Abstract
Nitrate reallocation to plant roots occurs frequently under adverse conditions and was recently characterized to be actively regulated by Nitrate Transporter1.8 (NRT1.8) in Arabidopsis (Arabidopsis thaliana) and implicated as a common response to stresses. However, the underlying mechanisms remain largely to be determined. In this study, characterization of NRT1.5, a xylem nitrate-loading transporter, showed that the mRNA level of NRT1.5 is down-regulated by salt, drought, and cadmium treatments. Functional disruption of NRT1.5 enhanced tolerance to salt, drought, and cadmium stresses. Further analyses showed that nitrate, as well as Na+ and Cd2+ levels, were significantly increased in nrt1.5 roots. Important genes including Na+/H+ exchanger1, Salt overly sensitive1, Pyrroline-5-carboxylate synthase1, Responsive to desiccation29A, Phytochelatin synthase1, and NRT1.8 in stress response pathways are steadily up-regulated in nrt1.5 mutant plants. Interestingly, altered accumulation of metabolites, including proline and malondialdehyde, was also observed in nrt1.5 plants. These data suggest that NRT1.5 is involved in nitrate allocation to roots and the consequent tolerance to several stresses, in a mechanism probably shared with NRT1.8.
Nitrate is the most important nitrogen source for terrestrial plants. Nitrate concentrations in soil vary widely; thus, plants have evolved both a high-affinity transport system and a low-affinity transport system (Crawford, 1995), which correspond to NRT2 and NRT1 transporter families that function at low and high external nitrate concentrations, respectively (Daniel-Vedele et al., 1998; Stitt, 1999; Tsay et al., 2007). In Arabidopsis (Arabidopsis thaliana), there are 53 members in the NRT1 family, nine of which were identified as nitrate transporters to date. NRT1.1, also known as CHL1 (for chlorate resistance1), was the first one identified as a dual-affinity nitrate uptake transporter (Tsay et al., 1993; Wang et al., 1998; Liu et al., 1999), while the other eight show solely low-affinity transport activity (Tsay et al., 2007; Li et al., 2010; Wang and Tsay, 2011). Recent studies revealed that NRT1.1 is also a nitrate receptor that can sense a wide range of nitrate concentrations through phosphorylation regulation, thus regulating the nitrate primary response process (Liu and Tsay, 2003; Ho et al., 2009). NRT1.1 and NRT1.2 genes are expressed mainly in roots and regulate nitrate uptake from soil (Tsay et al., 1993; Huang et al., 1999). Another two responsible transporters are NRT2.1 and NRT2.2, which possibly represent the major uptake mechanism, as suggested by the dramatic decrease of nitrate uptake in their mutant plants (Filleur et al., 2001; Li et al., 2007).
Once taken up into roots, most nitrate undergoes long-distance transport to leaves and is assimilated in chloroplasts (Smirnoff and Stewart, 1985; Andrews, 1986). NRT1.5 and NRT1.8 have been identified as two essential transporters in nitrate long-distance transport (Lin et al., 2008; Li et al., 2010). The NRT1.5 gene is expressed mainly in root pericycle cells and functions to load nitrate into xylem. Functional disruption of NRT1.5 did not abort nitrate transport to aerial tissues, indicating that other xylem-loading transporter(s) may exist (Lin et al., 2008). NRT1.8 is expressed predominantly in xylem parenchyma cells within the vasculature and functions to remove nitrate from xylem vessels. NRT1.5 works together with NRT1.8 to fine-tune nitrate long-distance transport from roots to shoots (Lin et al., 2008; Li et al., 2010).
Nitrate assimilation is an energy-intensive process, and many herbaceous plants tackle this problem by transporting nitrate to leaves, where energy and reductants derived from photosynthesis can be directly accessed by nitrate assimilation, thus rendering leaf nitrate assimilation more energy efficient than root assimilation (Canvin and Atkins, 1974; Smirnoff and Stewart, 1985; Andrews, 1986). However, it has long been observed that nitrate undergoes reallocation to roots when exposed to stresses, including low light and heavy metals (Smirnoff and Stewart, 1985; Hernandez et al., 1997). A recent study identified that the nitrate reallocation process is regulated by the induction of NRT1.8 and contributes essentially to Cd2+ stress tolerance (Li et al., 2010). NRT1.5 might also be involved in nitrate reallocation because it is down-regulated by Cd2+ stress (Zimmermann et al., 2004; Li et al., 2010); thus, nitrate retained in roots probably occurs based on the xylem nitrate-loading function of NRT1.5. Additionally, Web-based microarray analyses indicated that the coordinated opposite regulation patterns of NRT1.8 and NRT1.5 could be repeatedly observed under various biotic and abiotic stresses (Zimmermann et al., 2004; Li et al., 2010), leading to the hypothesis that nitrate reallocation in plants might be a common response to stresses (Gojon and Gaymard, 2010; Li et al., 2010), in which NRT1.5 might be another essential component. However, this hypothesis was established only with NRT1.8 under Cd2+ stress so far; whether it is physiologically relevant to the NRT1.5 gene or of general physiological significance remains to be determined.
In this study, several lines of experimental evidence are provided to demonstrate that NRT1.5 functions to mediate nitrate reallocation to roots, stress-responsive gene expression and metabolism, and consequently salt, drought, and Cd2+ tolerance, supporting the hypothesis that nitrate reallocation to roots might be a common response to stresses and is coordinately regulated by NRT1.8 and NRT1.5 genes.
