Abstract
The plant hormone jasmonic acid (JA) plays a pivotal role in plant-insect interactions. Herbivore attack usually elicits dramatic increases in JA concentrations, which in turn activate the accumulation of metabolites that function as defenses against herbivores. Although almost all enzymes involved in the biosynthesis pathway of JA have been identified and characterized, the mechanism by which plants regulate JA biosynthesis remains unclear. Calcium-dependent protein kinases (CDPKs) are plant-specific proteins that sense changes in [Ca2+] to activate downstream responses. We created transgenic Nicotiana attenuata plants, in which two CDPKs, NaCDPK4 and NaCDPK5, were simultaneously silenced (IRcdpk4/5 plants). IRcdpk4/5 plants were stunted and aborted most of their flower primordia. Importantly, after wounding or simulated herbivory, IRcdpk4/5 plants accumulated exceptionally high JA levels. When NaCDPK4 and NaCDPK5 were silenced individually, neither stunted growth nor high JA levels were observed, suggesting that NaCDPK4 and NaCDPK5 have redundant roles. Attack from Manduca sexta larvae on IRcdpk4/5 plants induced high levels of defense metabolites that slowed M. sexta growth. We found that NaCDPK4 and NaCDPK5 affect plant resistance against insects in a JA- and JA-signaling-dependent manner. Furthermore, IRcdpk4/5 plants showed overactivation of salicylic-acid-induced protein kinase, a mitogen-activated protein kinase involved in various stress responses, and genetic analysis indicated that the increased salicylic-acid-induced protein kinase activity in IRcdpk4/5 plants was a consequence of the exceptionally high JA levels and was dependent on CORONATINE INSENSITIVE1. This work reveals the critical roles of CDPKs in modulating JA homeostasis and highlights the complex duet between JA and mitogen-activated protein kinase signaling.
Many aspects of a plant’s physiology are regulated by a suite of phytohormones, that include auxin, gibberellins, brassinosteroids, jasmonic acid (JA), salicylic acid (SA), abscisic acid, ethylene, cytokinins, and strigolactones. These hormones modulate specific but yet overlapping aspects of plant development, growth, and stress resistance traits.
Among these, JA plays an important role in defense against herbivores and is also required for reproduction. Mutants or transgenic plants impaired in JA biosynthesis or signaling have greatly decreased levels of defensive secondary metabolites and suffer large losses of biomass and fitness under insect attack (Howe et al., 1996; Halitschke and Baldwin, 2003; Paschold et al., 2007), and these plants are also male sterile (Feys et al., 1994; Stintzi et al., 2001). JA is synthesized in chloroplasts and peroxisomes by at least eight enzymes, including phospholipase, 13-LIPOXYGENASE (13-LOX), ALLENE-OXIDE SYNTHASE (AOS), ALLENE-OXIDE CYCLASE (AOC), OPDA REDUCTASE (OPR), and ACYL-COENZYME A OXIDASE (ACX; Delker et al., 2006). Recent studies have revealed that a conjugate between JA and Ile, JA-Ile, plays a critical role in JA signaling: JA-Ile, but not JA, binds to an F-box protein, CORONATINE INSENSITIVE1 (COI1) and activates downstream responses by degrading the negative regulators of JA responses, namely, JASMONATE ZIM-DOMAIN proteins (Chini et al., 2007; Thines et al., 2007; Browse, 2009).
The transient accumulation of JA is one of the early responses after plants are attacked by herbivores (Howe and Jander, 2008; Wu and Baldwin, 2010). In addition to the wounding caused by herbivore feeding, certain plant species also recognize insect-derived elicitors and are able to deploy herbivory-specific responses, including JA biosynthesis (Howe and Jander, 2008; Wu and Baldwin, 2009, 2010). A wild tobacco plant, Nicotiana attenuata, can perceive fatty acid–amino acid conjugates (FACs) in the oral secretions (OS) of the specialist insect herbivore Manduca sexta (Halitschke et al., 2001), and compared with mechanical wounding, FACs elicit greatly amplified levels of JA and ethylene, which are important phytohormones in regulating the accumulation of defensive secondary metabolites against M. sexta, such as trypsin proteinase inhibitors (TPIs; Zavala et al., 2004), diterpene glycosides (Jassbi et al., 2008; Heiling et al., 2010), and caffeoylputrescine (CP; Kaur et al., 2010).
Although much is known about the biosynthetic pathway and the signal transduction of JA, how plants control the accumulation of JA is not well understood. Reverse genetic studies indicated that mitogen-activated protein kinases (MAPKs), SA-induced protein kinase (SIPK), and wound-induced protein kinase (WIPK), are rapidly activated by FACs and these MAPKs are necessary for the induction of JA in response to herbivory (Kandoth et al., 2007; Wu et al., 2007). In addition to MAPKs, Ca2+ also appears to be involved in herbivore resistance: Caterpillar feeding increases the levels of Ca2+ intracellularly in lima bean (Phaseolus lunatus) and Arabidopsis (Arabidopsis thaliana; Maffei et al., 2004; Kanchiswamy et al., 2010), suggesting that certain Ca2+-sensing proteins may be involved in herbivory-induced defense reactions.
Calcium is a ubiquitous second messenger in the signal transduction in eukaryotes, which controls a large number of signaling pathways and activates various cellular processes in response to developmental and stress-induced stimuli (Sanders et al., 2002; Hetherington and Brownlee, 2004; Lecourieux et al., 2006). In plants, cytosolic Ca2+ levels transiently arise due to Ca2+ fluxes from apoplast, chloroplast, vacuole, endoplasmic reticulum, mitochondrion, and nuclear envelope. Particular stimuli trigger characteristic Ca2+ oscillations by altering the activities of Ca2+ channels, H+/Ca2+ antiporters, and Ca2+- and H+-ATPases localized in different cell compartments (Hetherington and Brownlee, 2004; McAinsh and Pittman, 2009). To decode the messages conveyed by Ca2+, various Ca2+-specific sensors are specialized in recognizing and transducing Ca2+ signals, including calmodulins, calmodulin-binding proteins, calcium-dependent protein kinases (CDPKs or CPKs), and calcineurin B-like proteins (Cheng et al., 2002; Zhang and Lu, 2003). These Ca2+ sensors further interact with their downstream target proteins and in turn trigger Ca2+ signature-specific responses (Sanders et al., 1999; Harper et al., 2004).
Among the various Ca2+-sensing proteins, CDPKs comprise a unique group. CDPKs are found only in the plant kingdom and in some protozoans (Harmon et al., 2001; Cheng et al., 2002) and the Arabidopsis and rice (Oryza sativa) genome sequencing efforts have revealed that CDPKs form a large gene family in plants: 34 and 29 CDPKs were predicted in Arabidopsis and rice genome (Arabidopsis Genome Initiative, 2000; Asano et al., 2005). A typical CDPK consists of four domains: an N-terminal variable domain, a kinase domain, an autoinhibitory domain, and a calmodulin-like domain; among these, the N-terminal domain shows the highest sequence divergence among CDPKs. Several CDPKs are known to be involved in phytohormone signaling, such as abscisic acid (Choi et al., 2005; Mori et al., 2006; Zhu et al., 2007), ethylene (Ludwig et al., 2005), gibberellins (Ishida et al., 2008), auxin (Lanteri et al., 2006), and brassinosteroid (Yang and Komatsu, 2000), as well as in plant development (Ivashuta et al., 2005; Yoon et al., 2006). Furthermore, a growing body of evidence has identified roles of CDPKs in abiotic and biotic stress responses in plants. CPK10 and CPK23 are both involved in drought tolerance in Arabidopsis (Ma and Wu, 2007; Zou et al., 2010). Tobacco (Nicotiana tabacum) CDPK2 and CDPK3 function in pathogen-induced hypersensitive response (Romeis et al., 2000, 2001). CPK1 confers resistance to fungal and bacterial pathogens in Arabidopsis (Coca and San Segundo, 2010). CDPKs are activated by the bacterial elicitor, flg22, and CPK4/11 and CPK5/6 were found to be important for the resistance of Arabidopsis to Pseudomonas syringae pv tomato DC3000 in a MAPK-independent manner (Boudsocq et al., 2010). Potato (Solanum tuberosum) CDPK4 and CDPK5 play a critical role in stress-induced oxidative burst (Kobayashi et al., 2007). In Medicago truncatula CDPK1 is important for root development and both CDPK1 and CPK3 are involved in nodulation (Ivashuta et al., 2005; Gargantini et al., 2006).
Although very little is known about how CDPKs function in plant defense against herbivores, emerging evidence has pointed to the roles of CDPKs in wounding- and herbivory-induced reactions: CDPKs were found to have elevated transcript levels after wounding in maize (Zea mays) and tobacco, and after herbivore attack in N. attenuata (Yoon et al., 1999; Szczegielniak et al., 2005; Wu et al., 2007). Furthermore, CPK3 and CPK13 regulate the transcript accumulation of several stress-related genes in Arabidopsis after Spodoptera littoralis feeding (Kanchiswamy et al., 2010).
Here we show that two CDPKs in N. attenuata, NaCDPK4 and NaCDPK5, play redundant roles in negatively controlling wounding- and herbivory-elicited JA accumulation. N. attenuata silenced in NaCDPK4 and NaCDPK5 showed greatly elevated levels of secondary metabolites that function as defenses against the specialist herbivore, M. sexta. We found that the greatly elevated JA levels in NaCDPK4- and NaCDPK5-silenced plants also activated SIPK in a COI1-dependent manner.
RESULTS
NaCDPK4 and NaCDPK5 Are Expressed Mainly in Stems and Reproductive Organs and Are Induced by Wounding and Simulated Herbivory
In tobacco, sequence analysis of NtCDPK4 and NtCDPK5 indicated that they are CDPKs; further in vitro biochemical assays also confirmed that their kinase activity is dependent on Ca2+ (Wang et al., 2005; Zhang et al., 2005). We cloned NaCDPK4 and NaCDPK5 from N. attenuata and an unrooted neighbor-joining tree was constructed to examine the phylogenetic relationship of their deduced protein sequences with other CDPKs (Supplemental Fig. S1). NaCDPK4 and NaCDPK5 closely clustered with NtCDPK4 and NtCDPK5 in tobacco, respectively, indicating that they are close homologs of these tobacco CDPKs. Although with weak bootstrap support, they were in the same clade with Arabidopsis AtCPK16, AtCPK18, and AtCPK28, which belong to subgroup IV of CDPKs (Cheng et al., 2002). Searching public Nicotiana EST databases on National Center for Biotechnology Information and The Institute for Genomic Research Plant Transcript Assembly BLAST Server did not reveal any close homologs of NaCDPK4 and NaCDPK5, suggesting that Nicotiana may have only CDPK4 and CDPK5 in subgroup IV. Importantly, NaCDPK4 and NaCDPK5 showed high similarities on amino acid (89%) and nucleotide sequence (85%) level (Supplemental Fig. S2), suggesting that these two kinases may have derived from a common ancestral gene by gene duplication.
