Abstract
C4 photosynthesis has evolved in at least 66 lineages within the angiosperms and involves alterations to the biochemistry, cell biology, and development of leaves. The characteristic “Kranz” anatomy of most C4 leaves was discovered in the 1890s, but the genetic basis of these traits remains poorly defined. Oat × maize addition lines allow the effects of individual maize (Zea mays; C4) chromosomes to be investigated in an oat (Avena sativa; C3) genetic background. Here, we have determined the extent to which maize chromosomes can introduce C4 characteristics into oat and have associated any C4-like changes with specific maize chromosomes. While there is no indication of a simultaneous change to C4 biochemistry, leaf anatomy, and ultrastructure in any of the oat × maize addition lines, the C3 oat leaf can be modified at multiple levels. Maize genes encoding phosphoenolpyruvate carboxylase, pyruvate, orthophosphate dikinase, and the 2′-oxoglutarate/malate transporter are expressed in oat and generate transcripts of the correct size. Three maize chromosomes independently cause increases in vein density, and maize chromosome 3 results in larger bundle sheath cells with increased cell wall lipid deposition in oat leaves. These data provide proof of principle that aspects of C4 biology could be integrated into leaves of C3 crops.
C4 photosynthesis has at least 66 independent origins within the angiosperms (Sage et al., 2012) and involves alterations to the biochemistry, cell biology, and development of leaves. These modifications increase the efficiency of photosynthesis by reducing photorespiration, which in C3 individuals can reduce photosynthetic efficiency by up to 40% (Ehleringer and Monson, 1993). Attenuation of photorespiration in C4 species represents a considerable competitive advantage, particularly in environments where conditions of drought and exposure to high light intensities predominate and CO2 concentrations in the intercellular air spaces of the leaf are low. Accordingly, C4 grasses and sedges are dominant components of semiarid tropical and subtropical savannah, and some of the most productive agricultural crops (e.g. maize [Zea mays], sorghum [Sorghum bicolor], and sugarcane [Saccharum officinarum]) use C4 photosynthesis (Sage et al., 1999). The increased productivity of C4 plants has stimulated interest in placing the C4 pathway into C3 crops to raise yield potential (Matsuoka et al., 2001; Hibberd et al., 2008; Sage and Zhu, 2011).
The alterations to C4 leaves allow the initial fixation of HCO3− by phosphoenolpyruvate carboxylase (PEPC) to be separated from the release of CO2 in a separate compartment that contains Rubisco (Hatch, 1987). This release of CO2 around Rubisco can be catalyzed by three different four-carbon decarboxylases. Traditionally, this has led to the classification of C4 plants into one of three biochemical subtypes, depending on whether they primarily use NADP-dependent malic enzyme (NADP-ME), NAD-dependent malic enzyme (NAD-ME), or phosphoenolpyruvate carboxykinase (Hatch et al., 1975; Hatch, 1987; Leegood, 2002). However, there appears to be reasonable mechanistic flexibility that is determined both temporally and environmentally (Walker et al., 1997; Wingler et al., 1999; Furumoto et al., 1999), which suggests that the distinctions between these three biochemical subtypes are less rigid than originally thought (Furbank, 2011). As well as this diversity within the biochemistry of C4 photosynthesis, there is also variation in terms of the leaf anatomy associated with the C4 pathway. Although several examples of C4 photosynthesis operating in single cells have been described (Reiskind et al., 1997; Casati et al., 2000; Reinfelder et al., 2000; Voznesenskaya et al., 2002, 2003), most C4 species compartmentalize carboxylation and decarboxylation activities into two distinct cell types. At least 22 variations of two-celled “Kranz” anatomy have been documented (Dengler and Nelson, 1999; Edwards and Voznesenskaya, 2011; Sage et al., 2012) whereby different cell types have been coopted as the photosynthetic carbon assimilation and photosynthetic carbon reduction tissue. In the grasses, nine types of Kranz anatomy have been described (Edwards and Voznesenskaya, 2011). Five of these nine types have two sheaths that surround each vascular bundle. The inner sheath has endodermal characteristics and is called the mestome sheath (MS); it is surrounded by a parenchymal bundle sheath (PBS; Brown, 1958). In the other four types, the MS cell layer is absent and the PBS is adjacent to the metaxylem tissue of the vascular bundle (Brown, 1958). In all of these Kranz variants, Rubisco is localized in the PBS, where it is used to fix CO2.
The molecular basis underpinning the Kranz anatomy of most C4 leaves remains to be defined. In maize, which has “classical” NADP-ME anatomy, the leaf is organized in repeating units of mesophyll (M)-PBS-vein (V)-PBS-M (Langdale and Nelson, 1991) with no MS cell layer. Although mutants of maize that show perturbations to plastid morphology and expression of photosynthesis genes in M or PBS cells of C4 leaves have been identified (Langdale and Kidner, 1994; Brutnell et al., 1999; Smith et al., 2001), they have so far provided little insight into the genetic basis of Kranz anatomy.
In addition to our relatively poor understanding of the mechanisms underlying Kranz anatomy in C4 leaves, it is also unclear whether elements of Kranz anatomy can be introduced into leaves of C3 plants. As the most agronomically productive C4 species operate a two-celled system, this is a major hurdle that must be overcome if C3 crops such as rice (Oryza sativa) are to be modified to use the C4 pathway (Sage and Zhu, 2011). To address this question, we used oat × maize addition lines (OMAs) to test if determinants of Kranz anatomy from maize were functional in a C3 leaf. OMAs are oat (Avena sativa) plants with individual maize chromosomal additions to the oat genome (Riera-Lizarazu et al., 1996, 2000; Kynast et al., 2001). OMAs allow the effects of individual maize (C4) chromosomes to be considered within a C3 context, substantially reducing the complexity of the maize genome and permitting traits of interest to be mapped to individual chromosomes (Riera-Lizarazu et al., 2000). OMAs have also previously been used to investigate the expression of ZmPEPC and ZmPPDK (for pyruvate, orthophosphate dikinase) and the consequences of their expression on photosynthesis in oat (Kowles et al., 2008). OMAs, therefore, are an interesting system with which to investigate the extent to which leaf anatomy can be modified in C3 species. Here, we use OMA lines to study the impact of individual maize chromosomes on the control of leaf development in C3 oat as well as aspects of cell biology, gene expression underlying the biochemistry, and leaf photosynthetic characteristics. Overall, we aimed to assess the regulation of core C4 pathway genes when placed in a C3 species, to characterize the extent to which C4 characteristics could be introduced into oat, and to relate any potential C4-like changes to the presence of particular maize chromosomes. Specifically, we aimed to examine the effect of maize chromosomal introductions on leaf morphology, chloroplast development, and carbon assimilation in a C3 leaf.