RESULTS
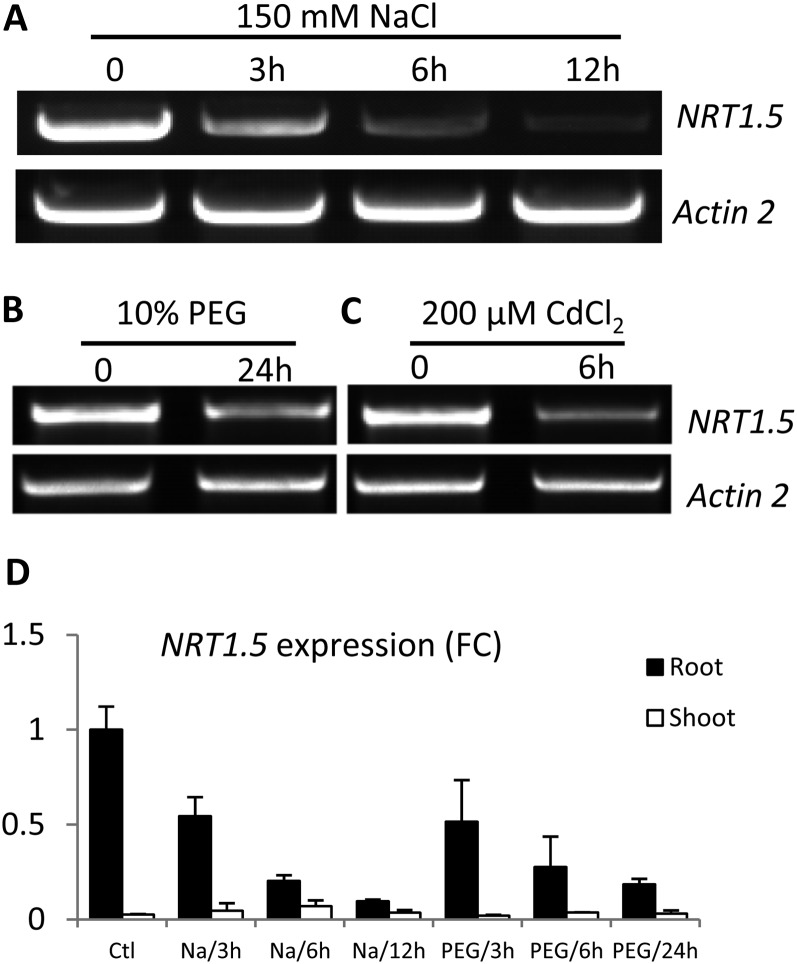
Down-Regulation of NRT1.5 by Various Stresses in Roots
To investigate if NRT1.5 might play a role in the nitrate-regulated stress tolerance, reverse transcription (RT)-PCR was first performed to characterize NRT1.5 expression using roots, as NRT1.5 shows steadily high expression in roots but is undetectable in shoots under control conditions (Li et al., 2010). When exposed to NaCl, a significant decrease in NRT1.5 expression level was observed 3 h after treatment in roots, and a further decrease occurred with prolonged treatment (Fig. 1A). Similar results were obtained for drought stress simulated by polyethylene glycol (PEG) application (Fig. 1B). Further quantitative RT-PCR analyses confirmed the down-regulation pattern of NRT1.5 by various stresses in roots. Specifically, NaCl resulted in the most significant inhibition: a 12-h treatment reduced NRT1.5 expression to one-tenth of the level under control conditions, while one-fifth of the control expression level was observed under 24-h drought stresses (Fig. 1D). Taken together, these data suggest that NRT1.5 expression in Arabidopsis roots is down-regulated by various stresses, including NaCl, drought, and Cd2+. Note that in shoots, the NRT1.5 expression level is very low and not affected by stress treatments (Fig. 1D).
Figure 1.
A to C, Down-regulation of NRT1.5 by various stresses. Plants were grown hydroponically for 4 weeks and then exposed to 150 mm NaCl (A), 10% PEG (B), or 200 μm CdCl2 (C) treatment for the indicated times. NRT1.5 mRNA levels in roots were determined by RT-PCR. D, Quantitative RT-PCR determination of NRT1.5 expression in both shoots and roots exposed to 200 mm NaCl or 10% PEG for the indicated times. The y axis shows fold change (FC) compared with the control (Ctl). Values are means ± sd (n = 3). Actin2 was used as a loading control.
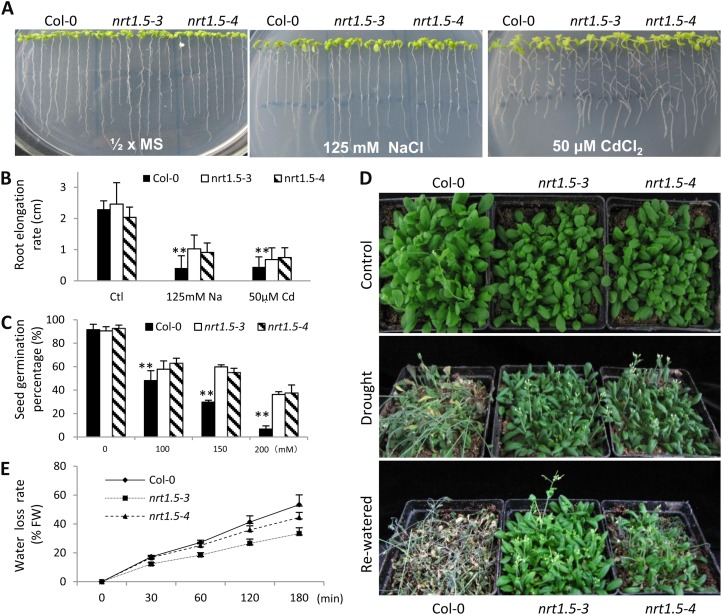
Enhanced Tolerance to Various Stresses in nrt1.5 Mutants
Our previous work indicated that NRT1.5 might contribute to Cd2+ tolerance and possibly other stress tolerance (Li et al., 2010). As expected, significantly enhanced Na+ as well as Cd2+ tolerance was observed in the functional disruption mutants nrt1.5-3 and nrt1.5-4 compared with the wild-type ecotype Columbia (Col-0; Fig. 2, A and B), while no significant difference was seen between them when grown under control conditions (Fig. 2, A and B). Furthermore, less reduction of germination rate was observed in nrt1.5 mutant seeds compared with the wild type, especially when under 200 mm NaCl treatment, where nrt1.5-3 and nrt1.5-4 seeds showed nearly 40% germination while Col-0 had less than 10% (Fig. 1C). Further analyses showed a significant increase in drought tolerance in nrt1.5 mutant plants compared with Col-0 (Fig. 2D, middle and bottom panels), while similar growth was observed when under control conditions (Fig. 2D, top panel). Correspondingly, decreased water loss rates were detected in nrt1.5 plants compared with the wild-type control (Fig. 2E). Furthermore, when replacing nitrate with ammonium, the enhanced stress tolerance in nrt1.5 mutants was abandoned (Supplemental Fig. S1). These data indicate that the down-regulation of NRT1.5 mediates tolerance to various stresses in a nitrate-dependent manner.