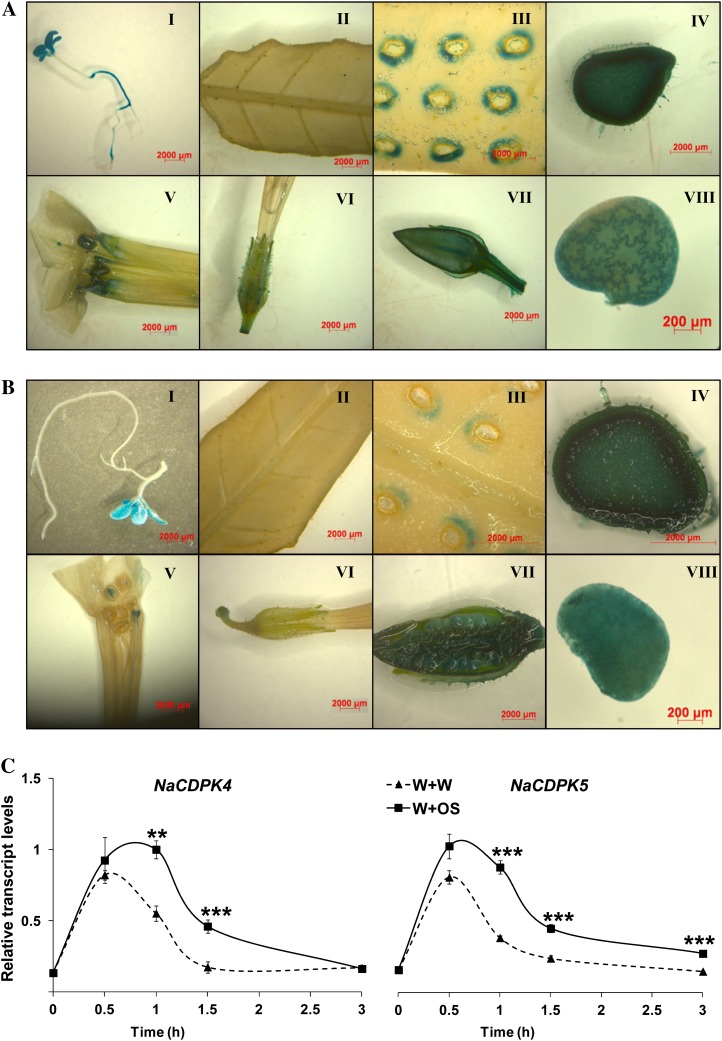
To further understand the function of these CDPKs, the promoters (about 1.3 kb including 5′-untranslated region) of NaCDPK4 and NaCDPK5 were isolated, and N. attenuata plants were transformed with constructs carrying promoters fused to GUS reporter gene to form NaCDPK4Pro:GUS and NaCDPK5Pro:GUS plants, respectively. At the seedling stage, both NaCDPK4 and NaCDPK5 were found to be expressed in cotyledons and true leaves, whereas NaCDPK4 was weakly expressed in hypocotyls and strongly in roots, no detectable levels of NaCDPK5 were found in these organs (Fig. 1, A and B). Staining was barely detectable in leaves of NaCDPK4Pro:GUS and NaCDPK5Pro:GUS plants. However, after being wounded, highly increased activity of NaCDPK4 and NaCDPK5 promoter was detected in the regions in vicinity of the wounds (Fig. 1, A and B). Furthermore, NaCDPK4 and NaCDPK5 were strongly expressed in stems, seed capsules, and developing seeds. GUS staining was also observed in the anthers, stigmas, and sepals of NaCDPK4Pro:GUS and NaCDPK5Pro:GUS plants (Fig. 1, A and B). The trichomes of NaCDPK4Pro:GUS plants also showed strong activity but not the trichomes of NaCDPK5Pro:GUS plants (Supplemental Fig. S3). Therefore, NaCDPK4 and NaCDPK5 might have functions in regulating the development of stems and reproductive organs, and may be involved in responses to wounding and herbivore attack.
Figure 1.
NaCDPK4 and NaCDPK5 are expressed in specific tissues and are induced by wounding and simulated herbivore feeding. Constructs harboring the promoters of NaCDPK4 and NaCDPK5 fused with GUS reporter gene were transformed into N. attenuata to create NaCDPK4Pro:GUS and NaCDPK5Pro:GUS plants, respectively. GUS histochemical assays were performed to examine the expression of NaCDPK4 (A) and NaCDPK5 (B) in seedlings (I), leaves (II), wounded leaves (III), stems (IV), flowers (V and VI), young seed capsules (VII), and developing seeds (VIII). N. attenuata was wounded with a pattern wheel and 20 μL of water (W + W) or M. sexta OS (W + OS) were immediately applied to wounds and relative transcript levels (mean ± se) of NaCDPK4 and NaCDPK5 (C) were determined with qRT-PCR in samples harvested at indicated times. Asterisks indicate significant differences between W + W- and W + OS-treated samples (n = 5; Student’s t test; **, P < 0.01; ***, P < 0.001).
To quantitatively examine whether wounding and herbivore feeding induce the accumulation of NaCDPK4 and NaCDPK5 transcripts, N. attenuata leaves were wounded with a fabric pattern wheel and 20 μL of either M. sexta OS (W + OS) or water (W + W) were immediately applied to the wounds to mimic M. sexta feeding or to generate mechanical wounding. Transcript levels of both genes were rapidly induced by both treatments: NaCDPK4 and NaCDPK5 mRNA levels increased 5- and 3-fold, respectively, 30 min after W + W treatment; in comparison, slightly higher levels of NaCDPK4 and NaCDPK5 transcripts were found in W + OS-treated plants (Fig. 1C).
Simultaneously Silencing NaCDPK4 and NaCDPK5 Impairs Plant Development
To study the function of NaCDPK4 and NaCDPK5, we used the RNA interference (RNAi) approach to obtain gene-silenced plants. CDPKs have similar kinase, autoinhibitory, and calmodulin-like domains, but their N-terminal variable domains show little similarities. Therefore, partial sequences from the 5′ of NaCDPK4 (201 bp) and NaCDPK5 (251 bp) open reading frame, where NaCDPK4 and NaCDPK5 have the greatest sequence difference (Supplemental Fig. S2), were cloned into a binary vector in an inverted repeat orientation to create RNAi constructs, pRESC5-CDPK4 and pRESC5-CDPK5, respectively. Agrobacterium-mediated transformation was used to transform these constructs into N. attenuata to produce the IRcdpk4 and IRcdpk5 lines. In the T1 generation, eight out of 10 independently transformed IRcdpk5 lines showed retarded growth and decreased seed production, whereas all IRcdpk4 plants of T1 generation were morphologically and developmentally similar to wild type. Two homozygous and independently transformed T2 lines of IRcdpk4 and IRcdpk5 were used for further studies.
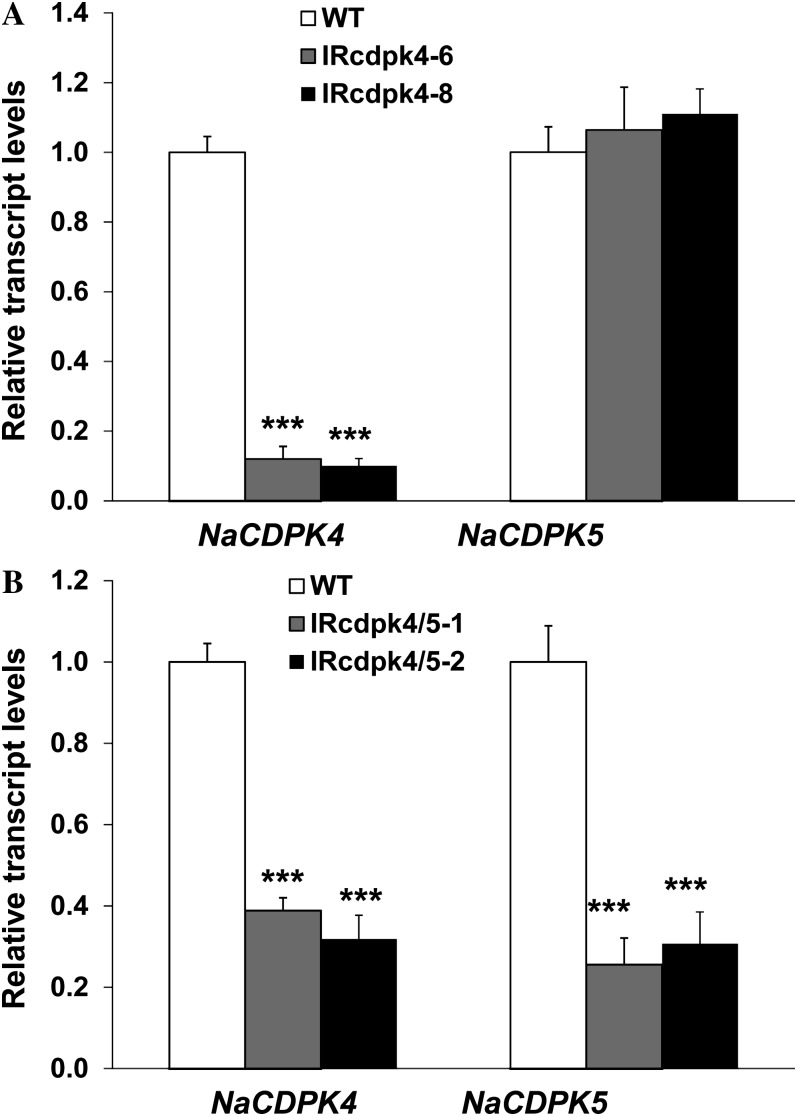
Quantitative real-time (qRT)-PCR was used to examine whether IRcdpk4 and IRcdpk5 lines had reduced transcript levels of NaCDPK4 and NaCDPK5. IRcdpk4 lines specifically silenced NaCDPK4 (approximately 90% decreased) with no changes in NaCDPK5 transcript levels (Fig. 2A). Unexpectedly, in IRcdpk5 lines, not only was NaCDPK5 (approximately 70% decreased) silenced but NaCDPK4 (approximately 60% decreased) was also cosilenced (Fig. 2B). Thus, IRcdpk5 was renamed IRcdpk4/5. Consistent with the large sequence distance between CDPKs in subgroup IV and those in other groups, qRT-PCR analyses indicated that IRcdpk4/5 did not have decreased levels of any other known CDPKs (NaCDPK1, NaCDPK2, and NaCDPK8) in N. attenuata (Supplemental Fig. S4; note: unlike tetraploid tobacco, the diploid N. attenuata genome has only one close homolog of NtCDPK2 and NtCDPK3, which is named NaCDPK2 here).
Figure 2.
NaCDPK4 is specifically silenced in IRcdpk4 plants, but both NaCDPK4 and NaCDPK5 are knocked down in IRcdpk4/5 plants. Relative transcript levels (mean ± se) of NaCDPK4 and NaCDPK5 in (A) wild type (WT) and IRcdpk4 (lines IRcdpk4-6 and IRcdpk4-8), and in (B) WT and IRcdpk4/5 (lines IRcdpk4/5-1 and IRcdpk4/5-2). Asterisks indicate significant differences between WT and IRcdpk4 or IRcdpk4/5 plants (n = 5; Student’s t test; ***, P < 0.001). Note: The levels of NaCDPK4 and NaCDPK5 in WT are designated to 1.
