Abstract
Quorum sensing plays a central role in regulating many community derived symbiotic and pathogenic relationships of bacteria, and as such has attracted much attention in recent years. Acyl-homoserine lactones (AHLs) are important signaling molecules in the quorum sensing gene regulatory processes found in numerous gram-negative species of bacteria that interact with eukaryotic organisms. AHLs are produced by acyl-homoserine lactone synthases. Bacteria can have multiple genes for AHL synthase enzymes, and such species are likely to produce several different types of AHLs. Determination of the types and the relative amounts of AHLs produced by AHL synthases in bacteria under varied conditions provides important insights into the mechanism of AHL synthase function and the regulation of transcriptional cascades initiated by quorum sensing signaling. This chapter describes a mass spectrometry method for determining the types and relative amounts of AHLs present in a sample.
Keywords: acyl-homoserine lactone, AHL, quorum sensing, mass spectrometry, AHL synthase
1. Introduction
Understanding the molecular mechanism of AHL–dependent signaling in bacterial quorum sensing requires knowledge of the recognition of AHLs by receptors as well as the AHLs that are present in the environment. The acyl-homoserine lactones AHLs differ in their length of acyl chain as well as the substituents at the 3-position of the acyl chain (Figure 1A) (1). Some species of bacteria have a single AHL synthase enzyme and produce predominantly one type of AHL, whereas others have multiple AHL synthases and produce many types of AHLs (2, 3). Furthermore, the amount and types of AHLs that are produced in bacteria can differ depending on the particular AHL synthases that are expressed and the availability of the substrates needed for AHL synthesis. The ability to identify populations of AHLs and to analyze changes in AHLs produced with mutant forms of AHL synthase genes or under different metabolic conditions is important for understanding quorum sensing signaling. However, it is difficult to identify all of the AHLs produced and to determine how much of each AHL species is present using conventional bioassay procedures (4, 5).
Figure 1.
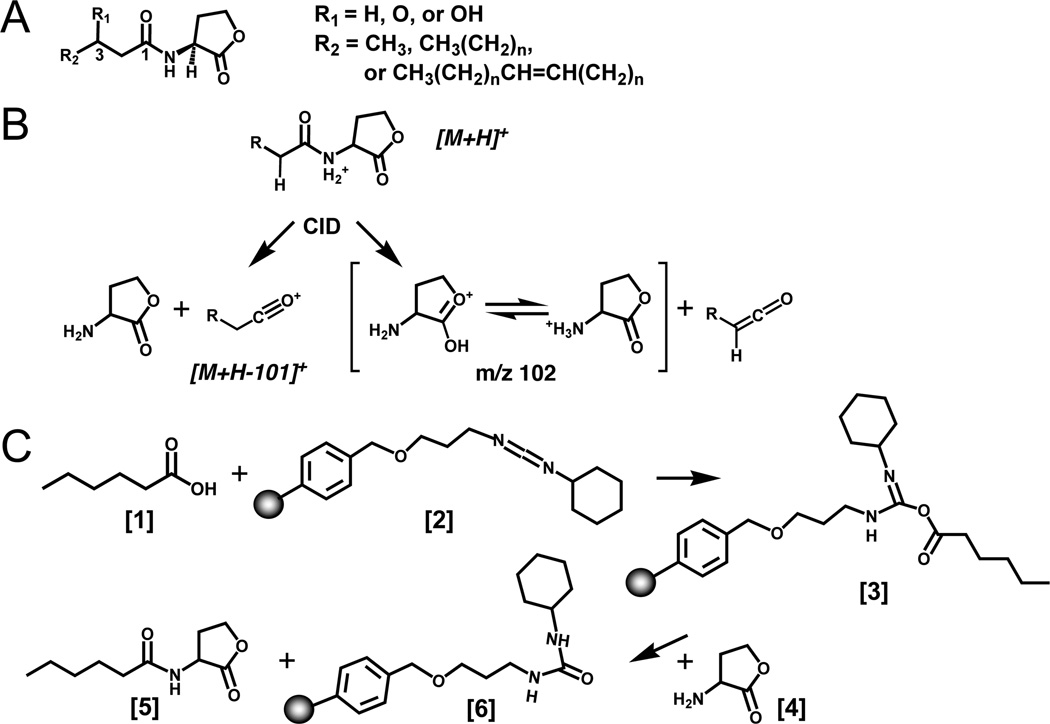
Scheme of AHL structure, mass spectrometry, and chemical synthesis. (A) Structure of AHLs. AHLs vary by substitution at the C3 position (R1) and the length and unsaturation of the acyl chain (R2). HSL refers to D/L-homoserine lactone; AHL refers to N-acyl-D/L-homoserine lactone, with any acyl chain length (indicated by the number of carbons CN) or degree of substitution, and the extent of deuterium labeling indicated by Dn. (B) Mass spectrometry scheme proposed for collisionally induced decomposition of AHLs into the two major fragment ions observed for the 3-oxo-HSLs (lactone and acyl). (C) Carbodiimide based chemical synthesis of D3-C6-HSL. The fatty acid [1] reacts with the resin bound carbodiimide reagent [2] to give an acylating intermediate [3]. Next, nucleophilic attack by the HSLhydrobromide amine [4] on the acylating intermediate releases the amide product AHL [5]. A urea-like byproduct remains coupled to the resin-bound carbodiimide [6]. Any excess or unreacted fatty acid was removed during filtration because it is left in the form of the acylating intermediate bound to the resin [6]. The final AHL product [5] was obtained by filtration of the reaction mixture and solid-phase extraction using a normal-phase SPE.