RESULTS
Maize Chromosomes Can Increase Vein Density in C3 Oat
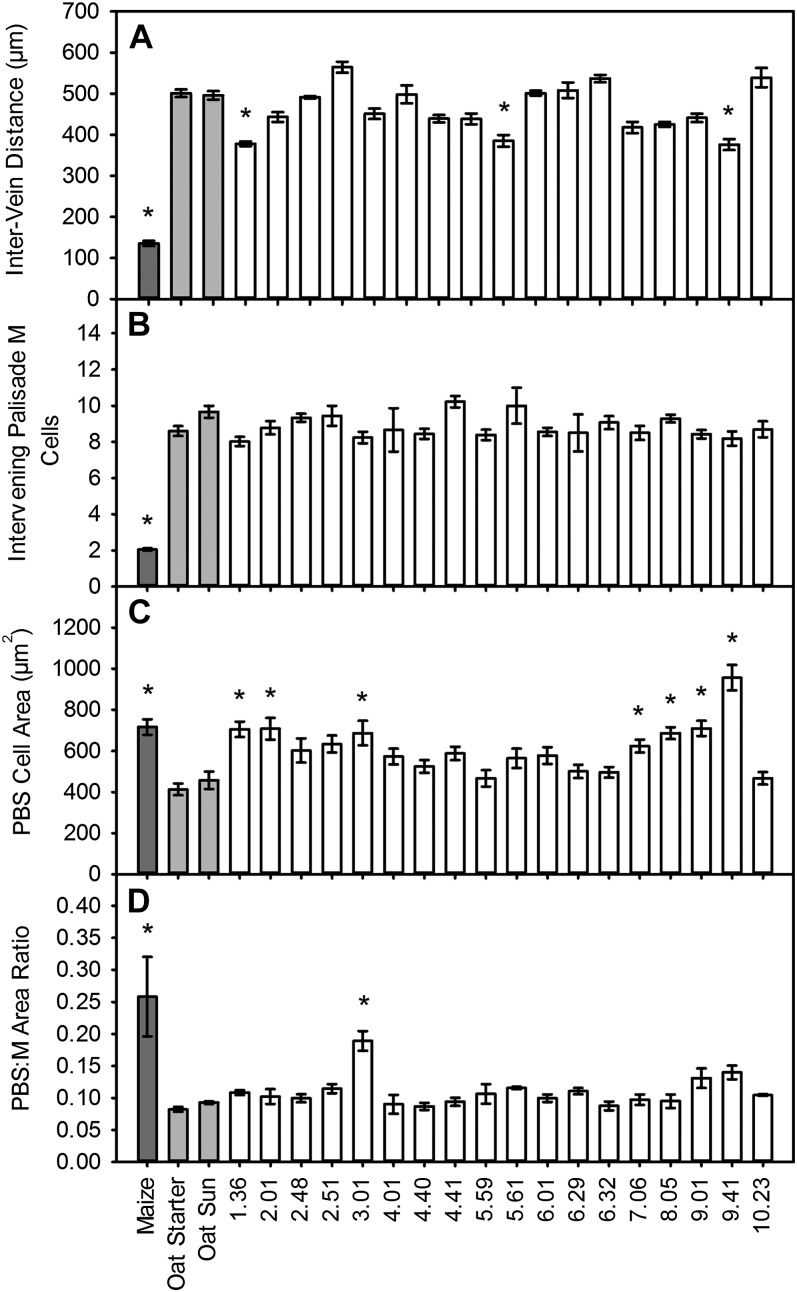
Little is known either about genes controlling the development of increased venation in C4 leaves or whether venation can be modified in C3 leaves. We used OMA lines generated from two oat parental backgrounds (sun II and starter) to investigate these two fundamental processes. Each maize chromosome was represented by at least one addition line, and in several cases an individual maize chromosome was represented by more than one line, with the lines sharing the same chromosome but differing in their parental backgrounds (Supplemental Table S1). In order to investigate whether the presence of individual maize chromosomes increased venation in oat, distances between the midpoints of adjacent veins were measured in transverse sections of fully expanded third leaves from OMA lines via light microscopy. While veins of maize were separated by approximately 130 µm, in both oat parental lines mean distances between adjacent veins were approximately 500 µm (Fig. 1A; Supplemental Fig. S1). Intervein distance (IVD) in OMA lines was variable, indicating that individual maize chromosomes in the oat background exerted an influence on vein spacing (Fig. 1A). In particular, the independent addition of maize chromosomes 1, 5, and 9 was associated with significantly reduced vein spacing compared with oat parents (Fig. 1A). Although none of the lines individually recapitulated maize-like values, additively the differences of OMA 1.36, OMA 5.59, and OMA 9.41 from wild-type oat would produce values similar to those of maize. To determine whether the proximity of adjacent veins in these three lines could be accounted for by a reduction in the number of intervening M cells, the number of palisade M cells separating adjacent vascular bundles was measured. Although the number of M cells between neighboring veins was variable in the OMA lines, none showed values that were significantly different from the oat parental lines (Fig. 1B; Supplemental Fig. S1).
Figure 1.
Alterations to leaf morphology in OMA lines. A and B, Distance between adjacent veins (A) and number of palisade M cells separating adjacent vascular bundles (B). C and D, PBS cell cross-sectional area (C) and PBS-M cross-sectional area ratio (D) in maize, oat, and OMA leaves. Results are expressed as means ± se of three biological replicates with a minimum of 10 measurements per leaf. Starred results are statistically significant relative to oat controls (P < 0.01).
Maize Chromosomes Can Increase PBS Cell Size in C3 Oat
C4 photosynthesis in maize is associated with large PBS cells that contain more chloroplasts than those of C3 leaves. Since very little is known about loci controlling PBS cell size, the cross-sectional area of the PBS cells in each of the OMA lines was measured and compared with oat and maize controls. Lines possessing maize chromosome 1, 2, 3, 7, or 8 and both lines with maize chromosome 9 had significantly larger PBS areas than either of the oat parental lines. This suggests that these chromosomal additions independently specified larger PBS cells (Fig. 1C; Supplemental Fig. S1).
In some lines, the increases in cell size observed in the OMA lines were not confined to the PBS, with M cells also larger compared with oat parental lines (Supplemental Fig. S2). In order to quantify the extent to which increases in cell size were specific to the PBS and not a consequence of constitutive cell expansion in the leaf, the area of PBS between the midpoints of two adjacent veins was expressed relative to the total intervening M area between the veins. Line 3.01 had a significantly larger PBS-M ratio than both of the oat parental lines, with a value 70% of the maize PBS-M ratio (Fig. 1D). This indicates that loci controlling PBS cell size are likely located on maize chromosome 3. However, the introduction of maize chromosome 3 was not sufficient to recapitulate other elements of maize PBS morphology, such as high chloroplast numbers and centrifugal chloroplast position, and in fact there was no evidence of increased chloroplast proliferation in any of the OMA lines (Supplemental Fig. S2).
Maize Chromosome 3 Introduction Causes Lipid Deposition in Oat PBS Cell Walls
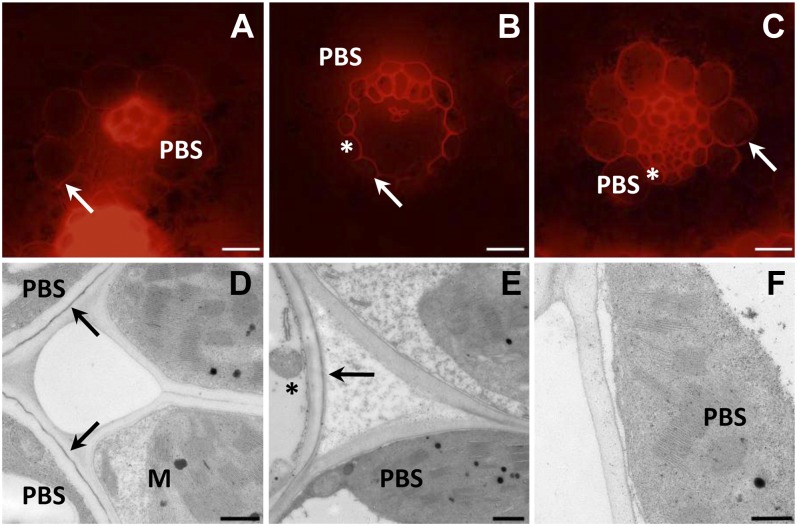
In order to determine the extent to which the enlarged PBS cells in OMA 3.01 resembled the maize PBS, transverse sections of leaves were stained with Nile red to detect fatty acid deposition. Nile red stains positively for lipids in plants (Fowler and Greenspan, 1985; Greenspan and Fowler, 1985; Greenspan et al., 1985; Pighin et al., 2004; Schmidt and Herman, 2008; Dietrich et al., 2009), fluorescing red in the presence of polar lipids and yellow when staining esterified cholesterol and triacylglycerols (Diaz et al., 2008). This approach showed polar lipids in the PBS of maize, but in the oat parental line, only the MS and some of the veinal cells stained (Fig. 2, A and B). In contrast, in OMA 3.01, the PBS stained positively when examined by fluorescence microscopy (Fig. 2C), indicating that PBS cells in line 3.01 have increased deposition of polar lipids in the PBS cell wall. Transmission electron microscopy (TEM) imaging of the leaves indicated the presence of suberin in the cell walls of the PBS in maize but only in the MS cell layer of OMA 3.01 and wild-type oat (Fig. 2, D–F). Taken together, these data indicate that chromosome 3 likely contains loci that can control PBS cell wall lipid deposition in oat but not the entire suberin deposition pathway.