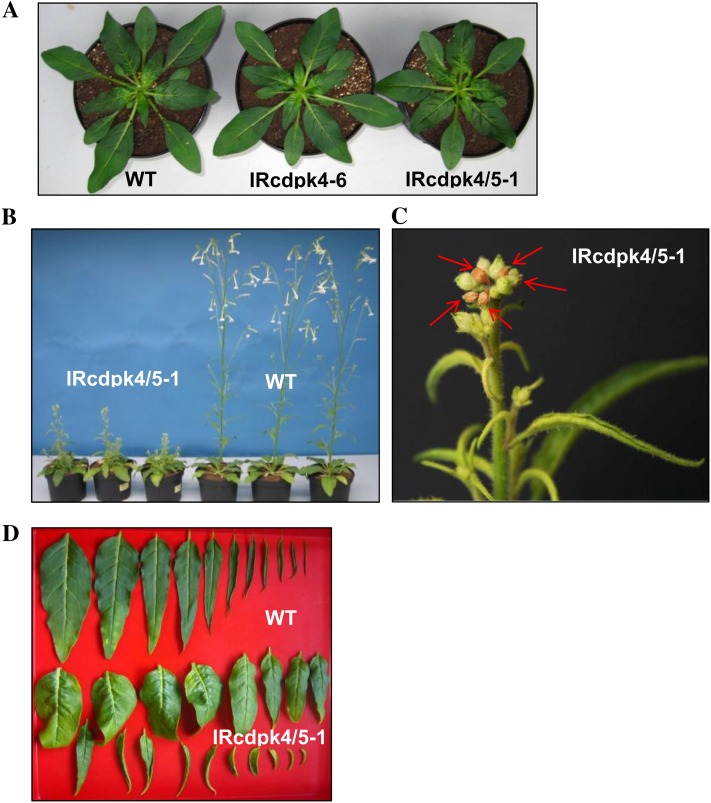
Similar to the T1 plants, the T2 generation showed no developmental differences between IRcdpk4 and wild type (Supplemental Fig. S5). IRcdpk4/5 was morphologically similar to wild type before bolting, although their rosette diameters were slightly smaller (Fig. 3A); however, during bolting, these plants showed highly stunted stems, and during the late elongation stage, IRcdpk4/5 showed defects in apical dominance, which resulted in multiple inflorescences and abortion of many stem leaf primordia (Fig. 3, B and C). The rosette leaves curled downwardly and most rosette leaves showed deformations probably due to uneven growth among different regions; in contrast, stem leaves were hyponastic (Fig. 3D). IRcdpk4/5 flowered much later than did wild type and during the flowering stage IRcdpk4/5 aborted more than 90% of its floral primordia and thus produced much less seeds than did wild type. All further experiments were therefore performed with rosette-staged plants.
Figure 3.
IRcdpk4/5 plants exhibit various growth defects. Plants were germinated at the same time and cultivated concurrently. A, Wild type (WT), IRcdpk4 (line IRcdpk4-6), and IRcdpk4/5 (line IRcdpk4/5-1) at rosette stage (30 d old). B, WT and IRcdpk4/5-1 at flowering stage (53 d old). C, IRcdpk4/5 plants aborted most of their leaf primordia during bolting. Red arrows indicate abscised and dried-out leaf primordia (53-d-old plant). D, Leaf morphology of WT (row 1) and IRcdpk4/5-1 (rows 2 and 3) plants (45 d old). Please note the curly rosette leaves (row 2) and the hyponastic stem leaves (row 3) of IRcdpk4/5.
IRcdpk4/5 Plants Have Substantially Elevated Levels of Wounding- and Simulated Herbivory-Induced JA
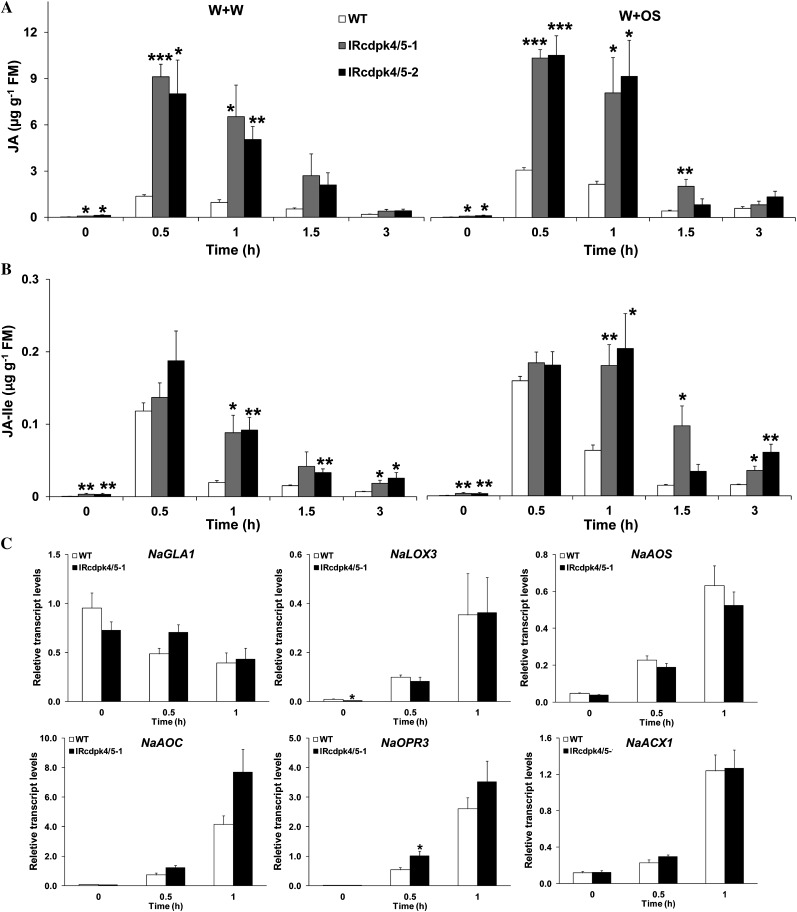
Given that JA plays a central role in plant defense against herbivores, we determined whether IRcdpk4/5 and IRcdpk4 plants have altered JA levels in response to wounding and herbivory. Plants were treated with W + W and W + OS and the JA contents were quantified (Fig. 4A). Before the treatments, IRcdpk4/5 already had 2- to 3-fold higher JA levels than did wild type (30, 90, and 120 ng/g JA in wild type, IRcdpk4/5-1, and IRcdpk4/5-2, respectively). By 30 min, W + W- and W + OS-treated wild-type plants accumulated 1,360 and 3,260 ng/g JA, indicating that N. attenuata recognized the FAC elicitors in OS and accumulated higher levels of JA to counteract herbivore attack (Halitschke et al., 2001; Wu et al., 2007). Strikingly, 8,500 and 11,100 ng/g JA (averages of two independent lines) were detected in W + W- and W + OS-elicited IRcdpk4/5 plants 30 min after treatments (Fig. 4A); these values are 5.25- and 2.4-fold (after W + W and W + OS, respectively) higher than those observed in wild-type plants. Similarly, 1 h after W + W and W + OS induction, the contents of the active form of jasmonates, JA-Ile, in IRcdpk4/5 were 4.5-fold and 3-fold higher than those in wild type, respectively (Fig. 4B). In contrast, both IRcdpk4 lines showed similar levels of JA and JA-Ile as those in wild type after W + W and W + OS treatments (Supplemental Fig. S6). Therefore, we inferred that silencing NaCDPK4 and NaCDPK5, but not NaCDPK4 alone, leads to dramatically elevated JA levels in response to wounding and herbivory. JA level is usually antagonized by SA (Spoel et al., 2003). However, we found that SA levels tended to be somewhat higher in IRcdpk4/5 lines than in wild-type plants (Supplemental Fig. S7), ruling out the possibility that the high JA levels in IRcdpk4/5 resulted from low SA contents.
Figure 4.
IRcdpk4/5 plants have dramatically increased JA and JA-Ile levels after wounding and simulated herbivory treatment. Wild type (WT) and IRcdpk4/5 (line IRcdpk4/5-1 and IRcdpk4/5-2) were wounded with a pattern wheel, and 20 μL of water (W + W) or M. sexta OS (W + OS) were immediately applied to wounds. Contents (mean ± se) of JA (A) and JA-Ile (B) and relative transcript levels (mean ± se) of genes involved JA biosynthesis (C) were determined in W + OS-treated samples after indicated times. Asterisks indicate significant differences between WT and IRcdpk4/5 plants (n = 5; Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
To further examine whether the greatly elevated W + W- and W + OS-induced JA accumulations in IRcdpk4/5 plants were associated with altered transcript levels of genes in the JA biosynthesis pathway, the transcript abundance of GLYCEROLIPASE A1, NaLOX3, NaAOS, NaAOC, NaOPR3, and NaACX1 was quantified by qRT-PCR. Compared with those in wild type, IRcdpk4/5-1 plants had similar levels of these genes before and either 0.5 or 1 h after W + OS treatment, except that 1 h after W + OS treatment the levels of NaAOC tended to be higher in irCDPK4/5 than in wild type (Fig. 4C). Given the rapid nature of JA biosynthesis and that the large differences in JA contents between wild-type and irCDPK4/5 plants appeared as early as 0.5 h after W + OS treatment, it is very unlikely that the elevated NaAOC levels in 1 h W + OS-treated irCDPK4/5 plants accounted for their high JA contents.
Thus, the large difference in W + W- and W + OS-induced JA levels between IRcdpk4/5 and wild-type plants probably resulted from modification of certain enzymes in the JA biosynthesis pathway at the posttranscriptional level, such as enzyme abundance and/or activity.
NaCDPK4 and NaCDPK5 Redundantly and Negatively Regulate Wounding- and Herbivory-Induced JA Accumulation
IRcdpk4 plants showed no growth phenotype and after wounding and simulated herbivory similar levels of JA were found in IRcdpk4 and wild type; in contrast, simultaneously silencing NaCDPK4 and NaCDPK5 in IRcdpk4/5 resulted in stunted growth and dramatically elevated JA levels in wounding- and herbivory-induced plants. Therefore, either NaCDPK5 itself is an important negative regulator of JA accumulation or these two CDPKs play redundant roles.
