we unexpectedly revealed an elevated level of Pol II in the promoter downstream region, with a peak at a distance of about 1.5 kb, at the (+) stage (Figure 2A and B). This peak was detected with antibodies against either total Pol II or its Ser5-(P) modification, which confirmed Pol II accumulation at the corresponding position. After ecdysone removal (30 min), the level of Pol II on the promoter remained unchanged (Figure 2A), whereas its second peak disappeared.
The distinctive feature of pausing is that Pol II starts transcription but it is aborted near the point of paused Pol II. To clarify the transcriptional status of the gene at the (+) stage, we measured the level of the synthesized transcript at different points downstream of the promoter by qRT–PCR (Figure 2D). As follows from this figure, the level of the ftz-f1 transcription close to promoter was fairly high, compared with the actively transcribed ras gene, but it dropped abruptly at a distance of ∼1.5 kb. This is evidence that the transcription of ftz-f1 at the (+) stage was initiated but halted at ∼1.5 kb downstream of the promoter, where Pol II was paused.
The run-on assay performed as described (3) showed that the transcription of ftz-f1 at the (+) stage could be stimulated in high salt or sarcosyl-containing buffer. This is evidence that the paused Pol II revealed on the ftz-f1 gene is transcriptionally engaged and competent for elongation (Supplement in Supplementary Data and Supplementary Figure S3).
Thus, DHR3 expression at the (+) stage provides for Pol II pausing. The latter is characterized by unusual features such as an extensive 1.5-kb region of abortive transcription and the second peak of Pol II accumulation at the point of transcription block, which disappears within a short period (30 min) following ecdysone removal, before the beginning of active transcription.
At the active transcription stage, Pol II transient pausing and CTD Ser2 phosphorylation take place at ∼1.5 kb downstream of the promoter
Next, we measured by ChIP the distribution of Pol II on actively transcribed ftz-f1 at different time points (30 min, 2 h and 3 h) after repression cessation (Figure 2E). Compared with the (+) stage, the Pol II level did not change on the promoter but increased on the body of the gene in a time course reflecting efficient transcription elongation. The second peak of Pol II was again observed at 1.5 kb as the level of transcription elongation increased. We considered that this peak could reflect the transient pausing of Pol II that correlates with its Ser2 phosphorylation.
To check this assumption, we analyzed the phosphorylation status of Pol II. The Ser5-(P) and Ser2-(P) profiles remained unchanged 30 min after ecdysone removal, compared with the repression stage (+), confirming that the active transcription did not start yet (Figure 2F and G). A high level of Ser5-(P) was detected on the promoter but not on the body of the gene, with the level of Ser2 phosphorylation being very low (Figure 2E). No second peak of Pol II at 1.5 kb was observed.
However, both Ser5-(P) and Ser 2-(P) profiles proved to change significantly upon with the activation of ftz-f1 transcription. Upon ecdysone removal, Ser5-(P) strongly increased on the promoter after 2 h and on the body of the gene after 3 h, reflecting a high level of elongation (Figure 2F). Within 2 h of active transcription, Ser2-(P) increased approximately twice on the promoter, with its high level being also detected on the body of the gene (Figure 2G). A distinct Ser2-(P) peak appeared at 1.5 kb and dramatically increased after 3 h of active transcription, as did the Ser2-(P) level on the body of the gene.
These results showed that rearrangements in the elongating Pol II complex and Pol II transient pausing on the ftz-f1 gene occurred at ∼1.5 kb downstream of its promoter. Comparison of Pol II distribution patterns along active hsp70 and ftz-f1 genes revealed significant differences between them. In the case of hsp70, Ser2-(P) level on the promoter was much higher in the active than in the repressed state (Supplementary Figure S2), indicating that Pol II CTD was phosphorylated close to promoter (9). In the case of ftz-f1, we revealed a specific site of Ser2-(P) accumulation, which was located 1.5 kb downstream of the promoter.
Thus, repressive Pol II pausing on ftz-f1 at the (+) stage and its transient pausing at the (+;–) stage occur at the same position, about 1.5 kb from the promoter.