Figure 2.
Enhanced tolerance to various stresses in nrt1.5 mutants. A, Five-day-old seedlings were transferred to one-half-strength Murashige and Skoog medium (½ × MS) or one-half-strength Murashige and Skoog medium supplemented with 125 mm NaCl or 50 μm CdCl2 and allowed another 6 d to grow. B, Root elongation between days 2 and 7 after transfer to the treatments in A. Values are means ± sd from three replicates, and each contained eight plants. ** P < 0.01. C, Seed germination rate on one-half-strength Murashige and Skoog medium supplemented with the indicated levels of NaCl. Values are means ± sd (n = 200 seeds). ** P < 0.01. D, Representative photographs for drought tolerance analysis of wild-type Col-0 and nrt1.5 mutant plants. Control (top panel), drought-stressed (middle panel), and rewatered (bottom panel) plants were treated as described in “Materials and Methods” before imaging. E, Water loss rate of leaves detached from 16-d-old plants. Values are means ± sd (n = 15). FW, Fresh weight.
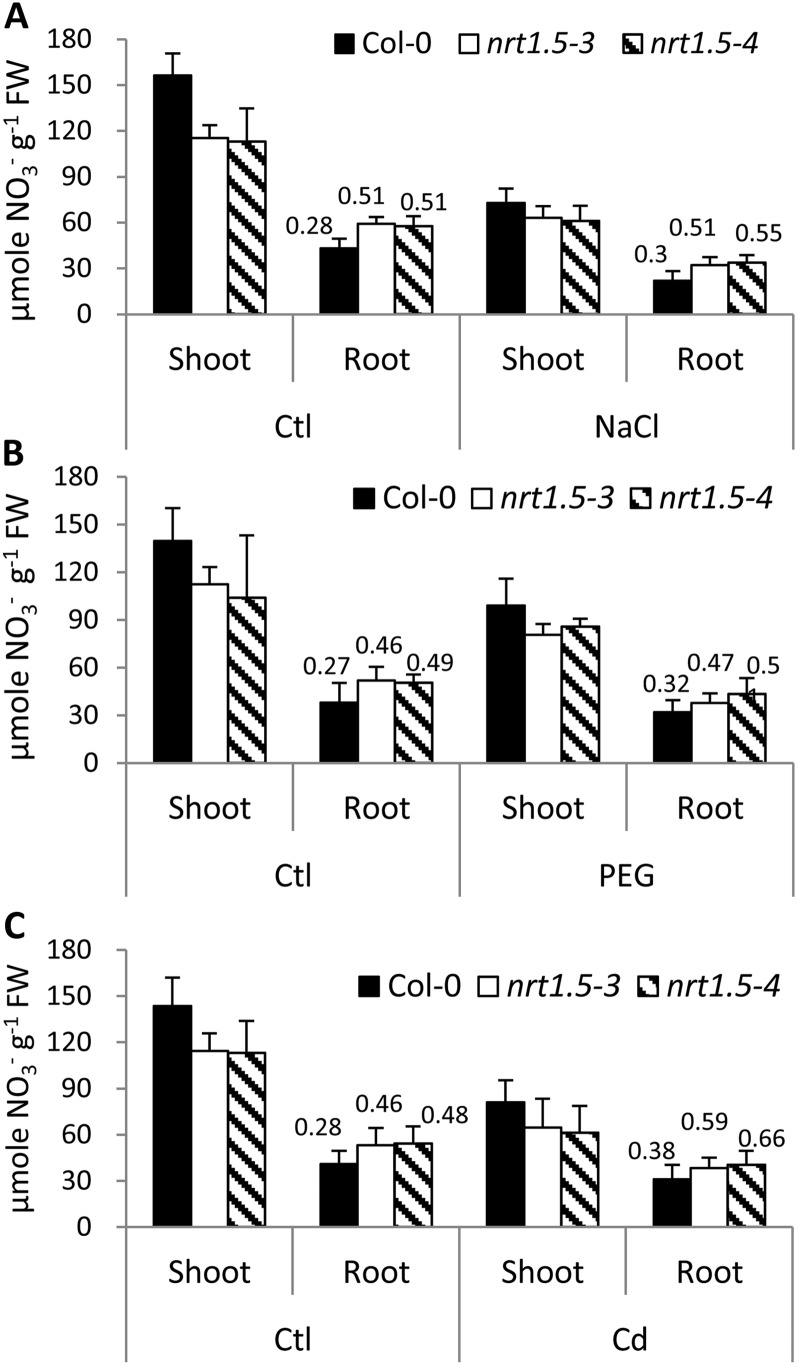
Increased Nitrate Allocation to Roots of nrt1.5 Mutants
It has been reported that NRT1.8 induction in Arabidopsis increases nitrate allocation to roots, thus enhancing Cd2+ tolerance (Li et al., 2010). Given that down-regulation of the nitrate transporter gene NRT1.5 in roots also enhanced tolerance to various stresses (Fig. 2), we tested whether a similar nitrate reallocation could be observed in nrt1.5 mutant plants.
Under control conditions, nitrate levels in shoots of nrt1.5-3 and nrt1.5-4 were significantly lower than in Col-0, while more were detected in roots (Fig. 3A), which translates to a root-shoot nitrate ratio of 0.28 in the wild type but 0.51 in nrt1.5 mutant plants, consistent with previous results (Lin et al., 2008). Salt stress significantly decreased the overall nitrate contents in both wild-type and mutant plants; however, proportionally more nitrate was still detected in roots of nrt1.5 than those of Col-0 (Fig. 3A). Similar results were obtained in plants grown under drought and Cd2+ stress conditions (Fig. 3, B and C). These data demonstrated that functional disruption of NRT1.5 constitutively impaired xylem nitrate loading, and thus more nitrate was retained in roots. Salt and drought stresses did not further increase nitrate reallocation to roots in nrt1.5 mutants, possibly because the disruption of NRT1.5 renders nitrate reallocation no longer a response to stresses but a constitutive alteration. In contrast, Cd2+ treatment increased the root-shoot nitrate ratio from 0.46 to 0.48 under control conditions to 0.59 to 0.66 under Cd2+ stress, indicating that more complicated mechanisms may be involved in regulating nitrate reallocation under Cd2+ stress.
Figure 3.