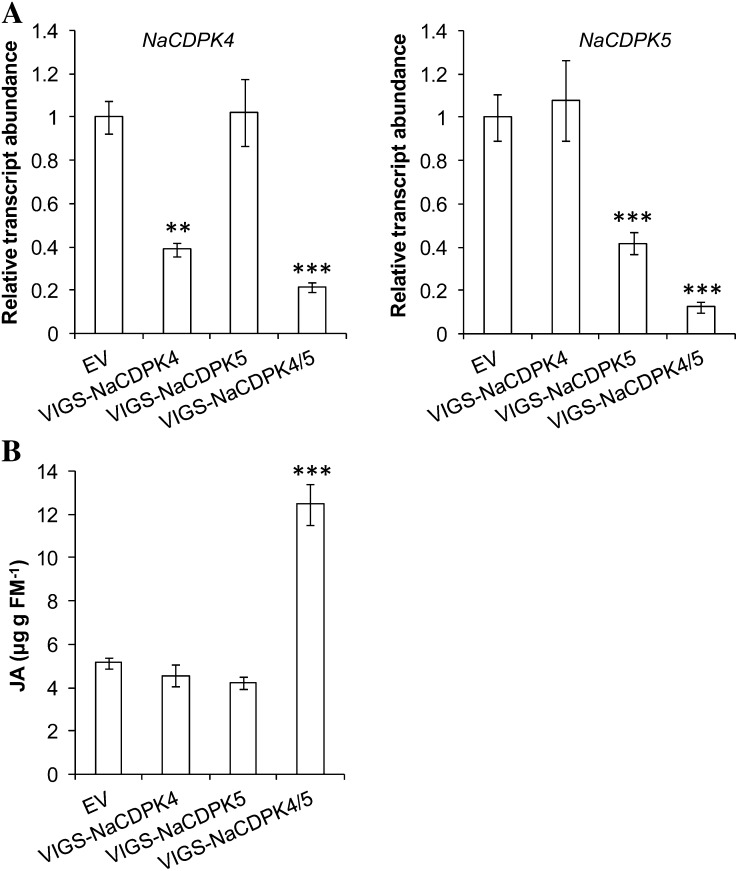
To examine whether these two CDPKs have a redundant function in JA regulation, we sought to create plants whose NaCDPK4 and NaCDPK5 were specifically silenced. To avoid the lengthy procedure required for creating stably transformed N. attenuata and the low success rate of transformation when both NaCDPK4 and NaCDPK5 were knocked down, a virus-induced gene silencing (VIGS) system was used (Ratcliff et al., 2001; Saedler and Baldwin, 2004). Constructs pTV-NaCDPK4 and pTV-NaCDPK5 were created to target the first 190 bp encoding the N termini of NaCDPK4 and NaCDPK5, respectively, where NaCDPK4 and NaCDPK5 showed the highest sequence difference; another VIGS construct (pTV-NaCDPK4/5) was prepared to silence both NaCDPK4 and NaCDPK5 using a region that shared high sequence similarity between these two kinases (Supplemental Fig. S8). Young N. attenuata plants were inoculated with Agrobacteria harboring these constructs to form VIGS-NaCDPK4, VIGS-NaCDPK5, and VIGS-NaCDPK4/5, and plants inoculated with the empty vector (EV) pTV00 served as comparisons. Using gene-specific primers, the transcript levels of NaCDPK4 and NaCDPK5 were determined. VIGS-NaCDPK4 plants showed unaltered NaCDPK5 levels but reduced NaCDPK4 transcripts by 62%; NaCDPK5 transcript levels were reduced by 58% in VIGS-NaCDPK5 plants without influencing NaCDPK4 transcript accumulations (Fig. 5A). VIGS-NaCDPK4/5 plants had attenuated levels of both NaCDPK4 and NaCDPK5 (78% and 88%, respectively; Fig. 5A). Notably, the pTV-NaCDPK4/5 construct had greater silencing efficiencies for NaCDPK4 and NaCDPK5 than did pTV-NaCDPK4 and pTV-NaCDPK5. This was likely due to the longer target sequence used in the transformation construct (Thomas et al., 2001). Compared with EV, VIGS-NaCDPK4 and VIGS-NaCDPK5 showed no obvious morphological changes, except that they were slightly shorter (Supplemental Fig. S9). We speculated that under the relatively low temperatures required for VIGS, NaCDPK4 and NaCDPK5 may have a minor function in stem elongation. Importantly, VIGS-NaCDPK4/5 plants were semidwarf, had more branches, and aborted many of their floral buds and flowers (Supplemental Fig. S9). These developmental phenotypes of VIGS-NaCDPK4/5 are highly reminiscent of those of IRcdpk4/5.
Figure 5.
NaCDPK4 and NaCDPK5 have redundant roles in controlling herbivory-induced JA levels. Agrobacterium carrying pTV00 (EV), pTV-NaCDPK4, pTV-NaCDPK5, and pTV-NaCDPK4/5 was inoculated into N. attenuata to create EV, VIGS-NaCDPK4, VIGS-NaCDPK5, and VIGS-NaCDPK4/5, respectively. A, Relative transcript levels (mean ± se) of NaCDPK4 and NaCDPK5 in untreated plants. Note: Levels NaCDPK4 and NaCDPK5 in EV are designated to 1. B, JA contents (mean ± se) in plants after simulated herbivore feeding. Plants were wounded with a pattern wheel and 20 μL of M. sexta OS were applied to wounds and samples were collected 1 h after treatment.
We further determined the JA levels in these plants, which were treated with W + W or W + OS 1 h before sample harvesting. Contents of JA in VIGS-NaCDPK4 and VIGS-NaCDPK5 plants were not significantly different from those in EV; in contrast, VIGS-NaCDPK4/5 showed around 1.3-fold greater JA levels (Fig. 5B). Thus, both NaCDPK4 and NaCDPK5 play redundant roles in negative regulating the JA levels in N. attenuata in response to herbivore feeding.
IRcdpk4/5 Plants Have Augmented Contents of Defensive Compounds and Increased Resistance to M. sexta
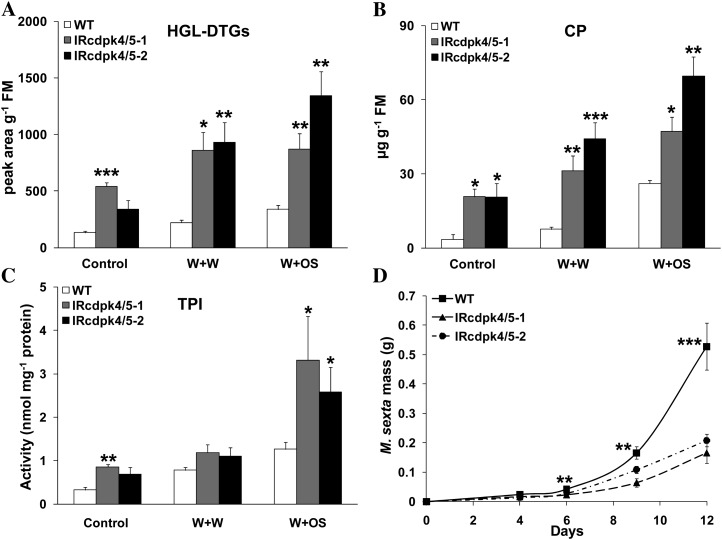
In N. attenuata, herbivore attack enhances the levels of JA and induces the accumulation of 17-hydroxygeranyllinalool diterpene glucosides (HGL-DTGs), TPIs, and CP, which are important plant secondary metabolites and serve as direct defenses against herbivores (Zavala et al., 2004; Paschold et al., 2007; Heiling et al., 2010; Kaur et al., 2010). To determine whether the highly increased JA contents in IRcdpk4/5 plants result in greater levels of defensive compounds and confer higher resistance to the specialist insect herbivore M. sexta, we applied W + W and W + OS treatment to wild type and IRcdpk4/5 and the concentrations of HGL-DTGs and CP and the activity of TPIs were quantified in samples harvested 3 d after treatments.
Even when not treated, compared with those in wild-type plants, IRcdpk4/5 plants had about 1- to 2-fold higher concentrations of HGL-DTGs, CP, and levels of TPI activity. After both W + W and W + OS treatments, 1- to 3-fold greater levels of HGL-DTGs, CP, and activity of TPIs were detected in IRcdpk4/5 plants (Fig. 6, A–C). Bioassays were performed to determine whether IRcdpk4/5 plants have increased resistance to M. sexta larvae. Consistent with the high contents of W + OS-induced defensive secondary metabolites, after feeding for 12 d, M. sexta larvae gained 60% less mass on IRcdpk4/5 than on wild type (Fig. 6D). In contrast, after W + W or W + OS treatment, no differences in the levels of defensive compounds (HGL-DTGs, CP, and TPIs) were found between wild type and IRcdpk4, and M. sexta growth rates were not affected by silencing NaCDPK4 alone (Supplemental Fig. S10).
Figure 6.
IRcdpk4/5 plants have elevated defensive secondary metabolites and enhanced defense against M. sexta. Wild type (WT) and IRcdpk4/5 (line IRcdpk4/5-1 and IRcdpk4/5-2) were wounded with a pattern wheel, and 20 μL of water or M. sexta OS were immediately applied to wounds (W + W and W + OS, respectively). Contents (mean ± se) of HGL-DTGs (A) and CP (B), and activity (mean ± se) of TPIs (C) were determined in samples harvested 3 d after treatments (nontreated plants served as controls). D, Masses (mean ± se) of M. sexta larvae grown on WT and IRcdpk4/5 plants. Asterisks indicate significant differences between WT and IRcdpk4/5 plants (n = 5 for A, B, and C; n = 30 for D; Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
The Increased Defense Levels of IRcdpk4/5 Plants Are Completely Dependent on JA and JA Signaling
Other pathways in addition to JA signaling may also confer resistance to M. sexta in IRcdpk4/5 plants. To examine this possibility, IRcdpk4/5-1 was crossed with IRcoi1 (silenced in COI1, the receptor for JA-Ile; Paschold et al., 2007) to abolish JA signaling (IRcdpk4/5-1 × IRcoi1 plants). Moreover, IRcdpk4/5-1 line was also crossed with a line ectopically expressing an Arabidopsis JASMONIC ACID CARBOXYL METHYLTRANSFERASE (ovJMT; Seo et al., 2001), which converts JA to methyl jasmonate, an inactive form of jasmonate in N. attenuata (Stitz et al., 2011; IRcdpk4/5-1 × ovJMT plants). Notably, the morphologies of both IRcdpk4/5-1 × IRcoi1 and IRcdpk4/5-1 × ovJMT plants were indistinguishable from those of IRcoi1 and ovJMT, respectively, including having similar stem lengths to those of IRcoi1 and ovJMT, normal leaf shape, and partial male sterility due to deficiencies in JA signaling and JA accumulation (M. Heinrich, C. Hettenhausen, I.T. Baldwin, and J. Wu, unpublished data). Thus, the developmental defects in IRcdpk4/5 resulted solely from overproduction of JA rather than other potential pathways regulated by NaCDPK4 and NaCDPK5.
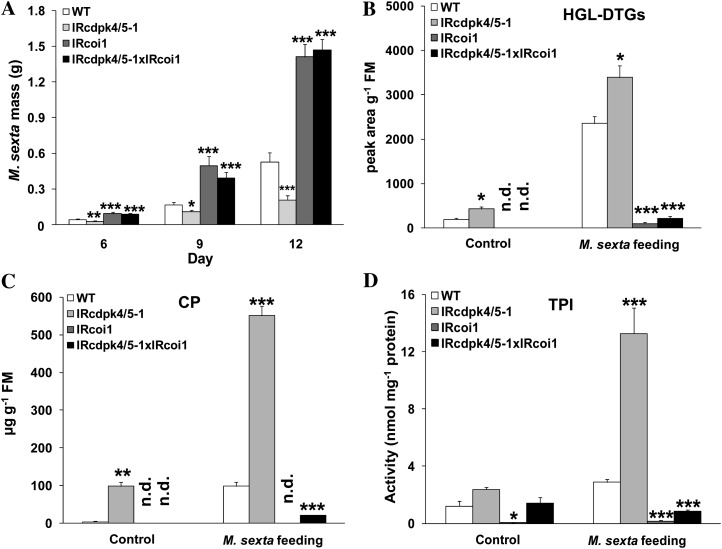
M. sexta bioassays were performed on wild-type, IRcdpk4/5-1, IRcdpk4/5-1 × IRcoi1, and IRcoi1 plants. We found that the growth rates of M. sexta on IRcdpk4/5-1 × IRcoi1 plants were very similar to those reared on IRcoi1 and after 9 d of feeding, M. sexta larvae fed on IRcoi1 and IRcdpk4/5-1 × IRcoi1 were 2- and 1.4-times heavier than those on wild-type and IRcdpk4/5-1 plants, respectively (Fig. 7A). Analyzing the contents of HGL-DTGs, CP, and the activity of TPIs confirmed that silencing COI1 in IRcdpk4/5-1 abolished the accumulation of these defensive metabolites (Fig. 7, B–D). Comparable bioassay results were obtained from wild-type, IRcdpk4/5-1, IRcdpk4/5-1 × ovJMT, and ovJMT plants (Supplemental Fig. S11, A–D). The levels of JA in W + OS-treated IRcdpk4/5-1 × ovJMT confirmed that ectopic expression of JMT effectively abolished JA accumulation (Supplemental Fig. S11E).