Figure 2.
Maize chromosome 3 causes lipid deposition in oat PBS cell walls. Fluorescence (A–C) and TEM (D–F) images of transverse sections of maize (A and D), oat (sun II; B and E), and OMA 3.01 (C and F) leaves are shown. Lipid detection was carried out by staining sections with Nile red (A–C). Suberin detection was by TEM (D–F). Asterisks indicate MS. In TEM images, arrows indicate suberin layers. Bars = 20 μm (A–C) and 500 nm (D–F).
Individual Maize Chromosomes Have Little Impact on Characteristics of Leaf Photosynthesis
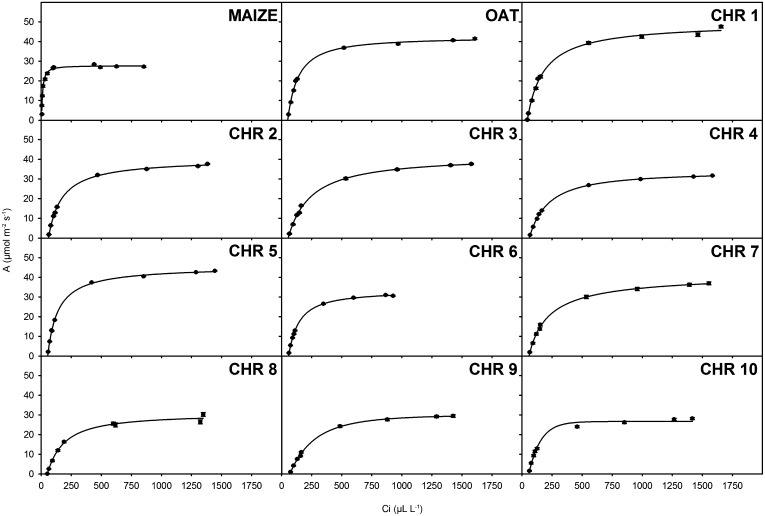
Previous work has shown that in OMA lines with maize chromosome 6 or 9, which contain ZmPPDK or ZmPEPC, respectively, there is little impact on CO2 compensation point (Kowles et al., 2008). However, it is possible that alterations to photosynthetic parameters could be induced in oat if a specific maize transcription factor or factors recognized and regulated a significant number of oat photosynthesis genes. In order to determine the extent to which photosynthetic parameters were influenced by the introduction of each maize chromosome, net photosynthesis of the OMAs, maize, and oat parental lines were measured over a range of external CO2 concentrations and photon flux densities (PFDs; Figs. 3 and 4). In C4 plants, photosynthesis responds more rapidly to CO2 supply than in C3 species, because PEPC displays a higher affinity for HCO3−. This leads to a steep initial slope (high carboxylation efficiency) and an intercept with the x axis close to zero (low CO2 compensation point). These characteristics were evident in maize compared with the oat parental lines starter and sun II (Fig. 3). Interestingly the two oat parental lines showed different maximum rates of photosynthesis at high CO2 concentration. When this is taken into account, CO2 responses in most OMAs were typical of C3 photosynthesis and for the most part unaffected by the presence of each maize chromosome, giving similar values to the oat parents (Fig. 3). However, three lines (6.32, 9.41, and 10.23) showed lower saturating levels of photosynthesis, and carboxylation was dramatically reduced at low internal CO2 values in line 9.41 (Fig. 3), indicating that the introduction of chromosome 9 had perturbed carbon assimilation. The introduction of maize chromosome 9 in line 9.01, however, did not show inhibited carbon assimilation.
Figure 3.
Photosynthetic assimilation rates are largely unchanged by maize chromosomal introduction. Light-saturated photosynthetic assimilation rates (A) over a range of leaf intercellular CO2 concentrations (Ci) for leaves of maize, oat, and OMA lines are shown. Each panel represents mean assimilation rates of maize and wild-type oat or chromosomal means of OMA lines. Results are expressed as means of at least three biological replicates ± se. Measurements were taken at 25°C, leaf vapor pressure deficit of less than 1.5 kPa, and PFD of 1,500 μmol m−2 s−1.
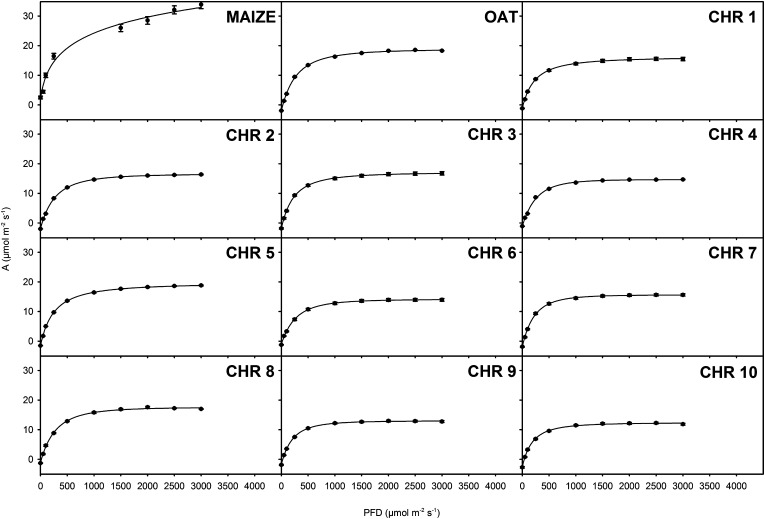
Figure 4.
Photosynthetic light responses are largely unchanged by maize chromosomal introduction. Photosynthetic assimilation rates (A) over a range of light intensities at ambient CO2 for leaves of maize, oat, and OMA lines are shown. Each panel represents mean assimilation rates of maize and wild-type oat or chromosomal means of OMA lines. Results are expressed as means of at least three biological replicates ± se. Measurements were taken at 25°C, leaf vapor pressure deficit of less than 1.5 kPa, and [CO2] of 380 µL L–1.
In C4 leaves, photosynthesis typically saturates at much higher light intensities compared with C3 leaves. In order to test whether any of the maize chromosomes resulted in C4-like light responses, photosynthetic rates of each line were measured over a range of light intensities. Whereas in maize, photosynthesis did not saturate below 3,000 μmol photons m−2 s−1, in all OMAs it saturated at values below 1,000 μmol photons m−2 s−1, not significantly different from those of oat parental lines (Fig. 4). Together, these data show that the light and CO2 responses in leaves from OMAs were typical of C3 plants.