Increased nitrate allocation to roots of nrt1.5 mutants. Four-week-old hydroponically grown plants were treated by 50 mm NaCl for 3 d (A), 10% PEG for 24 h (B), 20 μm CdCl2 for 3 d (C), or under control conditions (Ctl). Then, shoot and root tissues were harvested and subjected to determination of nitrate concentration by HPLC. The numbers above each bar represent root-shoot nitrate ratio. Values are means ± sd from three replicates, and each pooled more than nine plants. FW, Fresh weight.
Given that further nitrate allocation could not be easily observed in nrt1.5 mutants when exposed to stresses, we then tested the stress sensitivity in nrt1.8-1, which could enhance nitrate allocation to shoots, thus resembling NRT1.5 overexpression. As expected, significantly increased drought sensitivity was observed in nrt1.8-1 compared with the wild-type ecotype Wassilewskija (Ws), and crossing nrt1.8-1 to nrt1.5 neutralized the drought tolerance observed in nrt1.5 plants to a level comparable to the wild-type control (Supplemental Fig. S2A). Similar results were observed when nrt1.8-1 and the double mutants were exposed to salt stress (Supplemental Fig. S2, B–E). These data suggest that nitrate allocation regulated either by NRT1.5 or NRT1.8 is essential to stress tolerance. Note that a different growth medium was used for nrt1.5 and nrt1.8 mutants in the salt tolerance assay, as similar growth between nrt1.5 mutants and their wild type under control conditions could be obtained only when the nitrate level paralleled those of other nutrients, and stress tolerance in nrt1.5 mutants depended less on nitrate concentration (Figs. 2, A and B; Supplemental Fig. S5).
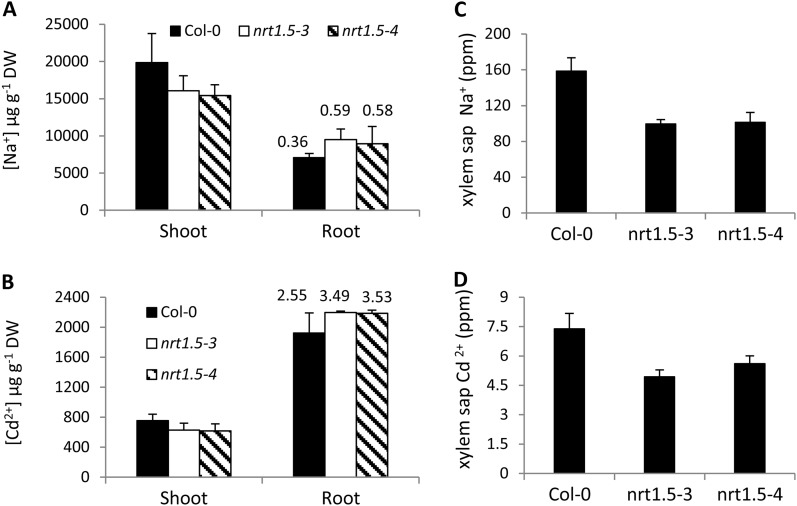
Altered Na+ or Cd2+ Distribution in nrt1.5 Mutants
A previous study showed that in the nrt1.8 mutant, proportionally more nitrate accumulated in shoots, where correspondingly more Cd2+ was detected (Li et al., 2010). Consistent with this observation, a similar positive correlation between nitrate and metal accumulation was also observed in nrt1.5-3 and nrt1.5-4 plants. Na+ accumulation in nrt1.5 shoots was significantly decreased compared with Col-0 while increased Na+ contents were detected in roots; the root-shoot Na+ content ratios were 0.59 and 0.58 in nrt1.5-3 and nrt1.5-4, respectively, in contrast to approximately 0.36 in Col-0 (Fig. 4A). Furthermore, the Na+ concentration in xylem sap from nrt1.5 mutant plants was significantly decreased compared with that from Col-0 (Fig. 4C). A similar Cd2+ distribution between roots and shoots was also observed in nrt1.5 mutant plants (Fig. 4, B and D). Given that functional disruption of NRT1.5 retains more nitrate in roots (Fig. 3) and decreases nitrate concentration in xylem sap (Li et al., 2010), these data suggest that whether regulated by NRT1.8 or NRT1.5, nitrate reallocation leads to increased metal accumulation in tissues where nitrate concentration increases. Note that although Na+ and Cd2+ were provided as chloride salts, a positive correlation between Cl− and NO3− could not be observed in nrt1.5 mutants (Supplemental Fig. S4), in contrast to that between Na+/Cd2+ and NO3− (Fig. 4), suggesting that the stress tolerance in nrt1.5 mutants might not be ascribed to Cl− allocation.
Figure 4.
Altered Na+ or Cd2+ distribution in nrt1.5 mutants. A and B, Increased Na+ or Cd2+ accumulation in nrt1.5 roots. Four-week-old plants were exposed to 50 mm NaCl (A) or 20 μm CdCl2 (B) for 3 d before shoots and roots were sampled. The numbers above each bar represent root-shoot ratio of Na+ or Cd2+. C and D, Decreased Na+ or Cd2+ concentration in xylem sap. Four-week-old plants were treated with 10 mm NaCl for 1 d (C) or 5 μm CdCl2 for 3 d (D) before xylem sap was collected. Values are means ± sd from three (A and B) or nine (C and D) replicates. DW, Dry weight.