Figure 7.
Silencing COI1 in IRcdpk4/5 plants abolishes herbivore defenses. IRcdpk4/5-1 was crossed with IRcoi1 to generate IRcdpk4/5-1 × IRcoi1. A, Masses of M. sexta larvae grown on wild type (WT), IRcdpk4/5-1, IRcoi1, and IRcdpk4/5-1 × IRcoi1. Concentrations (mean ± se) of HGL-DTGs (B) and CP (C), and activity (mean ± se) of TPIs (C) were quantified in plants that had been fed for 12 d. Asterisks indicate significant differences between WT and other plants (n = 30 for A; n = 5 for B, C, and D; t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). n.d., Not detected.
Thus, we concluded that the strong defense of IRcdpk4/5 against M. sexta was completely dependent on JA signaling and the highly enhanced JA levels.
Simultaneously Silencing NaCDPK4 and NaCDPK5 Increases the Levels of SIPK Activity Induced by Simulated Herbivory
In N. attenuata, tobacco, and tomato (Solanum lycopersicum), SIPK and WIPK (or their homologs) are required for JA accumulation in response to wounding and herbivory (Seo et al., 1999; Kandoth et al., 2007; Seo et al., 2007; Wu et al., 2007). To determine whether the increased JA levels in IRcdpk4/5 plants were associated with elevated MAPK activity, total proteins in wild-type and IRcdpk4/5-1 leaves that had been treated with W + W and W + OS were extracted and the MAPK activity levels were measured using an in-gel MAPK activity assay.
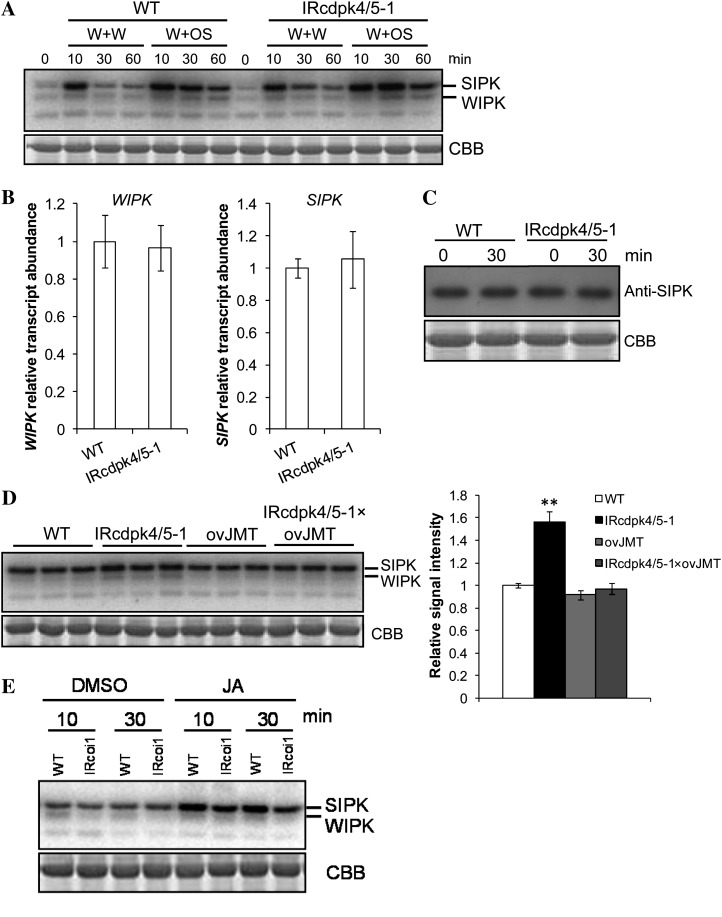
In wild-type plants, both W + W and W + OS rapidly activated SIPK and WIPK within 10 min of treatments (Fig. 8A). Thirty minutes after W + W treatments, SIPK activity declined to levels that were only slightly higher than those in nontreated plants; in contrast, strong SIPK activity was found in W + OS-treated wild-type plants even 60 min after W + OS treatment (Fig. 8A), indicating that N. attenuata recognized the FACs in M. sexta OS and responded with greater MAPK activity. Importantly, 30 and 60 min after both W + W and W + OS treatment, greater SIPK activity levels were seen in IRcdpk4/5-1 than in wild type (Fig. 8A). Elevated SIPK activity was also detected in line IRcdpk4/5-2, 30 and 60 min after W + OS (Supplemental Fig. S12A). Since biological replicates were pooled for these assays, in another independently performed experiment, we analyzed the kinase activity in four biological replicates that were harvested 30 min after W + OS treatment: Compared with those in wild type, both IRcdpk4/5-1 and IRcdpk4/5-2 showed elevated SIPK activity levels (Supplemental Fig. S12B). Notably, increased WIPK activity levels were observed in these kinase assays (Fig. 8A; Supplemental Fig. S12, A and B), although these could not be reliably quantified due to poor signal-noise ratios.
Figure 8.
IRcdpk4/5 plants have increased levels of SIPK activity caused by high JA accumulation. A, MAPK activity assay in wild-type (WT) and IRcdpk4/5-1 plants that were treated with W + W and W + OS. B, Transcript levels (mean ± se) of WIPK and SIPK in WT and IRcdpk4/5-1 plants 30 min after W + OS treatment (Note: Levels of SIPK and WIPK in WT are designated to 1). C, Protein-blotting analysis of SIPK protein abundance in WT and IRcdpk4/5-1 plants before and 30 min after W + OS treatment. D, MAPK activity levels in WT, IRcdpk4/5-1, ovJMT, and IRcdpk4/5-1 × ovJMT plants 30 min after being treated with W + OS (three biological replicates each). Right section: quantification of relative band intensities (mean ± se; the average intensity of WT samples was designated as 1); asterisks represent significant difference between WT and other plants (n = 3, Student’s t test; **, P < 0.01). E, Exogenous supplementation of JA to N. attenuata activates MAPKs. JA (200 μg/mL in 5% DMSO) or 5% DMSO was pressure infiltrated into N. attenuata WT or IRcoi1 leaves and the induced MAPK activity was determined in samples (pooled from three biological replicates) harvested at indicated times. CBB, Coomassie Brilliant Blue (photographs of the Rubisco large subunit that were visualized by staining the gels with CBB).
Two scenarios may account for the increased SIPK activity in IRcdpk4/5 plants: (1) NaCDPK4 and NaCDPK5 suppress the transcript abundance of SIPK and WIPK; (2) IRcdpk4/5 had normal levels of SIPK and WIPK protein but these kinases had a higher degree of phosphorylation/activity after wounding or herbivore attack. First, qRT-PCR was performed to determine the transcript levels of SIPK and WIPK after simulated herbivory treatment. No differences in SIPK and WIPK transcript levels between wild type and IRcdpk4/5 were found (Fig. 8B). Next, a SIPK-specific antibody (Zhang et al., 1998), Ab-p48C, was used to examine abundance of the SIPK protein in wild-type and IRcdpk4/5 plants. Similar levels of SIPK protein were found in IRcdpk4/5 and wild type, before and 30 min after W + OS treatment (Fig. 8C). This was also found in samples collected in another independently performed experiment (Supplemental Fig. S12C). Furthermore, the levels of SIPK protein in a line that had been stably silenced in SIPK by RNAi (IRsipk; Meldau et al., 2009) were almost undetectable, demonstrating the specificity of this antibody (Supplemental Fig. S12C). Given the similar transcript abundance of WIPK in wild-type and IRcdpk4/5 plants, it is likely that WIPK had also comparable protein levels in these plants. Therefore, silencing NaCDPK4 and NaCDPK5 leads to elevated activity of SIPK in plants challenged by herbivory through a pathway that regulates the phosphorylation levels of SIPK but not protein abundance.
Applying JA to Arabidopsis seedlings leads to a rapid activation of MPK6, which is the homolog of SIPK in this species (Takahashi et al., 2007). Thus, we speculated that the remarkably high levels of JA observed in IRcdpk4/5 plants might be the reason for the increased SIPK activity after wounding and simulated herbivore attack.
To examine this hypothesis, an in-gel kinase assay was conducted to measure MAPK activity in wild-type, IRcdpk4/5-1, ovJMT, and IRcdpk4/5-1 × ovJMT plants, which were treated with W + OS 30 min before harvesting. After crossing with ovJMT plants, SIPK activity levels in IRcdpk4/5-1 decreased to those in wild type and ovJMT (Fig. 8D). Similar results were obtained from W + OS-treated wild-type, IRcdpk4/5-1, IRcoi1, and IRcdpk4/5-1 × IRcoi1 plants (Supplemental Fig. S13A). To further examine the hypothesis that overaccumulated JA activates SIPK, we measured MAPK activity in the wounded dongle-D (dgl-D) mutant. In dgl-d, a chloroplast-localized galactolipase is overexpressed and thus this mutant has highly elevated JA contents after wounding (Hyun et al., 2008). Indeed, after wounding levels of MPK6 activity (the homolog of SIPK in Arabidopsis) in dgl-d plants were greater than in Columbia-0 (Col-0; Supplemental Fig. S13B), a result consistent with the substantially higher levels of JA in dgl-d (Supplemental Fig. S13C). In addition, we infiltrated a solution of JA (200 μg/mL in 5% dimethyl sulfoxide [DMSO]) into N. attenuata leaves and measured MAPK activity. Compared with those in solvent (5% DMSO)-inoculated plants, SIPK activity was significantly increased in JA-treated wild-type plants, whereas in IRcoi1 plants lower SIPK activity was found both 10 and 30 min after JA infiltration (Fig. 8E). All these results are consistent with the notion that highly elevated JA contents elicit the activation of SIPK and that COI1 is required for this activation pathway.