SAYP and Brahma are present on the ftz-f1 promoter and at the point of Pol II pausing in the repression stage
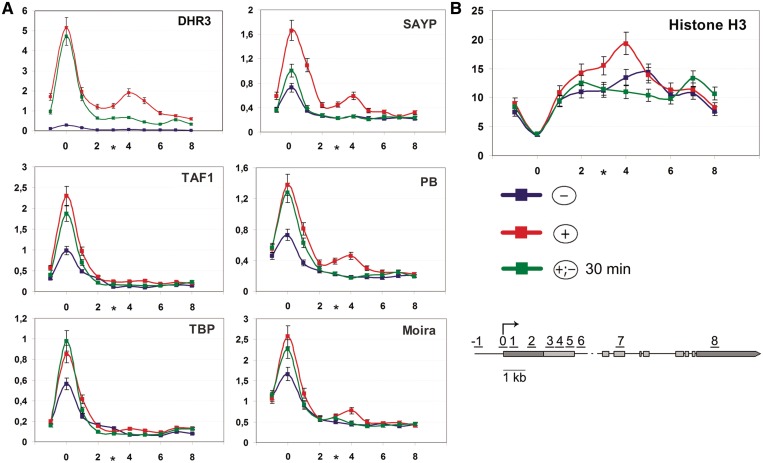
At the next step, we used ChIP to analyze the distribution of DHR3, SAYP and BTFly components on the ftz-f1 gene in the presence of ecdysone and after its removal (Figure 3). Certain amounts of the above factors were detected on the ftz-f1 promoter even in the absence of ecdysone (–). After ecdysone treatment (+) and consequent DHR3 expression, the levels of SAYP, Brahma and TFIID subunits increased considerably, indicating that DHR3 binding initiates the recruitment of coactivators onto the promoter prior to the onset of transcription (Figure 3A). Upon ecdysone removal (+;–), the levels of TFIID and Brahma subunits and DHR3 on the promoter remained almost unchanged after 30 min, whereas the level of SAYP decreased approximately by half at this stage (Figures 1E and 3A).
Figure 3.
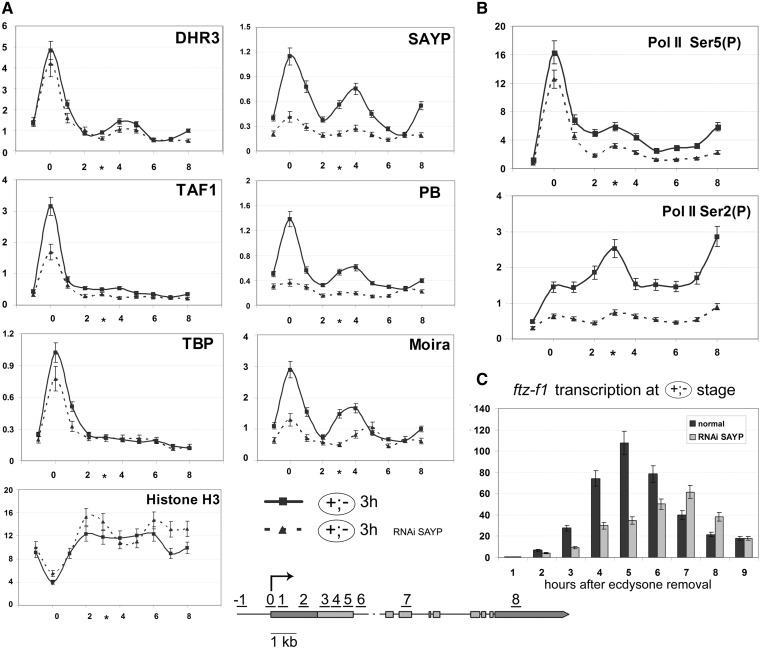
In the repressed state of the ftz-f1 gene, SAYP and Brahma are present on its promoter and at the point of Pol II repressive pausing where the region of high nucleosome density (nucleosome barrier) is formed. (A) Distribution profiles of transcription factors along the ftz-f1 gene without ecdysone (–) shown in blue, at a high ecdysone titer (+) shown in red, and 30 min after ecdysone removal (+;–) shown in light green. The levels of DHR3, SAYP, TFIID subunits (TAF1, TBP) and Brahma subunits (PB, Moira) are shown according to ChIP measurements. Here and in (B), each profile represents the average data of at least 3–5 experiments. (B) Nucleosome distribution along the ftz-f1 gene at the same stages as in (A) as determined by ChIP with α-histone H3 antibody.
It is noteworthy that SAYP, DHR3 and components of the Brahma complex, but not TFIID subunits (TAF1 and TBP), at the (+) stage had the second, smaller peak near the 1.5-kb position (Figure 3A), which almost coincided with the Pol II peak (Figure 2A) but was slightly shifted distally to the promoter (cf Figures 2A and 3A). This peak was absent at the (–) stage prior to DHR3 expression (Figure 3A) and disappeared within 30 min after ecdysone removal. Thus, the presence of SAYP and Brahma components at the 1.5-kb position correlated with that of paused Pol II (Figure 2A), suggesting that SAYP together with Brahma apparently have a role in Pol II pausing at the (+) stage.