Functional Disruption of NRT1.5 Alters the Expression of Stress-Related Genes
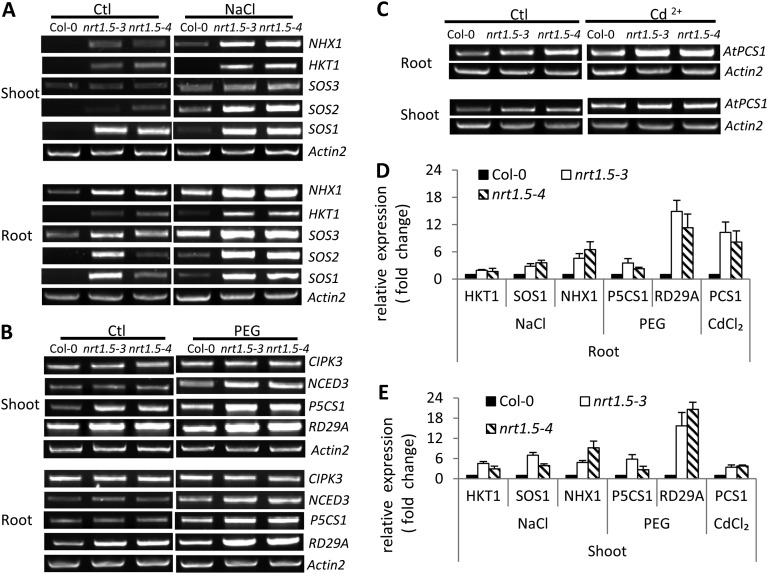
To investigate the underlying mechanisms of enhanced stress tolerance in nrt1.5 mutant plants (Fig. 2), the expression levels of marker genes in stress response pathways were determined. Under control conditions, NHX1, High-affinity K+ transporter1 (HKT1), Salt overly sensitive1 (SOS1), and SOS2 showed steadily higher expression in nrt1.5 than in Col-0 (Fig. 5A). Further increased expression of these genes, especially of NHX1, HKT1, and SOS1, which are responsible for vacuolar Na+ sequestration, long-distance Na+ transport, and Na+ efflux, respectively (Shi et al., 2000; Yokoi et al., 2002; Berthomieu et al., 2003), was observed in nrt1.5 plants than in the wild type when treated by salt stress (Fig. 5A).
Figure 5.
A to C, Functional disruption of NRT1.5 alters the expression levels of stress-related genes. Four-week-old hydroponically grown plants were treated with 200 mm NaCl for 6 h (A),10% PEG for 24 h (B), 200 μm CdCl2 for 6 h (C), or under control conditions (Ctl). D and E, RT-PCR was performed for selected stress-related marker genes. Quantitative RT-PCR determination of representative gene expression in roots (D) and shoots (E). Values are means ± sd (n = 3). Actin2 was used as a loading control.
Among the selected drought tolerance-related genes, P5CS1 and RD29A were most significantly affected by NRT1.5 (Fig. 5B). In shoots of nrt1.5, steadily higher expression of P5CS1 was observed than in Col-0 under control conditions, and further increases were detected in both shoots and roots of nrt1.5 when exposed to drought stress (Fig. 5B). Similar results were obtained for RD29A, in which drought stress further increased its relatively higher expression level in nrt1.5 than in Col-0 under control conditions (Fig. 5B). In terms of the Cd2+ response pathway, significantly higher expression of AtPCS1 was observed in nrt1.5 roots under Cd2+ stress (Fig. 5C). Further quantitative RT-PCR analyses confirmed these results (Fig. 5, D and E). Taken together, these data suggest that nitrate reallocation affects the expression levels of several genes in stress response pathways, and these genes, including SOS1, NHX1, P5CS1, RD29A, and AtPCS1, might be used as biomarkers to further investigate the interaction between nitrate reallocation and stress tolerance.
Interestingly, even under control conditions, significantly increased expression of NRT1.8 was observed in nrt1.5 mutants, and dramatic increases occurred in roots of both mutants and in shoots of nrt1.5-3 (Supplemental Fig. S3A). Stress treatments did not show any further effect on NRT1.8 expression in nrt1.5 mutants, except in nrt1.5-3 shoots (Supplemental Fig. S3A). In contrast to NRT1.5 expression in the nrt1.8-2 mutant, the nrt1.8-1 mutant with a replaced genetic background of Col-0 had similar expression to that in its wild-type control (Supplemental Fig. S3B) and appeared not to respond to stresses (data not shown). These data suggest that the down-regulation of NRT1.5 occurs prior to the induction of NRT1.8 in roots exposed to stresses.
Altered Accumulation of Stress-Related Metabolites in nrt1.5
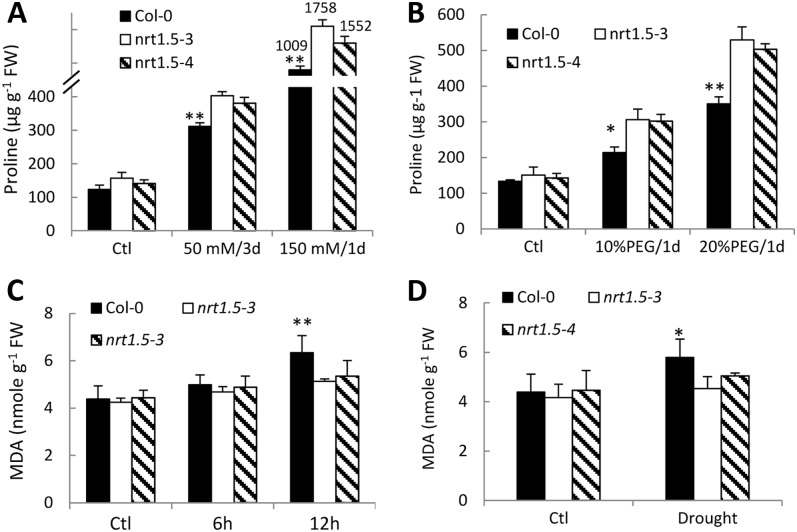
Pro functions as an important osmolyte and contributes essentially to stress tolerance in higher plants. In this study, the expression of P5CS1, the essential regulatory gene in the Pro biosynthesis pathway (Kishor et al., 2005; Székely et al., 2008), was significantly altered by functional disruption of NRT1.5 (Fig. 5B), which leads to the notion that the Pro level might also be changed in nrt1.5 mutants. Further analyses showed that under control conditions, Pro levels were slightly higher in nrt1.5 than in wild-type Col-0, while Na+ stress significantly increased Pro contents in nrt1.5 compared with Col-0, and the most significant increase was observed with 150 mm NaCl treatment for 1 d (Fig. 6A). Similar results were obtained in drought-treated plants (Fig. 6B). These data suggest that nitrate allocation to roots enhances Pro synthesis and accumulation.
Figure 6.
Stress-related metabolite accumulation in nrt1.5 mutants. Four-week-old plants were exposed to the indicated NaCl treatments (A), PEG treatments (B), or 150 mm NaCl (C) for the indicated times, 10% PEG for 1 d (D), or under control conditions (Ctl). Shoots were sampled for the determination of Pro or malondialdehyde (MDA) levels. Values are means ± sd (n = 6). ** P < 0.01. FW, Fresh weight.