SIPK and WIPK Are Required for Induced JA Accumulation in IRcdpk4/5 Plants
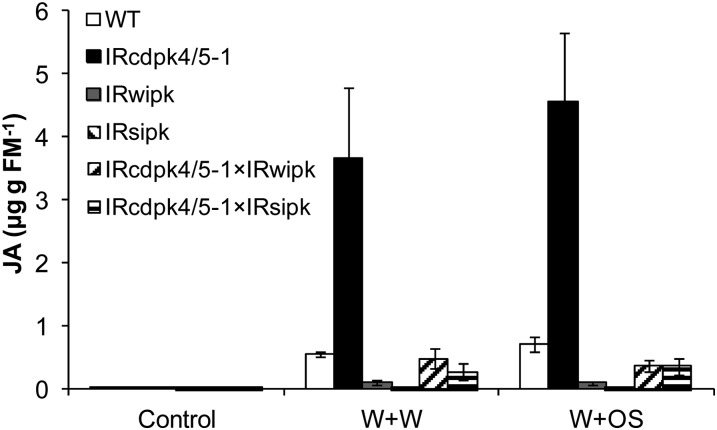
Overproduction of JA in IRcdpk4/5 plants results in increased SIPK and WIPK activity. However, previous studies also indicated that these MAPKs are needed for wounding- and herbivory-elicited JA accumulation (Kandoth et al., 2007; Wu et al., 2007). To examine whether SIPK and WIPK are still necessary for JA accumulation in IRcdpk4/5, IRcdpk4/5-1 was crossed with IRsipk and IRwipk (Meldau et al., 2009) to create IRcdpk4/5-1 × IRsipk and IRcdpk4/5-1 × IRwipk, respectively. After W + W and W + OS induction, the JA contents were quantified. By 30 min, both W + W and W + OS elicited more than 4-fold higher levels of JA in IRcdpk4/5-1 than in wild type, but silencing SIPK and WIPK in IRcdpk4/5-1 plants greatly reduced their JA contents (Fig. 9). From these results we conclude that the high levels of JA in IRcdpk4/5 plants elicited by wounding and herbivory stress require SIPK and WIPK activity.
Figure 9.
Knocking down SIPK or WIPK in IRcdpk4/5 plants decreases wounding- and simulated herbivory-induced JA levels. IRcdpk4/5-1 was crossed with IRwipk and IRsipk to create IRcdpk4/5-1 × IRwipk and IRcdpk4/5-1 × IRsipk, respectively. Plants were wounded with a pattern wheel and 20 μL of water or M. sexta OS were applied to wounds immediately (W + W and W + OS, respectively). Samples were harvested after 30 min and the JA contents (mean ± se) were determined on an HPLC-tandem mass spectrometer (n = 5).
DISCUSSION
JA is the most important hormone in regulating plant defense against herbivores (Wu and Baldwin, 2009). As in many other physiological processes, the transient accumulations of JA that elicit defense signaling are a result of a combination of biosynthesis and degradation, and the enzymes involved in JA anabolism or catabolism are likely controlled by both positive and negative signaling pathways: After herbivory, enhanced JA accumulation is important for increasing defense; however, negative regulatory pathways that suppress accumulation of JA are also needed to avoid overly high JA levels, which compromise plant development and growth. Although almost all the enzymes for JA biosynthesis have been cloned (Delker et al., 2006), the signaling pathways that control the production of JA remain largely unknown. Here we show that silencing two CDPKs, NaCDPK4 and NaCDPK5, resulted in remarkably high levels of JA in wounded or herbivore attacked N. attenuata. The high accumulations of JA not only conferred increased resistance to attack from M. sexta larvae but also resulted in enhanced SIPK and WIPK activity.
CDPKs and JA Accumulations
Tomato AOC, which encodes one of the important JA biosynthetic enzymes, is expressed in vascular tissues and reproductive organs (Stenzel et al., 2003, 2008) and plants deficient in JA biosynthesis or signaling have defects in fertility (Feys et al., 1994); thus JA biosynthesis is thought to be mainly in these tissue and organ types. NaCDPK4 and NaCDPK5 are mainly expressed in stems and reproductive organs and consistently, IRcdpk4/5 plants had stunted stems and aborted flower buds (Fig. 3). These phenotypes were consistent with their functions in (negatively) regulating JA biosynthesis: The overaccumulated JA in IRcdpk4/5 resulted in developmental abnormalities and remarkably high JA contents after wounding or herbivory. Furthermore, the developmental phenotype of IRcdpk4/5 is largely similar to the dgl-d mutant, which accumulates JA to exceptionally high levels and is stunted in stem elongation, and decreased in apical dominance and fertility (Hyun et al., 2008). These data are consistent with the finding that these NaCDPK4 and NaCDPK5 are involved in controlling JA accumulation.
However, how NaCDPK4 and NaCDPK5 function as strong suppressors of stress-induced JA accumulation is unclear. Transcriptional analysis revealed wild-type transcript levels of all important enzymes involved in JA biosynthesis in IRcdpk4/5. Although after wounding or simulated herbivore feeding, the JA contents in IRcdpk4/5 were remarkably higher than in wild type, the dynamics of JA were similar among these plants: Both reached their highest levels 0.5 h after either treatment and declined to almost basal levels after 3 h (Fig. 4). This implies that the overaccumulation of JA in IRcdpk4/5 probably did not result from impaired JA metabolism but from the enhanced JA biosynthesis activity. Due to the complexity of JA metabolism (many enzymes and rapid turnover of intermediate precursors of JA, and the less-known mechanisms of JA degradation), we were not able to determine the reason for the high JA accumulation in IRcdpk4/5 plants at an enzyme activity level. Tobacco NtCDPK5 was found to localize in cell membranes, and NtCDPK4 localizes in cytoplasm and nuclei (Wang et al., 2005). Although not totally ruled out, it is unlikely that these kinases enter chloroplasts or peroxisomes and directly phosphorylate JA biosynthesis enzymes to inhibit their activity.
In N. attenuata, a number of proteins have been identified that affect herbivory-induced JA accumulation. Knocking down BRI1-ASSOCIATED RECEPTOR KINASE1, SUPPRESSOR OF G-TWO ALLELE OF SKP1, S-NITROSOGLUTATHIONE REDUCTASE, SIPK, or WIPK compromises wounding- and herbivory-induced JA levels and the exact mechanisms by which these proteins influence JA biosynthesis remain unclear (Wu et al., 2007; Meldau et al., 2011; Wünsche et al., 2011; Yang et al., 2011). In Arabidopsis, in addition to dgl-d (Hyun et al., 2008), a mutation in cellulose synthase CeSA3 also results in increased production of JA and ethylene (Ellis et al., 2002). After wounding, Arabidopsis carrying a missense mutation in a Ca2+-permeable nonselective cation channel (the fou2 mutant) exhibits severalfold increased JA contents (Bonaventure et al., 2007). It was speculated that the high JA levels of fou2 mutant may result from altered cytosolic [Ca2+]. Wounding and herbivore feeding both induce changes in intracellular [Ca2+] (Maffei et al., 2004; Schäfer et al., 2011), suggesting that Ca2+ signaling is involved in plant responses to wounding and insect feeding. Studying whether NaCDPK4 and NaCDPK5 sense changes of [Ca2+] and how they translate Ca2+ signaling into altered activity of certain enzymes or transcription factors will provide important insight into how Ca2+ signaling modulates JA biosynthesis.
When COI1 was silenced in IRcdpk4/5-1 plants (IRcdpk4/5-1 × IRcoi1 plants), the highly elevated defense of IRcdpk4/5 was completely abolished and the performance of M. sexta larvae was equivalent to that on IRcoi1 plants (Fig. 7). We also obtained similar results when JA accumulation was suppressed by ectopically overexpressing JMT (Supplemental Fig. S11D). From these genetic analyses, we infer that although NaCDPK4 and NaCDPK5 might also function in other physiological processes, their involvement in plant-herbivore interactions is largely through JA signaling.
MAPKs and JA Accumulation
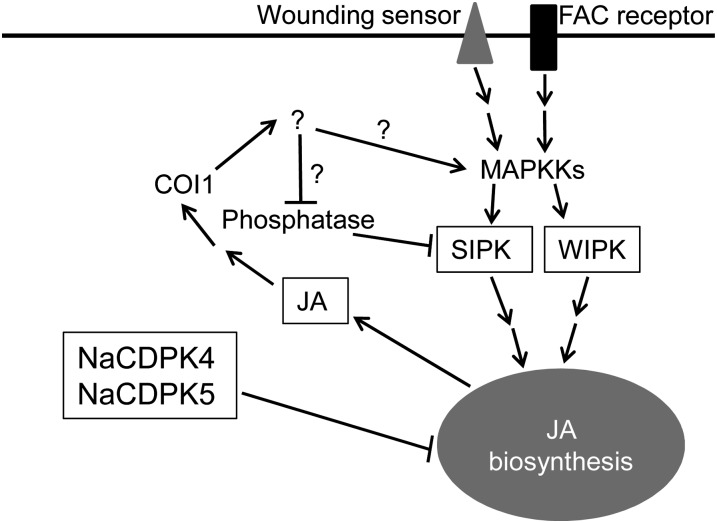
Our data indicated reciprocal interactions between MAPK and JA signaling. Silencing SIPK and WIPK in N. attenuata and their homologs in tomato, compromises wounding- and OS-elicited JA bursts (Kandoth et al., 2007; Wu et al., 2007), indicating that these MAPKs are required for JA biosynthesis. This is also supported by the fact that silencing SIPK and WIPK in IRcdpk4/5 plants greatly diminish W + W- and W + OS-induced JA levels. However, several lines of evidence also indicated that JA in high concentrations activates SIPK/MPK6. Treating Arabidopsis seedlings with exogenous JA activates MPK6 (Takahashi et al., 2007). After wounding, elevated levels of MPK6 activity were also found in the JA-overproducing dgl-d mutant compared with the levels found in wounded Col-0 plants (Supplemental Fig. S13B). Compared with wild-type plants, in IRcdpk4/5, after either W + W or W + OS, substantially larger quantities of JA were produced, which in turn resulted in elevated levels of SIPK activity. Consistently, directly infusing JA into N. attenuata enhanced the activity of SIPK. In IRcdpk4/5-1 × ovJMT plants, in which JA accumulation was compromised (because JA is rapidly converted to the inactive MeJA), SIPK activity levels were similar to those of wild type, confirming that the highly increased JA contents in IRcdpk4/5 plants were responsible for the overactivation of SIPK. Moreover, JA signaling is partly required for the activation of MPK6 by exogenously applied JA in Arabidopsis seedlings, given that the levels of JA-induced MPK6 activity were less in coi1 mutant than in wild type (Takahashi et al., 2007). Consistent with this observation, IRcdpk4/5-1 × IRcoi1 plants showed decreased SIPK activity levels compared with IRcdpk4/5-1 plants, and infiltrating JA directly into IRcoi1 resulted in lower SIPK activity levels than those in similarly treated wild type. All these data underscore that it is not JA itself but rather a signaling compound regulated by JA signaling that activates SIPK (Fig. 10).
Figure 10.
A working model summarizing early wounding- and herbivory-induced responses in N. attenuata. Wounding and FACs in M. sexta OS are perceived by wounding sensors and FAC receptors and in turn activate downstream MAPK signaling, including SIPK and WIPK. Using an unknown mechanism, NaCDPK4 and NaCDPK5 negatively control JA biosynthesis. Furthermore, when JA attains high concentrations, JA has a feedback function on SIPK activity through a COI1-mediated pathway by either activating the upstream MAPKKs (such as MEK2) or by reducing the activity of SIPK-specific phosphatases. Question marks indicate hypothetical signaling molecules or pathways.
Both qRT-PCR and protein-blotting analysis indicated that the increased SIPK (and likely WIPK) activity is not due to enhanced levels of transcripts or protein. We speculate that this JA-signaling-regulated compound can either activate upstream kinases of SIPK and WIPK, such as MEK2 (Yang et al., 2001; Heinrich et al., 2011), or decrease the activity of the phosphatases of SIPK and WIPK (Fig. 10). Compared with wild type, after W + OS treatment, ovJMT and IRcoi1 plants showed very little changes in SIPK activity (Fig. 8E; Supplemental Fig. S13A). Thus, when JA concentrations in plants are within physiological range (less than 3,000 ng/g fresh mass, levels commonly induced by W + OS in wild-type N. attenuata), this COI1-mediated pathway seems to have little impact on SIPK activity.