Several Maize Transcripts Associated with C4 Photosynthesis Are Detectable in OMAs
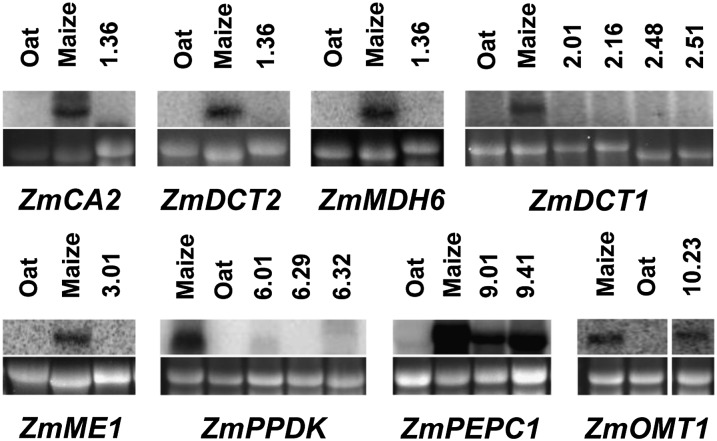
RNA blotting has shown that the maize genes encoding PPDK and PEPC are expressed in OMA lines 6 and 9, and the proteins that they encode were detectable by immunoblotting (Kowles et al., 2008). To investigate the extent to which other genes important for the biochemistry of C4 photosynthesis are expressed in maize, we undertook RNA blotting and then quantitative PCR for eight maize transcripts required for the core C4 cycle. This showed that in addition to ZmPPDK and ZmPEPC, the maize 2′-oxoglutarate/malate transporter (OMT1) was expressed in oat. So, three of the eight genes associated with the C4 biochemical pathway generated transcripts of the correct size in oat, indicating that these genes are expressed and their transcripts are correctly spliced (Fig. 5). To quantify the extent to which ZmOMT, ZmPPDK, and ZmPEPC were expressed in the oat background, mRNA levels were measured by real-time quantitative (RTq) PCR. For all genes, mRNA expression in OMA leaves was detectable but low compared with that in maize leaves (Supplemental Fig. S3), indicating that although transcripts of the correct size were generated, levels of expression in oat were low compared with those in maize.
Figure 5.
Detection of maize-derived transcripts associated with C4 photosynthesis in leaves of OMA plants. Total RNA was isolated from mature leaves of OMA plants and resolved on a 1.2% agarose-formaldehyde gel. RNA (10 µg per lane) was hybridized with a radiolabeled maize probe for the transcript of interest. RNA from maize, oat, and an OMA line with a different maize chromosome were probed as controls. rRNA (25S) from the gel used for blotting stained with ethidium bromide is shown as a loading control below each blot. Transcripts detected with maize-specific ZmCA2 (2.0 kb), ZmDCT2 (1.9 kb), ZmMDH6 (1.5 kb), ZmDCT1 (2.1 kb), ZmME1 (2.2 kb), ZmPPDK (3.5 kb), ZmPEPC1 (3.2 kb), and ZmOMT1 (1.9 kb) probes were of the expected sizes.
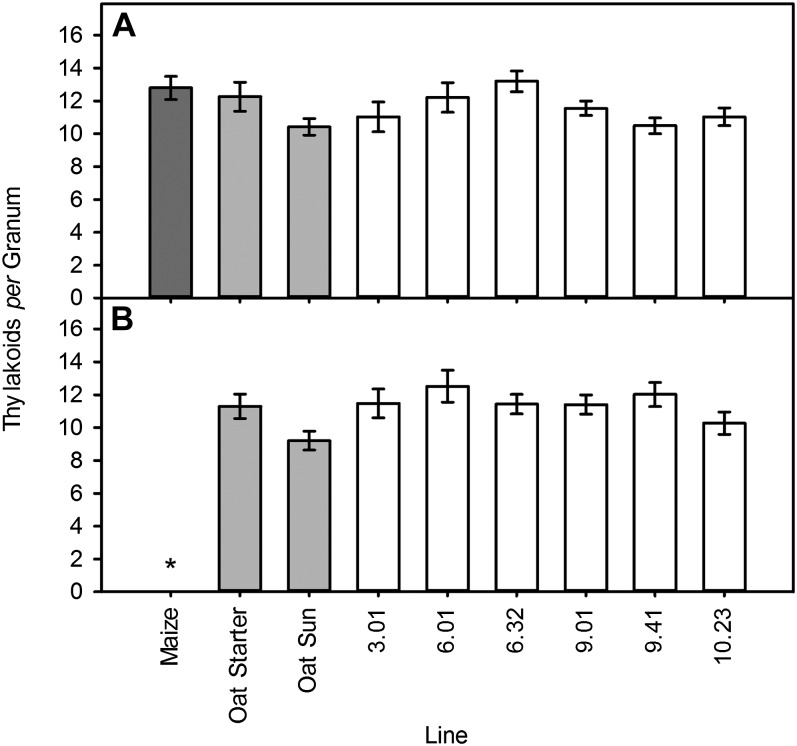
Thylakoid Stacking in Chloroplasts of Oat PBS Cells Is Unchanged by Individual Maize Chromosomes
C4 plants that belong to the NADP-ME subtype often show reduced granal stacking in PBS chloroplasts. In order to determine whether the OMA lines showing either an accumulation of C4 transcripts or changes to PBS cell development or carboxylation efficiency also possessed alterations to chloroplast ultrastructure, OMA leaves were examined using TEM. The number of thylakoid stacks per granum in M and PBS chloroplasts in lines 3.01, 6.01, 6.32, 9.01, 9.41, and 10.23 was quantified and compared with oat parental lines (starter and sun II) and maize (B73). Chloroplast morphology was normal in OMA lines (Supplemental Fig. S4). The level of thylakoid stacking in PBS cells was variable in the OMA lines but was not significantly different from that in oat parental lines (Fig. 6). This was also true of M cells, with similar numbers of thylakoids in the grana of both M and PBS cells of OMA lines, in contrast to the dimorphic chloroplast arrangement observed in maize (Fig. 6). This demonstrated that changes to chloroplast and PBS development are uncoupled in line 3.01 and that the reduction in carboxylation efficiency in line 9.41 was unlikely unrelated to granal structure (Fig. 6). Maize also has higher numbers of plasmodesmata between M and PBS cells; however, examination lines of 3.01, 6.01, 6.32, 9.01, 9.41, and 10.23 using TEM showed no evidence of this (Supplemental Fig. S4).
Figure 6.
Thylakoid stacking in grana of oat PBS cells is unchanged by maize chromosomal introduction. Numbers of thylakoid stacks per granum in M (A) and bundle sheath chloroplasts (B) in maize, oat, and OMA leaves are shown. Results are expressed as means ± se of three biological replicates with a minimum of 10 measurements per leaf. Starred results are statistically significant relative to oat controls (P < 0.01).
DISCUSSION
Expression of Maize Genes Important for C4 Photosynthesis in C3 Oat
The extent to which genes from C4 species are recognized and expressed in C3 species is fundamental to our understanding of gene regulation and its conservation between species. It is also important to establish which maize genes are expressed in C3 leaves to inform attempts being made to introduce characteristics of the C4 pathway into crops such as rice (Matsuoka et al., 2001; Hibberd et al., 2008; Kajala et al., 2011). Our data extend those of Kowles et al. (2008) to show that, in addition to the maize PPDK and PEPC genes, OMT1 is expressed. For all three genes, the size of the mRNA indicates correct splicing in oat. In the case of PEPC and PPDK, this is consistent with previous work that indicated that full-length maize PEPC and PPDK genes driven by their native promoters are expressed and generate transcripts of the correct size in transgenic rice leaves (Ku et al., 1999; Fukayama et al., 2001; Taniguchi et al., 2008). The most parsimonious explanation is that the same transacting factors recognize maize PEPC and PPDK in these two C3 grasses. However, it should be noted that transcript levels of PPDK, PEPC, and OMT were lower in each OMA line than in maize and, in the case of PEPC, lower than the endogenous oat PEPC. This suggests either that enhancer elements associated with the maize gene were not recognized in oat or that transcript stability was reduced. Consistent with our findings, Kowles et al. (2008) showed that the activity of PEPC was higher in OMAs than in wild-type oat but significantly lower than that in maize.