Malondialdehyde is one of the major lipid oxidation products (Esterbauer et al., 1991; Weber et al., 2004), which indicates how severely plants are stressed (Zheng et al., 2008). Corresponding to the increased Pro level in nrt1.5 plants (Fig. 6, A and B), relatively decreased malondialdehyde levels were detected in nrt1.5 plants compared with Col-0 under salt and drought stresses (Fig. 6, C and D), indicating that nitrate allocation to roots decreases oxidative stress in plants.
DISCUSSION
NRT1.5 Is Another Essential Component in Regulating Nitrate Reallocation to Roots and the Consequent Stress Tolerance
Our previous study showed that nitrate redistribution contributes essentially to Cd2+ tolerance in Arabidopsis, and this process is actively regulated by the xylem nitrate-unloading transporter NRT1.8. Interestingly, NRT1.5, the xylem nitrate-loading transporter, was significantly down-regulated by Cd2+, leading to the hypothesis that NRT1.5 may also contribute to the nitrate reallocation to roots, hence enhancing plant stress tolerance (Li et al., 2010). Here, we present data to show that in Col-0, the root-shoot nitrate ratios were 0.28 to 0.38, while in nrt1.5, the value varied from 0.46 to 0.66 (Fig. 3), indicating that proportionally more nitrate accumulated in nrt1.5 roots compared with wild-type Col-0. Moreover, nrt1.5 showed enhanced tolerance to Cd2+, salt, and drought stresses (Fig. 2). These data together suggested that down-regulation of NRT1.5 by various stresses helps to retain nitrate in roots and contributes essentially to stress tolerance in a similar mechanism to that proposed for NRT1.8 (Li et al., 2010).
The Role of Nitrate Reallocation in Regulating Stress Tolerance
It has been believed that nitrate long-distance transport to shoots allows plants more energy efficiency during evolutionary competition, although nitrate reallocation to roots is frequently observed under adverse conditions (Canvin and Atkins, 1974; Smirnoff and Stewart, 1985; Andrews, 1986; Ericsson, 1995; Hernandez et al., 1997). In our studies, either the up-regulation of NRT1.8 or the down-regulation of NRT1.5 enhances plant tolerance to Cd2+ stress, establishing that no matter the molecular basis, nitrate reallocation to roots represents an essential mechanism regulating Cd2+ tolerance (Li et al., 2010; Fig. 2) rather than a passive consequence of inhibited transpiration rate (Hernandez et al., 1997).
Furthermore, the opposite expression pattern of NRT1.8 and NRT1.5 is regulated not only by Cd2+ but by a wide range of stresses, including abiotic and biotic stresses (Zimmermann et al., 2004; Li et al., 2010), implying that nitrate reallocation may be a universal mechanism in regulating various stress tolerances than a specific response to Cd2+ (Gojon and Gaymard, 2010; Li et al., 2010). An extended assay on nrt1.5 mutant plants under salt and drought stresses provided experimental evidence for this hypothesis, because increased nitrate accumulation in nrt1.5 roots (Fig. 3) significantly enhanced drought and salt tolerance in nrt1.5 (Fig. 2). It is worth noting that, in contrast to the wild type, nrt1.5 mutants show a complete absence of NRT1.5; thus, nitrate allocation is no longer a response to stresses but a constant alteration (Fig. 3), which resembles stress pretreatment and would possibly lead to a wide range of physiological consequences, as was observed in seed germination and water loss rate in nrt1.5 mutants (Fig. 2). Further support for this hypothesis came from the result that, in addition to other important genes, NRT1.8 shows constitutive high expression in nrt1.5 mutants (Fig. 5; Supplemental Fig. S3A).
Possible Mechanisms of Stress Tolerance Regulated by Nitrate Reallocation, and Prospects of Future Research
Nitrate reallocation to roots was proposed to serve as a signal to regulate Cd2+ tolerance in Arabidopsis (Li et al., 2010), mainly based on the following observations: (1) nitrate represents only a minor proportion of all the nitrogen subjected to reallocation to roots, so the nutritional effect is subtle and insignificant; (2) extensive changes of gene expression in NRT1.8 overexpression lines; and (3) the Cd2+ distribution between roots and shoots was altered and showed a positive correlation with nitrate levels, although NRT1.8 appears unable to transport either Cd2+ or its major chelators, phytochelatins and glutathione.
Consistently, nitrate reallocation in nrt1.5 extensively altered the expression levels of many marker genes in drought, salt, and Cd2+ response pathways (Fig. 5), especially the levels of HKT1 and SOS1, which regulate Na+ distribution, and P5CS1 and AtPCS1, which regulate osmolyte synthesis and Cd2+ distribution, respectively (Strizhov et al., 1997; Shi et al., 2000; Gong et al., 2003, 2004; Sunarpi et al., 2005; Pomponi et al., 2006; Székely et al., 2008). Indeed, Cd2+ and Na+ distribution in roots and shoots was altered correspondingly (Fig. 4). Moreover, significant changes were also observed in Pro and malondialdehyde levels (Fig. 6), indicating that nrt1.5 mutants adapt to stresses more efficiently than the wild type by altering osmolyte and reactive oxygen species metabolism, consistent with the results that several genes in the stress response pathway show steadily higher expression in nrt1.5 than in Col-0 (Fig. 5). Given that these alterations derive from different environmental cues and are involved in gene expression (Fig. 5), solute transport (Fig. 4), and secondary metabolism (Fig. 6), the most simple interpretation would be that nitrate reallocation serves as a signal to bridge various stress cues and extensive physiological changes, consequently enhancing stress tolerance.