Redundant Roles of NaCDPK4 and NaCDPK5
NaCDPK4 and NaCDPK5 have a relatively high sequence similarity, suggesting that they may have evolved from a common ancestral gene. Their similar tissue- and induction-specific expression profiles also suggest that they may have similar functions. Compared with those in plants inoculated with Agrobacterium carrying pTV00 (EV), specifically silencing NaCDPK4 and NaCDPK5 using VIGS did not elevate W + OS-induced JA levels. With a construct that targets both NaCDPK4 and NaCDPK5, we created VIGS-NaCDPK4/5 plants. They exhibited largely similar phenotypes as those in IRcdpk4/5, such as stunted growth, abortion of flower primordia, and importantly, highly elevated W + OS-elicited JA levels. These results indicate that NaCDPK4 and NaCDPK5 function redundantly in suppressing the accumulation of JA. However, NaCDPK4 and NaCDPK5 have distinct N-terminal sequences, which are usually required for subcellular targeting. Wang et al. (2005) found that in onion (Allium cepa) epidermal cells, tobacco NtCDPK4 is localized in cytoplasm and nuclei, while NtCDPK5 localizes in cell membranes. Promoter activity analyses indicated that NaCDPK4 is expressed in roots and trichomes whereas NaCDPK5 is not. Thus, NaCDPK4 and NaCDPK5 may have distinct functions in other plant physiological processes apart from the regulation of JA biosynthesis.
Taken together, we show that two CDPKs in N. attenuata, NaCDPK4 and NaCDPK5, play an important role in suppressing wounding and OS-induced JA biosynthesis. Moreover, MAPK and JA signaling appear to have a complex interaction: SIPK is required for JA biosynthesis, while COI1-mediated JA signaling also activates SIPK, when cells experience very high JA levels (Fig. 10).
MATERIALS AND METHODS
Plant Growth, Sample Treatments, and Manduca sexta Growth Assays
Nicotiana attenuata (Solanaceae) seeds were from a line maintained in our laboratory that was originally collected in Utah and inbred for 30 generations in the greenhouse. Seed germination and plant cultivation followed Krügel et al. (2002). Seeds were germinated on petri dishes to synchronize their germination, and the seedlings were transferred to soil after 10 d. Four- to 5-week-old plants were used for all experiments except for growth observations.
For simulated herbivory treatments, leaves were wounded with a pattern wheel and M. sexta OS (20 μL of one-fifth diluted OS) were immediately rubbed onto wounded leaves (W + OS); for wounding treatment, leaves were wounded with a pattern wheel, and 20 μL of water were applied (W + W). Arabidopsis (Arabidopsis thaliana) Col-0 and dgl-d were grown under the long-day conditions, and rosette leaves of slightly elongated plants were wounded with a pattern wheel. For the JA infiltration experiment, a stock JA solution was prepared (2 mg/mL in 50% DMSO) and was 10-fold diluted before inoculating in to wild-type and IRcoi1 N. attenuata leaves using a 1-mL syringe; 5% DMSO was inoculated for comparisons. For M. sexta growth assays, freshly hatched neonates of M. sexta were placed on 30 replicated plants (one larva/plant), and the masses of these caterpillars were measured after different days of continuous feeding.
Generation of Transformed Plants, Plant Crossing, and VIGS
Partial sequences of NaCDPK4 and NaCDPK5 were cloned into pRESC5 vector in an inverted-repeat fashion (primer sequences are listed in Supplemental Table S1) to form pRESC5-CDPK4 and pRESC5-CDPK5. This vector was subsequently transformed into Agrobacterium tumefaciens (strain LBA4404) to transform N. attenuata (Krügel et al., 2002). The number of transferred DNA insertions was determined by Southern hybridization of genomic DNA using a PCR fragment of the HYGROMYCIN PHOSPHOTRANSFERASE (hptII) gene as a probe. Two T2 homozygous lines with single transferred DNA insertions were identified and used in subsequent experiments. Crossing IRcoi1 and ovJMT with IRcdpk5 plants was done by removing anthers from flowers of IRcoi1 and ovJMT plants before pollen maturation and pollinating the stigmas with pollen from IRcdpk4/5 plants.
For VIGS, partial sequences of NaCDPK4 and NaCDPK5 were cloned into pTV00 to form pTV-NaCDPK4, pTV-NaCDPK5, and pTV-NaCDPK4/5 using primers listed in Supplemental Table S1, and these plasmids were thereafter transformed into A. tumefaciens (strain GV3101). Twenty-five days after germination, following a procedure optimized for N. attenuata (Saedler and Baldwin, 2004), plants were inoculated with A. tumefaciens carrying these plasmids to create VIGS-NaCDPK4, VIGS-NaCDPK5, and VIGS-NaCDPK4/5, respectively. Plants silenced in PHYTOENE DESATURASE (NaPDS) were used to visually monitor the degree of VIGS, since these plants showed a photobleaching phenotype. About 14 d after inoculation, when the leaves of NaPDS-silenced plants were completely white, experiments were performed.
Histochemical GUS Assays
Sequences about 1.3-kb upstream of NaCDPK4 and NaCDPK5 coding sequences were isolated using a GenomeWalker kit (Clontech). Using primers listed in Supplemental Table S1, these sequences were cloned into pCAMBIA1301 vector (www.cambia.org) to create pCAM-NaCDPK4Pro:GUS and pCAM-NaCDPK5Pro:GUS. These binary vectors were thereafter transformed into A. tumefaciens (strain LBA4404) and to create stably transformed N. attenuata. Histochemical assays were done following Jefferson et al. (1987). After samples were fixed in ice-cold 90% acetone for 2 h, they were washed in phosphate-buffered saline buffer. Thereafter, the samples were immersed in the enzymatic reaction mixture (1 mg/mL of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 2 mm ferricyanide, and 0.5 mm of ferrocyanide in 100 mm phosphate buffer, pH 7.4). The reaction was performed at 37°C in dark for 4 h to overnight, and then the samples were cleared with pure ethanol. Photos were taken under a stereomicroscope (SV 11, Carl Zeiss), which was equipped with a CCD camera and a workstation.
RNA Extraction and qRT-PCR
Total RNA was extracted from ground leaf samples using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. For qRT-PCR analysis, five replicated biological samples were used. A total of 0.5 μg of total RNA sample were reverse transcribed using oligo(dT)18 and Superscript II reverse transcriptase (Invitrogen). qRT-PCR was performed on an ABI PRISM 7700 sequence detection system (Applied Biosystems) using qRT-PCR core kits (Eurogentec). For each analysis, a linear standard curve, threshold cycle number versus log (designated transcript level), was constructed using a series dilution of a specific complementary DNA standard; the levels of the transcript in all unknown samples were determined according to the standard curve. An N. attenuata actin2 gene, which is a housekeeping gene that had been shown to have constant levels of transcripts by microarray analysis, RNA gel blotting, and qRT-PCR after W + W and W + OS treatments (B. Bubner, J. Wu, and I.T. Baldwin, unpublished data), was used as an internal standard for normalizing complementary DNA concentration variations. Relative transcript levels of genes were obtained by dividing the extrapolated transcript levels of the target genes by the levels of actin2 from the same sample. Sequences of primers used for qRT-PCR are listed in Supplemental Table S2.
Sequence Alignment and Phylogeny Analysis
The protein sequences were retrieved from GenBank. Sequences were aligned in MegAlign (DNASTAR, Lasergene 8) using the Clustal W algorithm and verified manually. For phylogeny analysis, the unrooted neighbor-joining tree and bootstrap values were obtained using MEGA 4 software using default parameters and 1,000 replications (Tamura et al., 2007; www.megasoftware.net). Accession numbers of genes encoding these proteins were retrieved from GenBank, which are listed in Supplemental Table S3.
In-Gel Kinase Assays and Protein-Blotting Analyses
Tissues were ground in liquid nitrogen. About 100 mg of tissue were resuspended in 300 μL of extraction buffer (100 mm HEPES pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycerolphosphate, 1 mm phenylmethylsulfonyl floride, 10% glycerol, one proteinase inhibitor cocktail tablet per 10 mL extraction buffer [Roche]). Samples were then centrifuged at 4°C, 13,000g for 20 min and the supernatants were transferred to fresh tubes. Protein concentrations were measured using the Bio-Rad protein assay dye reagent (Bio-Rad) with bovine serum albumin (Sigma-Aldrich) as a standard. Ten micrograms of total protein from each sample were used for in-gel kinase activity assay according to a procedure described by Zhang and Klessig (1997). The image of in-gel kinase activity assays were obtained on a phosphorimager (FLA-3000 phosphor imager system, Fuji Photo Film), and the band intensities were quantified using the AIDA software (Raytest Isotopenmessgeräte GmbH). For protein-blotting analysis, five biologically replicated samples were pooled for protein extraction. Protein samples (10 μg) were separated in a 10% SDS-PAGE gel and electrotransferred to a polyvinylidene difluoride membrane (GE Healthcare). A westernBreeze chemiluminescent immunodetection kit (Invitrogen) was used to detect SIPK protein. The primary antibody, Ab-p48C (Zhang et al., 1998), was 1:2,000 diluted for immunoblotting analysis.
To examine equal loading, duplicated gels were run at the same time and were subsequently stained using the GelCode blue safe stain reagent (Thermo Scientific) to visualize proteins.
Analysis of JA, JA-Ile, and SA Concentrations
One milliliter of ethyl acetate spiked with 200 ng of D2-JA, D4-SA, and 40 ng of 13C6-JA-Ile, the internal standards for JA and JA-Ile, respectively, was added to each briefly crushed leaf sample (approximately 150 mg). Samples were then ground on a FastPrep homogenizer (Thermo Electron). After being centrifuged at 13,000g for 10 min at 4°C, supernatants were transferred to fresh Eppendorf tubes and evaporated to dryness on a vacuum concentrator (Eppendorf). Each residue was resuspended in 0.5 mL of 70% methanol (v/v) and centrifuged to remove particles. The supernatants were analyzed on an HPLC-tandem mass spectrometer (1200L LC-MS system, Varian).
Analyses of Herbivore Defense-Related Secondary Metabolites
TPI activity was analyzed with a radial diffusion assay described by van Dam et al. (2001). The accumulation of the direct defenses, CP and diterpene glycosides, were analyzed in samples harvested 3 d after treatments using an HPLC method described in Keinänen et al. (2001).
Statistical Analysis
Data were analyzed by Student’s t test using StatView, version 5.0 (SAS Institute).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF121307 (NaCDPK4) and EF121305 (NaCDPK5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of CDPKs.
Supplemental Figure S2. Alignment of NaCDPK4 and NaCDPK5 amino acid and nucleotide sequences and regions used for RNAi constructs.
Supplemental Figure S3. NaCDPK4, but not NaCDPK5, is expressed in trichomes of N. attenuata.
Supplemental Figure S4. Transcript levels of other CDPKs in IRcdpk4/5 plants.
Supplemental Figure S5. Morphology of IRcdpk4 plants at early flowering stage.
Supplemental Figure S6. JA and JA-Ile contents in IRcdpk4 plants.