Transcripts from a number of genes, such as CARBONIC ANHYDRASE2 (CA2), DICARBOXYLIC TRANSPORTER1 (DCT1) and DCT2, MALATE DEHYRODENASE6 (MDH6), and ME1, important for C4 photosynthesis in maize were not detectable by RNA blotting in the OMA lines. For example, we were not able to detect significant expression of the maize NADP-ME gene. This contrasts with work on the maize NADP-ME rice, in which the promoter of the maize NADP-ME gene was able to generate expression of the uidA reporter in rice (Nomura et al., 2005). The difference between the behavior of the maize NADP-ME gene in these C3 crops is interesting and could be due to differences in transacting factors or, alternatively, to large-scale silencing of maize genes in the OMA lines. If large-scale silencing of gene expression occurs in these OMA lines, it may well be possible for these five maize genes important for the C4 cycle to be expressed successfully in oat.
Consistent with the lack of expression of many of the C4 genes in oat, including those encoding transporter proteins important for flux across organelle membranes (Weber and von Caemmerer, 2010), we did not detect substantial alterations to the characteristics of gas exchange of the OMA leaves compared with controls. We did detect reductions in carboxylation efficiency, maximal photosynthetic rate, and stature in OMA 9.41 (Supplemental Figs. S5 and S6), indicating a disruption to photosynthesis. However, chloroplast development in this line was normal in both M and PBS cells. Our finding that photosynthesis in the line containing chromosome 6 on which PPDK resides responded to both light and CO2 supply in a similar way to controls is consistent with the fact that the expression of maize PPDK had no effect on net assimilation (Fukayama et al., 2001). It has been reported that expression of maize PEPC causes a slight reduction in net assimilation and stunting in transgenic rice plants (Taniguchi et al., 2008). Retarded growth that correlated with PEPC activity was also reported for potato (Solanum tuberosum; C3) plants overexpressing a modified C4 PEPC from Flaveria trinervia, and metabolite analysis of these plants revealed elevated levels of malate and total amino acids (Rademacher et al., 2002). Increases in malate and nitrogen content were also observed in maize PEPC-overexpressing rice plants (Agarie et al., 2002). All OMA lines containing chromosome 9 were stunted; however, we detected no consistent reduction in the rate of photosynthesis, and this may be due to the relatively low accumulation of maize PEPC in these plants.
The Plasticity of Oat Leaf Anatomy
Assessing the impact of individual maize chromosomes on oat allowed us to investigate the plasticity of oat leaves, specifically whether elements of Kranz anatomy can be introduced into them. None of the maize chromosomal introductions showed wholesale changes to leaf anatomy consistent with C4 photosynthesis, but three lines showed reductions in vein spacing, a key trait associated with C4 photosynthesis. In these three lines, IVDs were not as low as those in maize, nor did the cellular arrangement generate the repeated units of M-PBS-V-PBS-M cells typical of maize leaves. However, these data show that vein spacing can be altered in a C3 leaf. Interestingly, only one of two lines with maize chromosome 9 (OMA 9.41) showed reduced IVD compared with the wild type. These data show that maize chromosome 9 is capable of decreasing IVD in oat, but this appears to be dependent on the genomic context. Although maize chromosome 9 is common to both oat recipient lines, they differ in their parental backgrounds (Supplemental Table S1), so it is possible that the loci on maize chromosome 9 encoding this trait are differentially silenced depending on their interaction with the oat genome or that they interact differently with specific oat alleles. Interestingly, two of the lines showing reduced vein spacing are derived from crosses involving oat sun II as the recipient, implying that the genetic background of sun II may be more receptive to alterations in vein spacing. The difficulty of generating stable double and triple OMA lines (Kynast et al., 2001) means that testing the additive effects of chromosomes 1, 5, and 9 is not trivial.
To date, mutant screens have failed to identify genes that control the establishment of Kranz anatomy in leaves of C4 plants. Maize mutants with differential effects on PBS and M cells are mostly defective in the accumulation of photosynthesis proteins rather than Kranz anatomy per se (Langdale and Kidner, 1994; Smith et al., 1998), and mutants such as tangled1 with ectopic PBS cells are associated with pleiotropic effects (Smith et al., 2001) and, therefore, are unlikely to be informative in dissecting the molecular basis of Kranz anatomy. Our results indicate that there are loci that are able to regulate PBS size and cell wall lipid deposition in oat that are located on maize chromosome 3. PBS chloroplast number and development were unchanged in this line, suggesting that these traits are not coupled to cell size, and M cells and chloroplasts develop normally in line 3.01, indicating that M development is independent from PBS development. This is consistent with the lack of mutants identified to date with disruptions in M cell development. The transcription factor Golden2, which controls chloroplast development in maize PBS cells (Langdale and Kidner, 1994; Hall et al., 1998), resides on maize chromosome 3; however, the absence of changes to chloroplast morphology in either M or PBS cells in OMA 3.01 suggests that it is not active in oat. The argentia mutant impacts on the speed with which patterns of the C4 mRNAs develop (Langdale et al., 1987), so our analysis of mature OMA leaves is unlikely to have detected phenotypes relevant to determining whether the wild-type ARGENTIA gene product on maize chromosome 9 is active in oat.
Maize PBS cells are also characterized by a suberized cell wall (O’Brien and Carr, 1970); however, suberin was not detected in the OMA 3.01 PBS cell wall, suggesting that the complete maize suberin deposition pathway was not active. Many of the maize homologs of Arabidopsis genes required for suberin synthesis (Li et al., 2010) are located on other maize chromosomes (Supplemental Table S2), supporting this hypothesis. Interestingly, however, ZmLP1 (for lipid transfer protein 1) is located on chromosome 3 and, therefore, is an interesting candidate for the deposition of lipids in the PBS wall observed in OMA 3.01. Expression of ZmLP1 in maize leaves is high and confined to the developing transition zone (Li et al., 2010), supporting a role in laying down lipids in the walls of developing PBS cells. Similarly, SND2, a transcription factor known to be required for secondary cell wall synthesis in Arabidopsis (Zhong et al., 2008; McCarthy et al., 2009), is also expressed in the developing region of the maize leaf (Li et al., 2010) and is located on maize chromosome 3. In order to obtain a more complete picture of the genes involved in lipid deposition in OMA 3.01, analysis of PBS cell phenotypes in OMA radiation hybrid lines (Kynast et al., 2004) containing fragments of maize chromosome 3 in an oat background would permit finer mapping of this trait. Similarly, a comparison of leaf transcriptomes in maize and OMA 3.01 would reveal candidate genes from maize chromosome 3 for controlling lipid deposition, cell wall synthesis, and expansion.
In summary, we have shown that the introduction of maize chromosomes into an oat background can increase PBS cell size and reduce vein spacing in the C3 oat leaf, two key traits of C4 photosynthesis. In addition, some maize genes involved in the core C4 pathway produce transcripts of the correct size in oat. The polyphyletic distribution of the pathway (Sage et al., 2012) combined with evidence of substantial differences in gene expression and spatial control required for C3-C4 transitions (Bräutigam et al., 2011) have led to the proposal that the expression of C4 traits may be governed by a “master switch” (Westhoff and Gowik, 2010). Because we found that there was no coordinated change to gene expression, cell biology, and leaf anatomy, our results provide no evidence for the existence of a global C4 activator. However, a comprehensive assessment of the transcriptional activity of each maize chromosome in the oat background is needed before this can be confirmed. If a two-celled C4 pathway is to be introduced to C3 crop species, changes to leaf anatomy and gene expression will be necessary. Importantly, this study demonstrates that mechanisms underlying the expression of some key maize genes are conserved in oat and that the anatomy of the monocot C3 leaf can be modified.