As to how exactly nitrate reallocation regulates stress tolerance, it remains largely to be investigated. In tobacco (Nicotiana tabacum), altered nitrate allocation to shoots had been suggested as a signal to coordinately reprogram nitrogen and carbon metabolism and significantly decreased sugar contents in roots (Scheible et al., 1997a, 1997b), indicating that sugar allocation and signaling might be part of the nitrate signaling pathway. Furthermore, in studies of nitrate reallocation under stress, an interesting observation is that NRT1.8 and NRT1.5 are oppositely regulated by various stresses (i.e. NRT1.8 is up-regulated while NRT1.5 is reduced; Zimmermann et al., 2004; Li et al., 2010), which eventually results in the similar consequence of retaining nitrate in roots. This specific regulation pattern raises the concern that whether NRT1.8 and NRT1.5 share a common regulation mechanism, the down-regulation of NRT1.5 may be actively regulated, or just a passive consequence of stresses, or even a feedback response of sugar allocation. The fact that NRT1.8 expression is constitutively up-regulated in nrt1.5 mutants suggests that the down-regulation of NRT1.5 might be an active process that functions genetically upstream of NRT1.8 (Supplemental Figs. S2 and S3); thus, NRT1.5 and NRT1.8 might share a similar downstream mechanism to regulate stress tolerance. This hypothesis is consistent with the altered gene expression pattern and solute transport observed in both nrt1.5 mutants and NRT1.8 transgenic lines (Figs. 4 and 5; Li et al., 2010). Further study on how NRT1.5 is down-regulated by stresses might shed light on and essentially promote the understanding of the stress-initiated nitrate signaling pathway.
In summary, functional disruption of the NRT1.5 gene enhanced nitrate accumulation in roots and tolerance to Cd2+, Na+ and drought stresses, consistent with the model that the opposite regulation of NRT1.8 and NRT1.5 serves as the essential molecular basis to regulate nitrate reallocation to roots. Our research further supports the hypothesis that nitrate allocation to roots is a common mechanism in regulating a wide range of stresses.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) Col-0 plants were used as the wild-type control for nrt1.5 mutants, while Ws was used as the control for nrt1.8-1 mutants. Crossing of nrt1.8 into nrt1.5 was performed to generate the homozygous double mutants dm1 (nrt1.5-3/nrt1.8-1) and dm2 (nrt1.5-4/nrt1.8-1), and the hybrids (C×W) from Col-0 and Ws plants were used as the wild-type control. The mutant nrt1.8-2 in the Col-0 genetic background was generated by crossing nrt1.8-1 (in the Ws background) into Col-0 five times.
Growth Conditions and Stress Sensitivity Analyses
Arabidopsis plants were grown in one-quarter-strength sterile hydroponic solution at 22°C with a 16-h-light/8-h-dark cycle as described (Arteca and Arteca, 2000; Gong et al., 2003). At 3 to 4 weeks of age, plants were exposed to treatments as indicated in the legends of Figures 1 and 3 to 6 and Supplemental Figures S3 and S4. For sensitivity analyses of plants on plates, seedlings were grown for 5 to 6 d on one-half-strength Murashige and Skoog basal medium (Sigma-Aldrich) with 1 g L−1 MES, 0.8% (w/v) Suc, and 1.5% Bacto agar or on one-quarter-strength minimal medium (Arteca and Arteca, 2000; Gong et al., 2003) when indicated. They were then transferred to plates with basal medium or medium supplemented with NaCl or CdCl2 at the indicated concentrations and allowed to grow vertically for another 6 to 7 d, at which point root elongation was determined. For the seed germination assay, Arabidopsis seeds were plated and allowed 4 d of incubation before the germination rate (percentage of germinated seeds) was determined. For the drought tolerance assay, wild-type and mutant plants grown in soil were irrigated for 12 d (control) and then drought stressed by terminating irrigation for 12 d for Col-0 and nrt1.5, 8 d for nrt1.8 and Ws, or 10 d for the double mutants dm1 and dm2 and the control plant C×W. Photographs of rewatered plants were taken 3 to 5 d after rewatering. For the water loss assay, wild-type (Col-0) and nrt1.5 mutant plants were irrigated normally for 16 d and then fully expanded rosette leaves were sampled; plant fresh weight was determined at the indicated time intervals to calculate the percentage of fresh weight decrease.
Nitrate-Dependent Assay
In the nitrogen source-dependent assay, KNO3 and Ca(NO3)2 in one-quarter-strength minimal medium were replaced with 1.25 mm KCl and 0.5 mm CaCl2, and then 5 mm filter-sterilized ammonium succinate was added as the sole nitrogen source to make ammonium plates. Seedlings were germinated on ammonium plates with 0.8% (w/v) Suc and 1.5% Bacto agar and incubated vertically for 7 d; then they were transferred to the minimal ammonium plates (without Suc) or plates supplemented with either 50 mm NaCl or 50 μm CdCl2. Alternatively, for the nitrate concentration assay, wild-type and nrt1.5 plants were grown in one-quarter-strength minimal medium with 0.8% (w/v) Suc and 1.5% Bacto agar for 5 d, and then the seedlings were transferred to minimal medium with nitrate at the indicated concentration with or without 50 mm NaCl or 50 μm CdCl2. Photographs were taken 6 d later, and root elongation between days 2 and 7 after transfer was determined.
RT-PCR and Quantitative RT-PCR
Four-week-old hydroponically grown plants were exposed to NaCl, PEG (PEG6000), or CdCl2 (Fig. 1, Fig. 5, and Supplemental Fig. S3). Total RNA was extracted from roots and shoots using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. First-strand complementary DNA was synthesized from DNaseI-digested total RNA using Moloney murine leukemia virus reverse transcriptase (Promega), and PCR was performed on a Perkin-Elmer GeneAmp 9700 with the indicated cycles using Ex-Taq DNA polymerase (TaKaRa); PCR products were separated on a 1% agarose gel and stained with ethidium bromide. Quantitative RT-PCR was performed on a Corbett Research Rotor-Gene 3000 thermal cycler using SYBR Premix Ex-Taq (TaKaRa) according to the manufacturers’ protocols.
Determination of Nitrate, Chloride, Na+, Cd2+, and Metabolite Levels
Four-week-old hydroponically grown plants were treated as indicated in the legends of Figures 3, 4, and 6 and Supplemental Figure S4. Shoots and roots were sampled as described (Li et al., 2010). Nitrate was extracted in boiling water and determined by HPLC (Agilent 1200 series) using a PARTISIL 10 strong anion-exchange column (Whatman) as described (Chiu et al., 2004). Chloride was extracted by deionized water and filtered through a C18 precolumn filter (Sigma-Aldrich). Chloride content was determined by ion chromatography (Agilent IC5000 series) as described by Kong et al. (2011) with minor modifications. Metal accumulation was determined using inductively coupled plasma-mass spectrometry (Elan DRC-e; Perkin-Elmer) as described (Gong et al., 2003) with minor modifications. To collect enough xylem sap, plants were exposed to mild treatments (Fig. 4). Xylem sap was collected for 6 h as described (Sunarpi et al., 2005). Alternatively, plants were treated with salt and drought stresses before sampling for the determination of Pro concentration by ninhydrin colorimetry (Bates et al., 1973; Sharma and Dubey, 2005) or the determination of malondialdehyde using the thiobarbituric acid method with minor modifications (Kramer et al., 1991; Zheng et al., 2008).