Supplemental Figure S7. SA contents in wild-type and IRcdpk4/5 plants.
Supplemental Figure S8. Sequences used for preparing VIGS constructs.
Supplemental Figure S9. VIGS-NaCDPK4/5, but not VIGS-NaCDPK4 or VIGS-NaCDPK5, shows developmental defects.
Supplemental Figure S10. Silencing NaCDPK4 alone does not result in enhanced levels of wounding- and herbivory-induced defensive secondary metabolites and insect resistance.
Supplemental Figure S11. Compromising the accumulation of JA in IRcdpk4/5 plants abolishes herbivore defenses.
Supplemental Figure S12. IRcdpk4/5 plants have increased levels of SIPK activity but not abundance.
Supplemental Figure S13. SIPK/MPK6 and WIPK/MPK3 over-activation is dependent on the high JA levels and JA signaling.
Supplemental Table S1. Primers used for preparation of stable transformation vectors and VIGS constructs.
Supplemental Table S2. Sequences of primers used for qRT-PCR (SYBR Green analysis).
Supplemental Table S3. Accession numbers or locus numbers of genes whose deduced protein sequences were used for phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Dr. Klaus Gase, Susan Kutschbach, Wibke Kröber, and Antje Wissgott (Max Planck Institute for Chemical Ecology) for plant transformation and Dr. Tamara Krügel, Andreas Schünzel, and Andreas Weber (Max Planck Institute for Chemical Ecology) for plant cultivation. Dr. Shuqun Zhang (University of Missouri, Columbia) and Dr. Ilha Lee (Seoul National University) are thanked for offering the Ab-p48C antibody and dgl-D seeds. Maria Heinrich (Max Planck Institute for Chemical Ecology) is thanked for her help with screening NaCDPK4Pro:GUS plants.
Glossary
- JA
jasmonic acid
- FACs
fatty acid-amino acid conjugates
- OS
oral secretions
- RNAi
RNA interference
- qRT
quantitative real-time
- SA
salicylic acid
- VIGS
virus-induced gene silencing
- HGL-DTGs
17-hydroxygeranyllinalool diterpene glucosides
- TPIs
trypsin proteinase inhibitors
- CP
caffeoylputrescine
- Col-0
Columbia-0
- DMSO
dimethyl sulfoxide
- EV
empty vector
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S. (2005) Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46: 356–366 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Gfeller A, Proebsting WM, Hörtensteiner S, Chételat A, Martinoia E, Farmer EE. (2007) A gain-of-function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. Plant J 49: 889–898 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, Kim YW, Hwang I, Kim SY. (2005) Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol 139: 1750–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca M, San Segundo B. (2010) AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J 63: 526–540 [DOI] [PubMed] [Google Scholar]
- Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C. (2006) Jasmonate biosynthesis in Arabidopsis thaliana—enzymes, products, regulation. Plant Biol (Stuttg) 8: 297–306 [DOI] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG. (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargantini PR, Gonzalez-Rizzo S, Chinchilla D, Raices M, Giammaria V, Ulloa RM, Frugier F, Crespi MD. (2006) A CDPK isoform participates in the regulation of nodule number in Medicago truncatula. Plant J 48: 843–856 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT. (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J 36: 794–807 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol 125: 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Gubrium E, Harper JF. (2001) The CDPK superfamily of protein kinases. New Phytol 151: 175–183 [DOI] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. (2004) Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Heiling S, Schuman MC, Schoettner M, Mukerjee P, Berger B, Schneider B, Jassbi AR, Baldwin IT. (2010) Jasmonate and ppH systemin regulate key malonylation steps in the biosynthesis of 17-hydroxygeranyllinalool diterpene glycosides, an abundant and effective direct defense against herbivores in Nicotiana attenuata. Plant Cell 22: 273–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M, Baldwin IT, Wu J. (2011) Two mitogen-activated protein kinase kinases, MKK1 and MEK2, are involved in wounding- and specialist lepidopteran herbivore Manduca sexta-induced responses in Nicotiana attenuata. J Exp Bot 62: 4355–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Brownlee C. (2004) The generation of Ca(2+) signals in plants. Annu Rev Plant Biol 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. (1996) An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Choi S, Hwang HJ, Yu J, Nam SJ, Ko J, Park JY, Seo YS, Kim EY, Ryu SB, et al. (2008) Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev Cell 14: 183–192 [DOI] [PubMed] [Google Scholar]
- Ishida S, Yuasa T, Nakata M, Takahashi Y. (2008) A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor REPRESSION OF SHOOT GROWTH in response to gibberellins. Plant Cell 20: 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashuta S, Liu J, Liu J, Lohar DP, Haridas S, Bucciarelli B, VandenBosch KA, Vance CP, Harrison MJ, Gantt JS. (2005) RNA interference identifies a calcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell 17: 2911–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassbi AR, Gase K, Hettenhausen C, Schmidt A, Baldwin IT. (2008) Silencing geranylgeranyl diphosphate synthase in Nicotiana attenuata dramatically impairs resistance to tobacco hornworm. Plant Physiol 146: 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchiswamy CN, Takahashi H, Quadro S, Maffei ME, Bossi S, Bertea C, Zebelo SA, Muroi A, Ishihama N, Yoshioka H, et al. (2010) Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol 10: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW. (2007) Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc Natl Acad Sci USA 104: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur H, Heinzel N, Schöttner M, Baldwin IT, Gális I. (2010) R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol 152: 1731–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen M, Oldham NJ, Baldwin IT. (2001) Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J Agric Food Chem 49: 3553–3558 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H. (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183 [Google Scholar]
- Lanteri ML, Pagnussat GC, Lamattina L. (2006) Calcium and calcium-dependent protein kinases are involved in nitric oxide- and auxin-induced adventitious root formation in cucumber. J Exp Bot 57: 1341–1351 [DOI] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A. (2006) Calcium in plant defence-signalling pathways. New Phytol 171: 249–269 [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Saitoh H, Felix G, Freymark G, Miersch O, Wasternack C, Boller T, Jones JD, Romeis T. (2005) Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc Natl Acad Sci USA 102: 10736–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma SY, Wu WH. (2007) AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol Biol 65: 511–518 [DOI] [PubMed] [Google Scholar]
- Maffei M, Bossi S, Spiteller D, Mithöfer A, Boland W. (2004) Effects of feeding Spodoptera littoralis on lima bean leaves. I. Membrane potentials, intracellular calcium variations, oral secretions, and regurgitate components. Plant Physiol 134: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- Meldau S, Baldwin IT, Wu J. (2011) SGT1 regulates wounding- and herbivory-induced jasmonic acid accumulation and Nicotiana attenuata’s resistance to the specialist lepidopteran herbivore Manduca sexta. New Phytol 189: 1143–1156 [DOI] [PubMed] [Google Scholar]
- Meldau S, Wu JQ, Baldwin IT. (2009) Silencing two herbivory-activated MAP kinases, SIPK and WIPK, does not increase Nicotiana attenuata’s susceptibility to herbivores in the glasshouse and in nature. New Phytol 181: 161–173 [DOI] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT. (2007) Co(i)-ordinating defenses: NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J 51: 79–91 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC. (2001) Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JD. (2001) Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J 20: 5556–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JD. (2000) Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT. (2004) Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. J Exp Bot 55: 151–157 [DOI] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF. (1999) Communicating with calcium. Plant Cell 11: 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. (2002) Calcium at the crossroads of signaling. Plant Cell (Suppl) 14: S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M, Fischer C, Meldau S, Seebald E, Oelmüller R, Baldwin IT. (2011) Lipase activity in insect oral secretions mediates defense responses in Arabidopsis. Plant Physiol 156: 1520–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Katou S, Seto H, Gomi K, Ohashi Y. (2007) The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J 49: 899–909 [DOI] [PubMed] [Google Scholar]
- Seo S, Sano H, Ohashi Y. (1999) Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11: 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–77012615947 [Google Scholar]
- Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan CA, Wasternack C. (2003) Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato—amplification in wound signalling. Plant J 33: 577–589 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Proels R, Miersch O, Oka M, Roitsch T, Wasternack C. (2008) The AOC promoter of tomato is regulated by developmental and environmental stimuli. Phytochemistry 69: 1859–1869 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitz M, Gase K, Baldwin IT, Gaquerel E. (2011) Ectopic expression of AtJMT in Nicotiana attenuata: creating a metabolic sink has tissue-specific consequences for the jasmonate metabolic network and silences downstream gene expression. Plant Physiol 157: 341–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczegielniak J, Klimecka M, Liwosz A, Ciesielski A, Kaczanowski S, Dobrowolska G, Harmon AC, Muszyńska G. (2005) A wound-responsive and phospholipid-regulated maize calcium-dependent protein kinase. Plant Physiol 139: 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. (2007) The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Thomas CL, Jones L, Baulcombe DC, Maule AJ. (2001) Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J 25: 417–425 [DOI] [PubMed] [Google Scholar]
- van Dam NM, Horn M, Mares M, Baldwin IT. (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27: 547–568 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang M, Ke K, Lu YT. (2005) Cellular localization and biochemical characterization of a novel calcium-dependent protein kinase from tobacco. Cell Res 15: 604–612 [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. (2009) Herbivory-induced signalling in plants: perception and action. Plant Cell Environ 32: 1161–1174 [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wünsche H, Baldwin IT, Wu J. (2011) S-Nitrosoglutathione reductase (GSNOR) mediates the biosynthesis of jasmonic acid and ethylene induced by feeding of the insect herbivore Manduca sexta and is important for jasmonate-elicited responses in Nicotiana attenuata. J Exp Bot 62: 4605–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Hettenhausen C, Baldwin IT, Wu J. (2011) BAK1 regulates the accumulation of jasmonic acid and the levels of trypsin proteinase inhibitors in Nicotiana attenuata’s responses to herbivory. J Exp Bot 62: 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Komatsu S. (2000) Involvement of calcium-dependent protein kinase in rice (Oryza sativa L.) lamina inclination caused by brassinolide. Plant Cell Physiol 41: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Yang KY, Liu Y, Zhang S. (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon GM, Cho HS, Ha HJ, Liu JR, Lee HS. (1999) Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol 39: 991–1001 [DOI] [PubMed] [Google Scholar]
- Yoon GM, Dowd PE, Gilroy S, McCubbin AG. (2006) Calcium-dependent protein kinase isoforms in Petunia have distinct functions in pollen tube growth, including regulating polarity. Plant Cell 18: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Hui D, Baldwin IT. (2004) Manipulation of endogenous trypsin proteinase inhibitor production in Nicotiana attenuata demonstrates their function as antiherbivore defenses. Plant Physiol 134: 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu YT. (2003) Calmodulin-binding protein kinases in plants. Trends Plant Sci 8: 123–127 [DOI] [PubMed] [Google Scholar]
- Zhang M, Liang S, Lu YT. (2005) Cloning and functional characterization of NtCPK4, a new tobacco calcium-dependent protein kinase. Biochim Biophys Acta 1729: 174–185 [DOI] [PubMed] [Google Scholar]
- Zhang S, Du H, Klessig DF. (1998) Activation of the tobacco SIP kinase by both a cell wall-derived carbohydrate elicitor and purified proteinaceous elicitins from Phytophthora spp. Plant Cell 10: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JJ, Wei FJ, Wang C, Wu JJ, Ratnasekera D, Liu WX, Wu WH. (2010) Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol 154: 1232–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.