A region of high nucleosome density is formed at the 1.5-kb position in the repression stage
Considering the possible role of SAYP and Brahma chromatin remodeler in impeding Pol II elongation, we analyzed the nucleosome distribution in the promoter downstream region by means of ChIP with antibodies to histone H3. At all stages of gene activation, the nucleosome density showed a drop at the promoter (Figure 3B). The nucleosome distribution in the body of the gene was relatively uniform at the (–) stage. At the (+) stage, however, a noticeable peak of histone H3 was detected ∼1.5 kb downstream of the promoter. As in the case of SAYP and Brahma, it almost coincided with the paused Pol II peak but was slightly shifted distally to it (Figure 2F).
The H3 level at 1.5 kb was about twice as high as in the body of the gene, indicating that the region of unusually high nucleosome density (below, referred to as nucleosomal barrier) was formed at this site. The H3 peak disappeared within 30 min after ecdysone removal, correlating with the disappearance of SAYP and Brahma. This is evidence that its organization may be a function of Brahma.
The analysis of the ftz-f1 gene region around the nucleosomal barrier using software described in (35) revealed its significant enrichment in sequences with a high nucleosome-positioning probability (Supplementary Figure S4), indicating that these sequences could contribute to organization of the nucleosomal barrier.
Thus, the (+) stage is specifically characterized by the formation of a nucleosomal barrier downstream of the promoter, the location of this barrier coinciding with the peaks of coactivators and Pol II and with the point of transcription halt. Both these peaks and the nucleosomal barrier disappear soon after the removal of transcription block.
SAYP is essential for organization of chromatin barrier and Pol II pausing in the repression stage
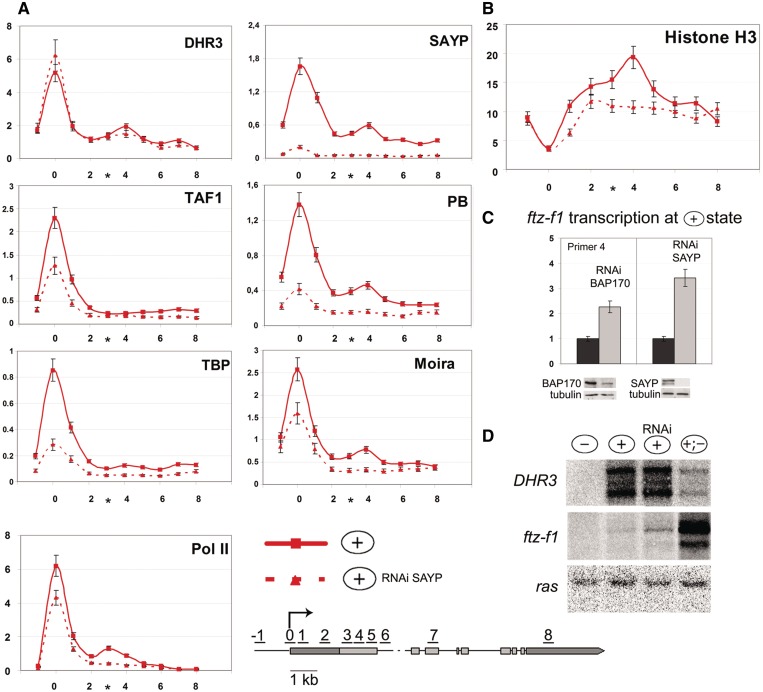
To directly check the role of SAYP in organization of the nucleosomal barrier and Pol II pausing, we used its RNAi knockdown. The SAYP knockdown at the (+) stage had no effect on the DHR3 level on the promoter or at the 1.5-kb position (Figure 4A), indicating that SAYP was not necessary for DHR3 recruitment to the gene. However, the level of TFIID and Brahma components on the ftz-f1 promoter dropped significantly (Figure 4A) with the level of promoter-bound Pol II decreasing as well. The level of Ser5-(P) Pol II decreased proportionally to the level of total Pol II (data not shown). These results suggest that SAYP is essential for the assembly of the paused Pol II preinitiation complex on the promoter prior to the start of transcription.
Figure 4.
In the repressed state of the ftz-f1 gene, SAYP and Brahma are essential for organization of nucleosome barrier and Pol II pausing. Distribution profiles determined by ChIP at the (+) stage against the background of SAYP RNAi knockdown (red dotted line) are compared with those in normal cells (red solid line) at the same stage (shown in Figure 3). Sham-treated cells were used as a control of RNAi in each experiment. (A) Distribution profiles of DHR3, SAYP, Brahma (PB and Moira) and TFIID (TAF1 and TBP) subunits, and Pol II along ftz-f1 after SAYP RNAi knockdown (red dotted line) compared with those in normal cells (red solid line) (Figure 3A). (B) Nucleosome distribution along the ftz-f1 gene after SAYP knockdown (red dotted line) compared with the H3 profile in normal cells (red solid line) (Figure 3B). (C) SAYP and BAP170 (Brahma subunit) knockdown leads to ftz-f1 transcription in the repressed (+) state. The levels of ftz-f1 transcription in the (+) state (dark bars) and upon its induction by RNAi knockdown of SAYP and BAP170 (light bars) were measured by qPCR and normalized relative to the level of ras transcription. The low level of ftz-f1 transcription in wild-type cells was taken as 1. The efficiency of knockdown was tested by western blot analysis. (D) Northern blot analysis of DHR3 and ftz-f1 transcription in S2 cells at different stages of ftz-f1 induction. At the (+) stage, RNA was prepared either from cells after SAYP RNAi knockdown or from sham-treated cells. Hybridization with the ras probe was used as a loading control.