Statistical Analyses
Two-tailed Student’s t tests were performed to compare fresh weights and nitrate, Na+, Cd2+, and metabolite contents between mutant and wild-type plants. Differences were deemed significant at P < 0.05 and extremely significant at P < 0.01.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g32450 (NRT1.5), At4g21680 (NRT1.8), At1g12110 (CHL1), At1g69850 (NRT1.2), At1g08090 (NRT2.1), At1g08110 (NRT2.2), AT5G44070 (PCS1), AT5G52310 (RD29A), AT2G39800 (P5CS1), AT2G26980 (CIPK3), AT3G14440 (NCED3), AT5G27150 (NHX1), AT4G10310 (HKT1), AT2G01980 (SOS1), AT5G35410 (SOS2), AT5G24270 (SOS3), and AT3g18780 (Actin2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Enhanced stress tolerance in nrt1.5 mutants is nitrate dependent.
Supplemental Figure S2. nrt1.8 is sensitive to drought and salt stresses and neutralizes phenotypes observed in nrt1.5 mutants.
Supplemental Figure S3. Increased NRT1.8 expression in nrt1.5 mutants.
Supplemental Figure S4. Chloride allocation in nrt1.5 mutants.
Supplemental Figure S5. Nitrate-dependent growth of nrt1.5 mutants.
Supplementary Material
Acknowledgments
We thank Laisheng Ji (Core Facility, Shanghai Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences) for helping with quantitative RT-PCR and Yi Zhang (Agilent) for technical support and helpful discussions.
Glossary
- RT
reverse transcription
- PEG
polyethylene glycol
- Col-0
ecotype Columbia
- Ws
ecotype Wassilewskija
References
- Andrews M. (1986) The partitioning of nitrate assimilation between root and shoot of higher plants. Plant Cell Environ 9: 511–519 [Google Scholar]
- Arteca RN, Arteca JM. (2000) A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol Plant 108: 188–193 [Google Scholar]
- Bates L, Waldren R, Teare I. (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39: 205–207 [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canvin D, Atkins C. (1974) Nitrate, nitrite and ammonia assimilation by leaves: effect of light, carbon dioxide and oxygen. Planta 116: 207–224 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Lin CS, Hsia AP, Su RC, Lin HL, Tsay YF. (2004) Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol 45: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Crawford NM. (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel-Vedele F, Filleur S, Caboche M. (1998) Nitrate transport: a key step in nitrate assimilation. Curr Opin Plant Biol 1: 235–239 [DOI] [PubMed] [Google Scholar]
- Ericsson T. (1995) Growth and shoot: root ratio of seedlings in relation to nutrient availability. Plant Soil 168-169: 205–214 [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11: 81–128 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F. (2001) An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489: 220–224 [DOI] [PubMed] [Google Scholar]
- Gojon A, Gaymard F. (2010) Keeping nitrate in the roots: an unexpected requirement for cadmium tolerance in plants. J Mol Cell Biol 2: 299–301 [DOI] [PubMed] [Google Scholar]
- Gong JM, Lee DA, Schroeder JI. (2003) Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc Natl Acad Sci USA 100: 10118–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JM, Waner DA, Horie T, Li SL, Horie R, Abid KB, Schroeder JI. (2004) Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez LE, Gárate A, Carpena-Ruiz R. (1997) Effects of cadmium on the uptake, distribution and assimilation of nitrate in Pisum sativum. Plant Soil 189: 97–106 [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Huang NC, Liu KH, Lo HJ, Tsay YF. (1999) Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishor PBK, Sangam S, Amrutha R, Laxmi PS, Naidu K, Rao K, Rao S, Reddy K, Theriappan P, Sreenivasulu N. (2005) Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Curr Sci 88: 424–438 [Google Scholar]
- Kong XQ, Gao XH, Sun W, An J, Zhao YX, Zhang H. (2011) Cloning and functional characterization of a cation-chloride cotransporter gene OsCCC1. Plant Mol Biol 75: 567–578 [DOI] [PubMed] [Google Scholar]
- Kramer GF, Norman HA, Krizek DT, Mirecki RM. (1991) Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry 30: 2101–2108 [Google Scholar]
- Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Zhang Y, Li HM, Huang J, et al. (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass AD. (2007) Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Huang CY, Tsay YF. (1999) CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11: 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF. (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J 22: 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomponi M, Censi V, Di Girolamo V, De Paolis A, di Toppi LS, Aromolo R, Costantino P, Cardarelli M. (2006) Overexpression of Arabidopsis phytochelatin synthase in tobacco plants enhances Cd(2+) tolerance and accumulation but not translocation to the shoot. Planta 223: 180–190 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M. (1997a) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Lauerer M, Schulze ED, Caboche M, Stitt M. (1997b) Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco? Plant J 11: 671–691 [Google Scholar]
- Sharma P, Dubey RS. (2005) Modulation of nitrate reductase activity in rice seedlings under aluminium toxicity and water stress: role of osmolytes as enzyme protectant. J Plant Physiol 162: 854–864 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Stewart G. (1985) Nitrate assimilation and translocation by higher plants: comparative physiology and ecological consequences. Physiol Plant 64: 133–140 [Google Scholar]
- Stitt M. (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Strizhov N, Abrahám E, Okrész L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L. (1997) Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J 12: 557–569 [DOI] [PubMed] [Google Scholar]
- Sunarpi HT, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, et al. (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al. (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53: 11–28 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. (2007) Nitrate transporters and peptide transporters. FEBS Lett 581: 2290–2300 [DOI] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM. (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM. (1998) The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc Natl Acad Sci USA 95: 15134–15139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Tsay YF. (2011) Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Chételat A, Reymond P, Farmer EE. (2004) Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J 37: 877–888 [DOI] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM. (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30: 529–539 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jia A, Ning T, Xu J, Li Z, Jiang G. (2008) Potassium nitrate application alleviates sodium chloride stress in winter wheat cultivars differing in salt tolerance. J Plant Physiol 165: 1455–1465 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.