MATERIALS AND METHODS
Plant Material
Oat (Avena sativa; starter and sun II) and OMA line seeds were dehulled and then germinated on saturated filter paper with the application of 200 µL of 75 mg L−1 solution of GA3 in tap water for 2 to 3 d at room temperature before placing them in 3:1 Levington M3 potting compost (Scotts Miracle-Gro):vermiculite treated with intercept (200 mg L−1; Scotts Miracle-Gro). Plants were grown in day/night temperatures of 20°C/13°C at relative humidity of 65% and under a photosynthetic PFD of 500 μmol photons m−2 s−1 for a 16-h light period. Leaves were defined as fully expanded on ligule emergence (Greenland and Lewis, 1981).
Maize (Zea mays) B73 was grown from seeds imbibed overnight in tap water before planting in soil potting mix and grown at 28°C and 50% relative humidity under a photosynthetic PFD of 500 photons μmol m−2 s−1 for a 16-h light period. Samples for DNA or RNA extraction were taken from fully expanded third leaves flash frozen in liquid nitrogen and stored at −80°C.
Microscopy
To quantify anatomical arrangements, leaf material was harvested from the widest points (8–10 cm from the leaf base) of fully expanded third leaves and prepared for light microscopy and TEM (Sage and Williams, 1995). Light microscope images of transverse sections were used to quantify the distance between adjacent veins, the number of palisade M cells between veins, the cross-sectional areas of M and PBS tissue, and the ratio of PBS to M tissue (Supplemental Fig. S7). Morphometric analyses were performed using the software packages QCapture Pro (QImaging) and ImageJ (National Institutes of Health). Three TEM images of PBS and M cells from three representative individuals for each line were used to examine the chloroplast number and quantify the number of thylakoid stacks per granum in M and PBS cells. All anatomical measurements were performed in biological triplicate and subjected to statistical analysis by two-tailed Student’s t test to determine their significance relative to oat parental lines (P < 0.01). Nile red (0.01% in 85% lactic acid saturated with chloral hydrate) was used to determine the presence of lipids using epifluorescence (Greenspan et al., 1985; Waduwara et al., 2008).
Gas-Exchange Measurements
The net assimilation rates of fully expanded leaves of oat (starter and sun II), maize (B73), and 20 OMA lines under varying internal leaf [CO2] and incident PFD were analyzed in order to screen for characteristic C4 physiological features. Net assimilation rates were measured over a range of external [CO2] using a LI-6400 portable open gas-exchange system (Li-Cor Biosciences) with a photosynthetic leaf cuvette. Leaf temperature was constant at 25°C, and PFD (provided by a Walz FL440 Fiber Illuminator with a 400-F fiberglass connection and heat-filtering projector) was set at 1,500 μmol m−2 s−1. Leaves were allowed to acclimatize to saturating light (1,500 µmol m–2 s–1) at ambient CO2 for 30 min. Air flow through the photosynthetic leaf cuvette was varied between 200 and 500 μmol s−1 to ensure that the leaf vapor pressure deficit remained less than 1.5 kPa. After reaching steady state, photosynthetic assimilation rate (A) as a function of internal CO2 concentration (Ci) was measured by varying external [CO2]. CO2 compensation points and carboxylation efficiencies were calculated by regression analysis of A/Ci (in the linear range), with CO2 compensation point equivalent to the x intercept and carboxylation efficiency corresponding to the initial slope gradient. Photosynthetic responses to a range of external light levels at ambient [CO2] were also measured. Leaf temperature and [CO2] were constant at 25°C and 380 µL L–1, respectively, and leaf vapor pressure deficit was maintained below 1.5 kPa. Leaves were equilibrated at 3,000 μmol photons m−2 s−1 prior to curve initiation (Kubiske and Pregitzer, 1996). CO2 and light response measurements are displayed as means of three biological replicates with 1 se.
RNA Extractions
Total RNA was extracted from leaves of 3-week-old oat, maize, and OMA plants using TriPure Isolation Reagent (Roche; Chomczynski and Sacchi, 1987). Approximately 200 mg of leaf tissue was ground in liquid nitrogen using a mortar and pestle. Ground tissue was subsequently transferred to an Eppendorf tube containing 1 mL of Tripure Isolation Reagent and mixed by inversion. Chloroform at 200 µL was added after 5 min of incubation at room temperature, and the sample was vortexed (15 s) and centrifuged at 13,000 rpm at 4°C (15 min). The aqueous phase (approximately 600 µL) was transferred to a clean Eppendorf tube containing 500 µL of isopropanol and centrifuged at 13,000 rpm at 4°C for 10 min. The supernatant was discarded, and the pellet was washed with 1 mL of 75% (v/v) ethanol and centrifuged at 7,500 rpm at 4°C for 5 min. Once again, the supernatant was removed, and the RNA pellet was resuspended in 40 µL of RNAsecure (Ambion) preheated to 60°C. To aid RNA resuspension, the sample was incubated at 60°C for 10 min, pulse centrifuged, and transferred to a clean Eppendorf tube for subsequent storage at −80°C.
The quantity of RNA was determined spectrophotometrically, and the quality of RNA was validated via agarose gel electrophoresis. RNA (1 µg) and 5 µL of RNA loading buffer (50% [v/v] glycerol, 1 mm EDTA, 0.4% [w/v] bromphenol blue, 0.005× Tris-borate-EDTA buffer, and 48% [v/v] formamide) were heated to 60°C for 10 min and subsequently run on a 1.5% (w/v) agarose gel containing 0.5 µg mL−1 ethidium bromide using 0.5× Tris-borate-EDTA buffer (44.5 mm Tris, 44.5 mm boric acid, and 1 mm EDTA [pH 8.0]).
RNA Blotting
Total RNA was separated on the basis of molecular weight via gel electrophoresis on a 1.2% formaldehyde agarose gel and transferred onto a nitrocellulose membrane (Genescreen Plus; Perkin-Elmer) by capillary blotting as described previously (Brown et al., 2005). Maize-specific oligonucleotide probes were amplified by PCR from maize cDNA made using an oligo(dT) primer (Invitrogen Life Technologies) and SuperScript II reverse transcriptase (Promega). Primer sequences are recorded in Supplemental Table S3. Radiolabeled probes were prepared by random priming reactions in the presence of [α-32P]dATP using T4 polynucleotide kinase (New England Biolabs) after Feinberg and Vogelstein (1983). All probes were used in 10 µm working concentrations. Prehybridization was conducted at 37°C in 3 mL of prehybridization buffer (Ambion). Removal of unincorporated nucleotides and verification of α-32P incorporation into probe 5′ PO4 was carried out by centrifugation through a Microspin S-200 HR column (GE Healthcare) prior to their addition to the prehybridization solution. [α-32P]dATP cDNA probes were added to the solution, and hybridization was carried out overnight at 42°C. Membrane washes to remove any unbound probe and blot hybridization analysis were carried out after Brown et al. (2005). Blots were analyzed using a Typhoon 8600 Variable Mode Imager (GE Healthcare).