Moreover, the SAYP knockdown eliminated the peak of Brahma components at the 1.5-kb position (Figure 4A). The nucleosomal barrier disappeared as well, and the level of histone H3 dropped to the average along the gene (Figure 4B).
The SAYP knockdown also resulted in the disappearance of the downstream Pol II peak at the 1.5-kb position (Figure 4A). This implies that the corresponding transcription block was removed. We revealed a severalfold induction of the ftz-f1 transcription by qPCR (Figure 4C). Moreover, the level of induction increased in downstream regions, compared with promoter proximal regions, reaching a maximum in the region of nucleosomal barrier (Supplementary Figure S5). Thus, SAYP depletion reduced the decrease of transcription in the downstream region of the gene.
Northern blot analysis showed that the full-length ftz-f1 transcript was slightly induced upon SAYP knockdown (Figure 4D). Transcription leakage at this stage was also detected upon the knockdown of Brahma component BAP170 (Figure 4C).
To check whether the observed effect was due to a drop in the level of repressive factors E75 or Usp, which were suggested to control DHR3-activated transcription (36,37), we measured the transcription of the corresponding genes. The results showed that its level remained unchanged upon the SAYP knockdown (Supplementary Figure S6), and, therefore, the derepression of ftz-f1 was not due to the removal of repressive factors. Thus, SAYP is important for the formation of the downstream Pol II pausing site, ensuring a strong block of transcription at the repression stage.
At the active transcription stage, SAYP is essential for Ser2 phosphorylation and transcription elongation
We checked the distribution of SAYP, Brahma and TFIID on the promoter and downstream region at the stage of active transcription, 3 h after its onset (Figure 5A). These factors proved to remain at a high level on the promoter, but they also showed a significant peak at 1.5 kb downstream of it. A noteworthy fact is that the above factors proved to disappear from the 1.5-kb position 30 min after the transcription onset (Figure 3A) but were recruited there again after 3 h, whereas the repeated formation of the nucleosomal barrier did not take place (Figure 5A). Checking the nucleosome density, we found it to be uniform downstream of the promoter. This finding correlated with the nucleosome distribution observed on the β-actin gene with promoter-distal pausing: the region upstream of the pausing point was free of nucleosomes, while their distribution in more distal regions was uniform (38). These results indicate that SAYP and Brahma may be involved in Pol II pausing at the active transcription stage, but the mechanism of this process should be different from that at the repression stage.
Figure 5.
In the transcriptionally active state of ftz-f1, SAYP is essential for the assembly of Brahma at the point of transient pausing as well as for Ser2 phosphorylation and transcription elongation. (A) Distribution profiles of SAYP, Brahma (PB and Moira) and TFIID (TAF1 and TBP) subunits, Pol II and nucleosomes (α-histone H3 antibody) along the ftz-f1 gene at the (+;–) stage (3 h after ecdysone removal) as determined by ChIP in normal cells (solid line) and after SAYP knockdown (dotted line). (B) Distribution profiles of Pol II Ser5-(P) and Pol II Ser2-(P) along the ftz-f1 gene at the (+;–) stage (3 h after ecdysone removal) in normal cells (solid line) and after SAYP knockdown (dotted line). (C) Changes in the level of ftz-f1 transcription at the (+;–) stage at different time points after ecdysone removal in cells after SAYP RNAi knockdown (light bars) and in normal cells (dark bars (Figure 1C). Transcription was detected with two pairs of primers corresponding to positions 1 and 4 in Figure 1A), which produced the same results.
To clarify the role of SAYP in organization of active transcription, we performed experiments with its knockdown. After 30 min following the drop of ecdysone titer at the (+;–) stage, the SAYP knockdown proved to have no effect on Pol II, Brahma and TFIID occupancy of the promoter (Supplementary Figure S7). This fact correlated with the observation that the SAYP level on the promoter dropped within 30 min after ecdysone removal (Figure 3A). Changes resulting from the SAYP knockdown manifested themselves 3 h after the onset of transcription (Figure 5A): the level of the promoter-bound TFIID complex decreased, the levels of Brahma components on the promoter and at the 1.5-kb position also decreased significantly, while the histone H3 level in the body of the gene slightly increased. However, SAYP depletion had no appreciable effect on the DHR3 level (Figure 5A), as was the case at the (+) stage.
We also checked the importance of SAYP for Pol II modifications at the active transcription stage. Upon the SAYP knockdown, the level of Ser5-modified Pol II on the promoter decreased only slightly (Figure 5B, upper panel), indicating that SAYP was not necessary for Ser5 phosphorylation. A more serious drop of Ser5-(P) level (approximately by half) was observed in the body of the gene, indicating that SAYP depletion disturbs transcription elongation.
A much stronger effect was observed on the Ser2-(P) level, which dropped severalfold in all regions of the gene (Figure 5B, lower panel), with its peak at the 1.5-kb position disappearing almost completely. These changes were not due to SAYP RNAi influence on the transcription level of the cdk9 and cycT genes encoding the components of p-TEF complex responsible for Ser2 phosphorylation (Supplementary Figure S8). Thus, SAYP in the active transcription stage is important for Ser2 phosphorylation and proper elongation of Pol II.
SAYP depletion not only significantly reduced the general level of the ftz-f1 transcription but also altered its time course (Figure 5C): in normal cells, the level of ftz-f1 mRNA reached a peak within 5 h, whereas the transcription of this gene against the background of SAYP depletion proceeded much more slowly and reached a peak after 7 h, when its level in normal cells had already decreased significantly. These data suggest that Pol II pausing is important for accurate activation of ftz-f1.
SAYP is important for proper timing of ftz-f1 transcription during Drosophila development
To verify the data on the role of SAYP in ftz-f1 regulation obtained with the cell culture model, we analyzed the influence of SAYP mutation on the ftz-f1 expression during fly development in experiments with the line carrying the mutant allele e(y)3u1 of the SAYP-encoding e(y)3 gene (with a reduced SAYP transcription level). The late third instar larvae from the mutant and control lines were collected at equal time points to measure the ftz-f1 transcription level. The results showed that SAYP depletion altered the time course of ftz-f1 transcription during metamorphosis, with its initiation taking place earlier in e(y)3u1 mutants than in wild-type larvae (Figure 6A). This could be explained by the role of SAYP in repressive pausing of the ftz-f1 gene. Moreover, the ftz-f1 transcription in e(y)3u1 mutants did not reach the wild-type level and proved to decline at the time point corresponding to its peak in normal larvae. These results are consistent with our cell culture data and confirm the important role of SAYP in Pol II transient pausing and elongation.
Figure 6.
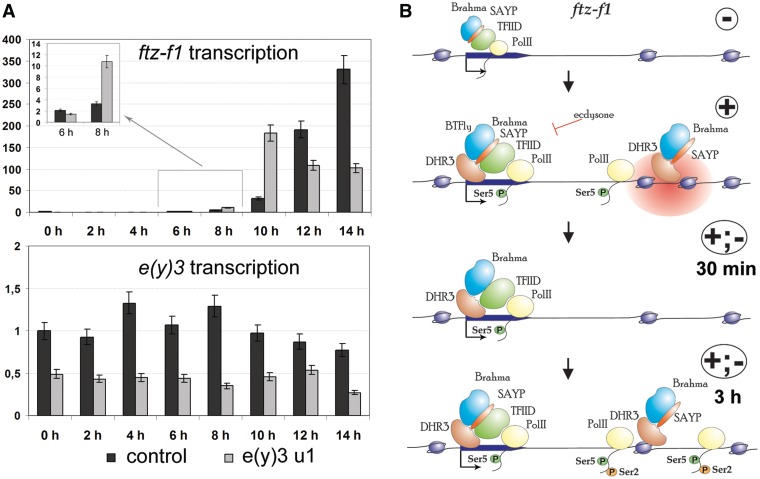
SAYP is important for ftz-f1gene transcription. (A) SAYP is important for proper timing of ftz-f1 transcription during Drosophila metamorphosis. The levels of ftz-f1 and e(y)3 transcription in the control line (dark bars) and the mutant e(y)3u1 line (light bars) during puparium formation were measured by qPCR at 2-h intervals, starting from the late third instar larvae. Each expression profile represents the average data of three experiments. (B) Model of ftz-f1 activation (only transcription factors considered in this study are shown). Activation of this gene occurs in several steps. The ftz-f1 promoter and downstream region in cells not treated with ecdysone. Certain amounts of Brahma, SAYP and TFIID are found on the promoter. Ecdysone induces the expression of DHR3, which facilitates the assembly of SAYP, Brahma, TFIID and Ser5-(P) Pol II into a complex on the promoter. SAYP is essential for TFIID and Brahma recruitment. Brahma and SAYP form the nucleosomal barrier 1.5 kb downstream of the promoter. DHR3 is present at this point and facilitates the recruitments of these two factors. Active transcription of the region proximal to promoter takes place at the (+) stage, but Pol II is paused at the nucleosomal barrier. Within 30 min after ecdysone removal, SAYP level on the promoter decreases, but the levels of other factors remain unchanged. The peaks of DHR3, SAYP and Brahma downstream of the promoter and the nucleosomal barrier are eliminated. Pol II pausing ceases, but transcription remains at the same level. After several hours following ecdysone removal, the transcription level strongly increases, as does Pol II level on the promoter. Pol II pausing is observed at ∼1.5 kb downstream of the promoter, correlating with Ser2 Pol II phosphorylation. DHR3, SAYP and Brahma are again present at the point of downstream pausing, but the nucleosomal barrier is not restored. At this stage, SAYP is necessary for Ser2 phosphorylation and transcription elongation.