RTq-PCR
Total RNA for RTq-PCR was extracted from maize, oat, and OMA leaves using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. To remove contaminating genomic DNA, samples were treated with 10 units μL−1 RNase-free DNase (Qiagen) for 30 min at 20°C and for 15 min at 65°C. RNA quality was determined spectrophotometrically. One microgram of RNA was reverse transcribed using oligo(dT) primer and SuperScript II (Invitrogen Life Technologies). The total cDNA volume of 20 μL was stored at −20°C overnight. RTq-PCR was carried out using SYBR Green JumpStart Taq Ready Mix (Sigma-Aldrich) and 5-fold dilution of the template and primers at 0.2 µm final concentration. Primers were designed using Primer 3 software (http://frodo.wi.mit.edu/primer3) to have melting temperatures of 60°C. Sequences of primers used to detect ZmPEPC, ZmOMT, AsPEPC, AsACTIN, and MAIZE ACTIN95 (ZmMAZ95; Lin et al., 2008) are listed in Supplemental Table S4. In the absence of oat actin sequence information, degenerate primers (Supplemental Table S4) were used to amplify a 427-bp fragment corresponding to AsACTIN from oat starter cDNA into a PJet 1.2 plasmid vector (Fermentas). This fragment was sequenced and verified by BLAST search against the EST database available at the National Center for Biotechnology Information, and RTq-PCR primers were subsequently designed to this fragment. RTq-PCR was performed in a Rotor-Gene thermal cycler (Qiagen). Cycling conditions were as follows: 94°C for 2 min, followed by 40 cycles of 94°C for 20 s, 60°C for 30 s, 72°C for 30 s, and 75°C for 5 s. The fluorescence threshold was set to a constant value of 0.04, which was manually determined to be as early as possible into the exponential phase of fluorescence for all transcripts. Cycle threshold values were calculated from means of three technical replicates for three independent biological replicates of each line. The relative abundance of transcripts (to AsACTIN or ZmMAZ95) was calculated using the 2−ΔΔCT method after Livak and Schmittgen (2001). se values were calculated from 2−ΔΔCT values of each combination of (biological and technical) replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf anatomical characteristics in the three individual lines of maize used.
Supplemental Figure S2. Leaf morphology in OMA lines.
Supplemental Figure S3. mRNA transcripts of ZmPEPC and ZmOMT are expressed at lower levels in OMA leaves than in maize.
Supplemental Figure S4. Chloroplast morphology in OMA lines.
Supplemental Figure S5. Phenotypes resulting from maize chromosomal introduction to the oat genome in 2-week-old OMA plants.
Supplemental Figure S6. Phenotypes resulting from maize chromosomal introduction to the oat genome in 4-week-old OMA plants.
Supplemental Figure S7. Schematic of vein, MS, PBS, and M cells in oat, and illustration of these cells in transverse sections.
Supplemental Table S1. Plant material used for analysis.
Supplemental Table S2. Chromosomal location of putative maize suberin pathway genes.
Supplemental Table S3. Maize-specific oligonucleotide probe primer sequences.
Supplemental Table S4. RTq-CR primer sequences.
Supplementary Material
Acknowledgments
We thank Prof. Ron Phillips and Howard Rines (University of Minnesota) for the generous gift of OMA seed. Thanks are due to Samart Wanchana (International Rice Research Institute) for providing maize sequence information. Thanks are also due to Susan Stanley (University of Cambridge) and Julie Bull (University of Oxford) for practical help.
Glossary
- OMA
oat × maize addition
- MS
mestome sheath
- M
mesophyll
- PBS
parenchymal bundle sheath
- IVD
intervein distance
- TEM
transmission electron microscopy
- PFD
photon flux density
- RTq
real-time quantitative
References
- Agarie S, Miura A, Sumikura R, Tsukamoto S, Nose A, Arima S, Matsuoka M, Miyao-Tokutomi M. (2002) Overexpression of C4 PEPC caused O2-insensitive photosynthesis in transgenic rice plants. Plant Sci 162: 257–265 [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, Sommer M, Gagneul D, Weber KL, Carr KM, Gowik U, Mass J, Lercher MJ, et al. (2011) An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol 155: 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NJ, Sullivan JA, Gray JC. (2005) Light and plastid signals regulate the expression of the pea plastocyanin gene through a common region at the 5′ end of the coding region. Plant J 43: 541–552 [DOI] [PubMed] [Google Scholar]
- Brown WV. (1958) Leaf anatomy in grass systematics. Bot Gaz 119: 170–178 [Google Scholar]
- Brutnell TP, Sawers RJH, Mant A, Langdale JA. (1999) BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Lara MV, Andreo CS. (2000) Induction of a C4-like mechanism of CO2 fixation in Egeria densa, a submersed aquatic species. Plant Physiol 123: 1611–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Dengler NG, Nelson T. (1999) Leaf structure and development in C4 plants. In RF Sage, RK Monson, eds, C4 Plant Biology. Academic Press, San Diego, pp 133–172
- Diaz G, Melis M, Batetta B, Angius F, Falchi AM. (2008) Hydrophobic characterization of intracellular lipids in situ by Nile red red/yellow emission ratio. Micron 39: 819–824 [DOI] [PubMed] [Google Scholar]
- Dietrich D, Schmuths H, De Marcos Lousa C, Baldwin JM, Baldwin SA, Baker A, Theodoulou FL, Holdsworth MJ. (2009) Mutations in the Arabidopsis peroxisomal ABC transporter COMATOSE allow differentiation between multiple functions in planta: insights from an allelic series. Mol Biol Cell 20: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards GE, Voznesenskaya EV. (2011) C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants. In AS Raghavendra, RF Sage, eds, C4 Photosynthesis and Related CO2 Concentrating Mechanisms. Springer, Dordrecht, The Netherlands, pp 29–61
- Ehleringer JR, Monson RK. (1993) Evolutionary and ecological aspects of photosynthetic pathway variation. Annu Rev Ecol Syst 24: 411–439 [Google Scholar]
- Feinberg AP, Vogelstein B. (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Fowler SD, Greenspan P. (1985) Application of Nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem 33: 833–836 [DOI] [PubMed] [Google Scholar]
- Fukayama H, Tsuchida H, Agarie S, Nomura M, Onodera H, Ono K, Lee BH, Hirose S, Toki S, Ku MS, et al. (2001) Significant accumulation of C4-specific pyruvate, orthophosphate dikinase in a C3 plant, rice. Plant Physiol 127: 1136–1146 [PMC free article] [PubMed] [Google Scholar]
- Furbank RT. (2011) Evolution of the C(4) photosynthetic mechanism: are there really three C(4) acid decarboxylation types? J Exp Bot 62: 3103–3108 [DOI] [PubMed] [Google Scholar]
- Furumoto T, Hata S, Izui K. (1999) cDNA cloning and characterization of maize phosphoenolpyruvate carboxykinase, a bundle sheath cell-specific enzyme. Plant Mol Biol 41: 301–311 [DOI] [PubMed] [Google Scholar]
- Greenland AJ, Lewis DH. (1981) The acid invertases of the developing third leaf of oat. I. Changes in activity of invertase and concentrations of ethanol-soluble carbohydrates. New Phytol 88: 265–277 [Google Scholar]
- Greenspan P, Fowler SD. (1985) Spectrofluorometric studies of the lipid probe, Nile red. J Lipid Res 26: 781–789 [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD. (1985) Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LN, Rossini L, Cribb L, Langdale JA. (1998) GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD. (1987) C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 895: 81–106 [Google Scholar]