Thus, SAYP plays a major role in ftz-f1 transcription regulation during Drosophila metamorphosis. However, the effect observed in flies appears to reflect the sum of SAYP functions at different stages of ftz-f1 activation. From this standpoint, the cell culture model used in our study is more expedient, since it allows fine discrimination between the functions of SAYP in ftz-f1 gene repression and at the active transcription stage.
DISCUSSION
The results show that SAYP, a component of the Brahma remodeling complex, is a new factor important for Pol II pausing.
The mechanism of ftz-f1 transcription activation has been analyzed in S2 cells. Sequential addition and removal of ecdysone allows the DHR3 and ftz-f1 genes in these cells to be activated in accordance with their expression pattern in vivo (39). This system is of considerable interest, since only a few Drosophila models of activated transcription are available. It also provides the possibility of studying the mechanism of pausing in the active and repressed transcription states of the same gene, whereas previous such studies have been performed with different genes (8).
Pol II pausing on ftz-f1 occurs at about 1.5 kb downstream of the promoter, i.e. at a much greater distance than that described for other genes (from +30 to +100 nt) (6). Future studies will show how widespread is this mode of pausing. It is of interest in this context that a case of Pol II pausing at 800 bp downstream of the promoter was described for the β-actin gene (38).
The ftz-f1 activation at the molecular level is a several-stage process (Figure 6B). At the first stage, when the ecdysone titer and DHR3 expression are high, DHR3, SAYP, TFIID, Brahma and Pol II accumulate at the promoter. Transcription is initiated, but Pol II is paused 1.5 kb downstream of the promoter; DHR3, SAYP and Brahma are also present at this site, where a nucleosomal barrier is formed. At the next stage, ∼1 h after ecdysone removal, promoter-bound factors remain at the same levels, except for SAYP (its level on the promoter decreases). Pol II and associated factors disappear from the site of pausing, and the nucleosomal barrier is eliminated, but the transcription level does not increase. The following stage is characterized by rapid intensification of transcription, which reaches a maximum within several hours; the level of Pol II increases in the body of the gene, and its pausing is observed again, with SAYP and Brahma being present at the corresponding position. In addition, the level of SAYP on the promoter is recovered, indicating that it is highly regulated at different transcription stages. The DHR3 activator is present at the site of pausing, and its level does not change upon SAYP knockdown. This is evidence that DHR3 may participate in SAYP recruitment for subsequent nucleosomal barrier formation and Pol II pausing.
The region of high nucleosome density (nucleosomal barrier) is specific for the repression stage, at which the DHR3 activator induces the assembly of the Pol II preinitiation complex on the promoter and makes paused Pol II competent for transcription initiation. Nucleosomal barrier disruption by SAYP knockdown leads to the full-length transcript synthesis, indicating that the nucleosomal barrier contributes to preventing the entry of Pol II to the transcribed region. Our data show that SAYP and Brahma play the crucial role in organization of the nucleosomal barrier: this barrier coincides in location with the peak of these coactivators and disappears after SAYP knockdown, which leads to elimination of Brahma from the gene. Thus, SAYP and Brahma at the stage of repressed transcription have an important role in blocking the synthesis of full-length transcripts. Although the transcription increases upon SAYP depletion and elimination of the nucleosomal barrier, its level remains low, compared with that in the permissive state. This is evidence for the existence of different mechanisms of Pol II pausing regulation, which also correlates with the fact that the depletion of NELF, an important factor of Pol II pausing, causes a 2.5-fold increase in the transcription of hsp70 or hsp26 gene in the repressed state, which, however, does not reaches the level characteristic of a fully activated gene (5).
The question arises as to the structure of the nucleosomal barrier. As shown previously, the human SWI/SNF complex can not only erase nucleosomes from the template but also produce a stable remodeled dimer of mononucleosome core, with this complex being also needed for converting this product back to the cores (40). One may suggest that the Drosophila Brahma complex operates in the same way. In our experiments, the level of histone H3 increased ∼2-fold in the region of the nucleosomal barrier, compared with its general level on the gene, which agrees with the assumption concerning the presence of a nucleosome dimer. The fact that the region of nucleosomal barrier is significantly enriched in sequences with a high nucleosome-positioning probability indicates that DNA sequences probably contribute to organization of this barrier.
Previous experiments have revealed a relationship between Pol II pausing and the nucleosomal structure of the template (5,41). It has been shown that Pol II stops at the site where the nucleosome density is restored to the average level characteristic of the gene (38). However, no specific nucleosome-dense regions preventing Pol II transcription have been described as yet.
The transition to the transcription-permissive state correlates with significant rearrangements in the promoter-distal region (disappearance of Brahma, SAYP, Pol II and nucleosomal barrier at the site of Pol II pausing). However, no increase in the ftz-f1 transcription level has been observed within the first 30 min after this transition. As shown in the study on estradiol (ER)-mediated gene expression, productive transcription is preceded by an unproductive cycle (∼40 min) that is necessary for promoter preparation to this process (42). This may be the case for ftz-f1, with a certain period of time being required for rearrangements preceding its active transcription.
At the (+;–) stage, the level of SAYP on the promoter is recovered within 2–3 h after the onset of transcription, with SAYP RNAi influencing the Brahma and TFIID levels on the promoter. Pol II pausing correlating with its Ser2 modification is again observed as the transcription level increases. Although SAYP and Brahma occur again together with paused Pol II, their function appears to be different from that at the repression stage. The nucleosomal barrier is not restored, and SAYP depletion has only a slight effect on chromatin structure.
However, SAYP depletion severely disturbs transient pausing, interfering with Ser2 phosphorylation. This impairs proper transition to productive elongation and leads to a decrease in Pol II level on the body of the gene. Thus, SAYP knockdown not only affects the level of ftz-f1 activation but also shifts the timing of its expression. The slower kinetics of transcription induction together with the slight decrease in the Pol II level on the promoter upon SAYP knockdown are evidence for the retarded Pol II passage in the coding region of the gene and, hence, for disturbances in the elongation mechanisms. Similar consequences are observed for other genes regulating on pausing mechanisms (7,43).
The results of this study show that SAYP is important for proper timing of ftz-f1 transcription during Drosophila metamorphosis. The ftz-f1 gene is a major regulator of metamorphosis, that is why its precise activation in time is crucial during development. On the whole, our data provide evidence for the important role of pausing in sequential activation of genes in cascades and indicate that this mechanism may have a general role in development.
In addition, these results also support the idea that Pol II pausing may require not only NELF and DSIF but also other factors, such as nucleosome-remodeling complexes (13,15). Interestingly, the depletion of NELF proved to result in an increased nucleosome occupancy at the promoters of some genes (5).
In summary, we have found that Pol II pausing is dependent on the interplay of several molecular mechanisms, including the formation of a specific chromatin structure via the action of coactivators. Together with data by other authors, our results indicate that, although Pol II pausing is a genome-wide phenomenon, the specific molecular mechanism controlling paused Pol II activity on individual genes may vary significantly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplement, Supplementary Figures 1–8 and Supplementary Reference [44].
FUNDING
Program ‘Molecular and Cell Biology’ of the Russian Academy of Sciences, CRP–ICGEB Research [CRP/RUS10-02]; RFBR [10-04-00257 and 11-04-91339]; Scientific School Support [No. 2814.2012.4]; University of Oslo, Centre for Medical Studies in Russia (fellowship to Y.S. and A.K.); RF Presidential Program in Support of Young Scientists [MK-5961.2012.4 and MD-4874.2011.4 to N.V. and A.K.]. Dmitry Zimin Dynasty Foundation (fellowship to N.V.). Funding for open access charge: Dmitry Zimin Dynasty Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to N. A. Gorgolyuk for his help in preparing the manuscript.