- Hatch MD, Kagawa T, Craig S. (1975) Subdivision of C4-pathway species based on differing C4 decarboxylating systems and ultrastructural features. Aust J Plant Physiol 2: 111–128 [Google Scholar]
- Hibberd JM, Sheehy JE, Langdale JA. (2008) Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr Opin Plant Biol 11: 228–231 [DOI] [PubMed] [Google Scholar]
- Kajala K, Covshoff S, Karki S, Woodfield H, Tolley BJ, Dionora MJ, Mogul RT, Mabilangan AE, Danila FR, Hibberd JM, et al. (2011) Strategies for engineering a two-celled C(4) photosynthetic pathway into rice. J Exp Bot 62: 3001–3010 [DOI] [PubMed] [Google Scholar]
- Kowles RV, Minnerath JM, Walch MD, Bernacchi C, Stec AO, Rines HW, Phillips RL. (2008) Expression of C4 photosynthetic enzymes in oat-maize chromosome addition lines. Maydica 53: 69–78 [Google Scholar]
- Ku MS, Agarie S, Nomura M, Fukayama H, Tsuchida H, Ono K, Hirose S, Toki S, Miyao M, Matsuoka M. (1999) High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nat Biotechnol 17: 76–80 [DOI] [PubMed] [Google Scholar]
- Kubiske ME, Pregitzer KS. (1996) Effects of elevated CO(2) and light availability on the photosynthetic light response of trees of contrasting shade tolerance. Tree Physiol 16: 351–358 [DOI] [PubMed] [Google Scholar]
- Kynast RG, Okagaki RJ, Galatowitsch MW, Granath SR, Jacobs MS, Stec AO, Rines HW, Phillips RL. (2004) Dissecting the maize genome by using chromosome addition and radiation hybrid lines. Proc Natl Acad Sci USA 101: 9921–9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira SB, Chen G, Ananiev EV, Odland WE, Russell CD, Stec AO, et al. (2001) A complete set of maize individual chromosome additions to the oat genome. Plant Physiol 125: 1216–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Kidner CA. (1994) bundle sheath defective, a mutation that disrupts cellular differentiation in maize leaves. Development 120: 673–681 [Google Scholar]
- Langdale JA, Metzler MC, Nelson T. (1987) The argentia mutation delays normal development of photosynthetic cell-types in Zea mays. Dev Biol 122: 243–255 [DOI] [PubMed] [Google Scholar]
- Langdale JA, Nelson T. (1991) Spatial regulation of photosynthetic development in C4 plants. Trends Genet 7: 191–196 [DOI] [PubMed] [Google Scholar]
- Leegood RC. (2002) C(4) photosynthesis: principles of CO(2) concentration and prospects for its introduction into C(3) plants. J Exp Bot 53: 581–590 [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Lin C, Shen B, Xu Z, Köllner TG, Degenhardt J, Dooner HK. (2008) Characterization of the monoterpene synthase gene tps26, the ortholog of a gene induced by insect herbivory in maize. Plant Physiol 146: 940–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Furbank RT, Fukayama H, Miyao M. (2001) Molecular engineering of C4 photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 52: 297–314 [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH. (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50: 1950–1964 [DOI] [PubMed] [Google Scholar]
- Nomura M, Higuchi T, Ishida Y, Ohta S, Komari T, Imaizumi N, Miyao-Tokutomi M, Matsuoka M, Tajima S. (2005) Differential expression pattern of C4 bundle sheath expression genes in rice, a C3 plant. Plant Cell Physiol 46: 754–761 [DOI] [PubMed] [Google Scholar]
- O’Brien TP, Carr DJ. (1970) A suberized layer in the cell walls of the bundle sheath of grasses. Aust J Biol Sci 23: 275–287 [Google Scholar]
- Pighin JA, Zheng HQ, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Rademacher T, Häusler RE, Hirsch HJ, Zhang L, Lipka V, Weier D, Kreuzaler F, Peterhänsel C. (2002) An engineered phosphoenolpyruvate carboxylase redirects carbon and nitrogen flow in transgenic potato plants. Plant J 32: 25–39 [DOI] [PubMed] [Google Scholar]
- Reinfelder JR, Kraepiel AML, Morel FMM. (2000) Unicellular C4 photosynthesis in a marine diatom. Nature 407: 996–999 [DOI] [PubMed] [Google Scholar]
- Reiskind JB, Madsen TV, Van Ginkel LC, Bowes G. (1997) Evidence that inducible C4-type photosynthesis is a chloroplastic CO2-concentrating mechanism in Hydrilla, a submersed monocot. Plant Cell Environ 20: 211–220 [Google Scholar]
- Riera-Lizarazu O, Rines HW, Phillips RL. (1996) Cytological and molecular characterization of oat x maize partial hybrids. Theor Appl Genet 93: 123–135 [DOI] [PubMed] [Google Scholar]
- Riera-Lizarazu O, Vales MI, Ananiev EV, Rines HW, Phillips RL. (2000) Production and characterization of maize chromosome 9 radiation hybrids derived from an oat-maize addition line. Genetics 156: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF, Li M, Monson RK. (1999). The taxonomic distribution of C4 photosynthesis. In RF Sage, RK Monson, eds, C4 Plant Biology. Academic Press, San Diego, pp 551–585
- Sage RF, Sage TL, Kocacinar F. (2012) Photorespiration and the evolution of C(4) photosynthesis. Annu Rev Plant Biol 63: 19–47 [DOI] [PubMed] [Google Scholar]
- Sage RF, Zhu XG. (2011) Exploiting the engine of C(4) photosynthesis. J Exp Bot 62: 2989–3000 [DOI] [PubMed] [Google Scholar]
- Sage TL, Williams EG. (1995) Structure, ultrastructure, and histochemistry of the pollen-tube pathway in the milkweed Asclepias exaltata L. Sex Plant Reprod 8: 257–265 [Google Scholar]
- Schmidt MA, Herman EM. (2008) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Smith LG, Gerttula SM, Han SK, Levy J. (2001) Tangled1: a microtubule binding protein required for the spatial control of cytokinesis in maize. J Cell Biol 152: 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LH, Langdale JA, Chollet R. (1998) A functional Calvin cycle is not indispensable for the light activation of C4 phosphoenolpyruvate carboxylase kinase and its target enzyme in the maize mutant bundle sheath defective2-mutable1. Plant Physiol 118: 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Ohkawa H, Masumoto C, Fukuda T, Tamai T, Lee K, Sudoh S, Tsuchida H, Sasaki H, Fukayama H, et al. (2008) Overproduction of C4 photosynthetic enzymes in transgenic rice plants: an approach to introduce the C4-like photosynthetic pathway into rice. J Exp Bot 59: 1799–1809 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Edwards GE, Kiirats O, Artyusheva EG, Franceschi VR. (2003) Development of biochemical specialization and organelle partitioning in the single-cell C4 system in leaves of Borszczowia aralocaspica (Chenopodiaceae). Am J Bot 90: 1669–1680 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya EV, Franceschi VR, Kiirats O, Artyusheva EG, Freitag H, Edwards GE. (2002) Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae). Plant J 31: 649–662 [DOI] [PubMed] [Google Scholar]
- Waduwara CI, Walcott SE, Peterson CA. (2008) Suberin lamellae of the onion root endodermis: their pattern of development and continuity. Can J Bot 86: 623–632 [Google Scholar]
- Walker RP, Acheson RM, Tecsi LI, Leegood RC. (1997) Phosphoenolpyruvate carboxykinase in C4 plants: its role and regulation. Aust J Plant Physiol 24: 459–468 [Google Scholar]
- Weber APM, von Caemmerer S. (2010) Plastid transport and metabolism of C3 and C4 plants: comparative analysis and possible biotechnological exploitation. Curr Opin Plant Biol 13: 257–265 [DOI] [PubMed] [Google Scholar]
- Westhoff P, Gowik U. (2010) Evolution of C4 photosynthesis: looking for the master switch. Plant Physiol 154: 598–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Walker RP, Chen Z-H, Leegood RC. (1999) Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol 120: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.