REFERENCES
- 1.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 3.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 4.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien T, Lis JT. RNA polymerase II pauses at the 5' end of the transcriptionally induced Drosophila hsp70 gene. Mol. Cell Biol. 1991;11:5285–5290. doi: 10.1128/mcb.11.10.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris DP, Michelotti GA, Schwinn DA. Evidence that phosphorylation of the RNA polymerase II carboxyl-terminal repeats is similar in yeast and humans. J. Biol. Chem. 2005;280:31368–31377. doi: 10.1074/jbc.M501546200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba K, Yamamoto J, Yamaguchi Y, Handa H. Promoter-proximal pausing and its release: Molecular mechanisms and physiological functions. Exp. Cell Res. 2010;316:2723–2730. doi: 10.1016/j.yexcr.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 13.Tao Y, Xi S, Briones V, Muegge K. Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS One. 2010;5:e9163. doi: 10.1371/journal.pone.0009163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 15.Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc. Natl Acad. Sci. USA. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgiev PG, Gerasimova TI. [Detection of new genes participating in the trans-regulation of locus yellow of MDG-4 in Drosophila melanogaster] Genetika. 1989;25:1409–1419. [PubMed] [Google Scholar]
- 17.Shidlovskii YV, Krasnov AN, Nikolenko JV, Lebedeva LA, Kopantseva M, Ermolaeva MA, Ilyin YV, Nabirochkina EN, Georgiev PG, Georgieva SG. A novel multidomain transcription coactivator SAYP can also repress transcription in heterochromatin. EMBO J. 2005;24:97–107. doi: 10.1038/sj.emboj.7600508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, Georgieva SG, Shidlovskii YV. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc. Natl Acad. Sci. USA. 2009;106:11049–11054. doi: 10.1073/pnas.0901801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalkley GE, Moshkin YM, Langenberg K, Bezstarosti K, Blastyak A, Gyurkovics H, Demmers JA, Verrijzer CP. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol. Cell. Biol. 2008;28:2920–2929. doi: 10.1128/MCB.02217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vorobyeva NE, Soshnikova NV, Kuzmina JL, Kopantseva MR, Nikolenko JV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. The novel regulator of metazoan development SAYP organizes a nuclear coactivator supercomplex. Cell Cycle. 2009;8:2152–2156. doi: 10.4161/cc.8.14.9115. [DOI] [PubMed] [Google Scholar]
- 22.Panov VV, Kuzmina JL, Doronin SA, Kopantseva MR, Nabirochkina EN, Georgieva SG, Vorobyeva NE, Shidlovskii YV. Transcription co-activator SAYP mediates the action of STAT activator. Nucleic Acids Res. 2012;40:2445–2453. doi: 10.1093/nar/gkr1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vorobyeva NE, Nikolenko JV, Krasnov AN, Kuzmina JL, Panov VV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. SAYP interacts with DHR3 nuclear receptor and participates in ecdysone-dependent transcription regulation. Cell Cycle. 2011;10:1821–1827. doi: 10.4161/cc.10.11.15727. [DOI] [PubMed] [Google Scholar]
- 24.Thummel CS. Dueling orphans–interacting nuclear receptors coordinate Drosophila metamorphosis. Bioessays. 1997;19:669–672. doi: 10.1002/bies.950190806. [DOI] [PubMed] [Google Scholar]
- 25.Lam GT, Jiang C, Thummel CS. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- 26.Broadus J, McCabe JR, Endrizzi B, Thummel CS, Woodard CT. The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell. 1999;3:143–149. doi: 10.1016/s1097-2765(00)80305-6. [DOI] [PubMed] [Google Scholar]
- 27.Ruaud AF, Lam G, Thummel CS. The Drosophila nuclear receptors DHR3 and betaFTZ-F1 control overlapping developmental responses in late embryos. Development. 2010;137:123–131. doi: 10.1242/dev.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodard CT, Baehrecke EH, Thummel CS. A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell. 1994;79:607–615. doi: 10.1016/0092-8674(94)90546-0. [DOI] [PubMed] [Google Scholar]
- 29.Yamada M, Murata T, Hirose S, Lavorgna G, Suzuki E, Ueda H. Temporally restricted expression of transcription factor betaFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development. 2000;127:5083–5092. doi: 10.1242/dev.127.23.5083. [DOI] [PubMed] [Google Scholar]
- 30.Kal AJ, Mahmoudi T, Zak NB, Verrijzer CP. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 2000;14:1058–1071. [PMC free article] [PubMed] [Google Scholar]
- 31.Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, Verrijzer CP. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 2004;24:3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgieva S, Kirschner DB, Jagla T, Nabirochkina E, Hanke S, Schenkel H, de Lorenzo C, Sinha P, Jagla K, Mechler B, et al. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol. Cell. Biol. 2000;20:1639–1648. doi: 10.1128/mcb.20.5.1639-1648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White KP, Hurban P, Watanabe T, Hogness DS. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubiger M, Truman JW. The RXR ortholog USP suppresses early metamorphic processes in Drosophila in the absence of ecdysteroids. Development. 2000;127:1151–1159. doi: 10.1242/dev.127.6.1151. [DOI] [PubMed] [Google Scholar]
- 37.Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, et al. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Egloff S, Al-Rawaf H, O'Reilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol. Cell. Biol. 2009;29:4002–4013. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan AA, Thummel CS. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol. Endocrinol. 2003;17:2125–2137. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- 40.Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 41.Kulaeva OI, Hsieh FK, Studitsky VM. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc. Natl Acad. Sci. USA. 2010;107:11325–11330. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 43.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.