This chapter describes a relatively unbiased method for determining the types and relative amounts of AHLs present in a sample. Mass spectrometry provides a complementary approach to traditional bioassays. It permits the semi-quantitative analysis of the changes to the distribution of AHLs by a system either grown under various conditions, or one in which the AHL synthase activity had been changed. The distribution of AHLs from experimental samples can be determined by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC/MS/MS) using a triple-quadrupole mass spectrometer (5). It is necessary first to extract the AHLs from the system of interest with an added internal standard and this is described for laboratory bacterial cultures in Sections 3.2 and 3.3. It is important to have appropriate reference standards and internal standards and to determine the HPLC retention times for each AHL. Therefore, the approach for synthesizing several types of AHLs and stable-isotope labeled internal standards is described in Section 3.1, since several AHLs, including internal standards for mass spectrometry, are not commercially available. Second, the AHLs require purification prior to this mass spectrometry analysis (Section 3.4). Finally, in section 3.5, the methods used to characterize the AHLs using LC/MS/MS are described. This approach is based on monitoring multiple major fragment ions that are produced during the collision induced decomposition process in the mass spectrometer (Figure 1B) (5).
A common approach for dissecting the AHL-mediated quorum sensing pathways is to examine the AHLs that exist in a wildtype bacterium and in bacteria that have been engineered bacteria harboring individual components of the signaling system in order to determine their specificity (5, 6, 7, 8). The simple example used to illustrate the method here is the analysis of the AHLs produced by the enzyme LasI expressed recombinantly in E. coli. The extracted and purified samples (Figure 4) contain 3-oxo-C12-HSL and other 3-oxo-HSLs that are made under the conditions of expression in the two independent experiments (5). These approaches can also be adapted to examine biological samples from the environment and patients.
Figure 4.
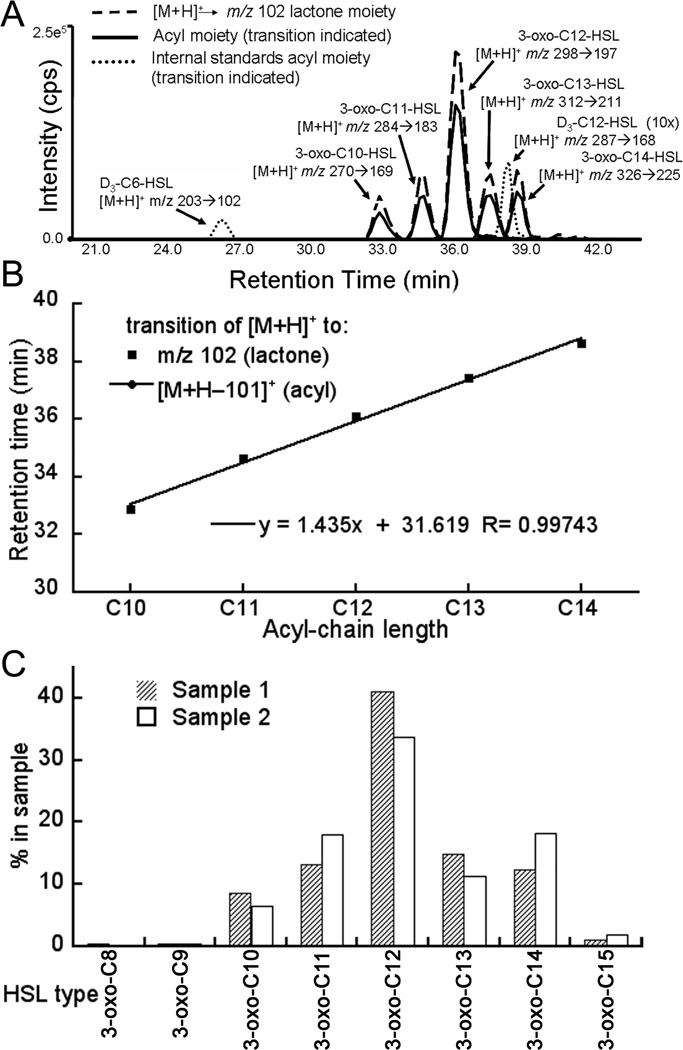
Example of AHL analysis of AHLs produced by LasI expressed in E. coli. (A) Total ion current observed in the MRM analysis of reversed phase chromatographic separation of AHLs. AHLs were examined using the MRM mode that was monitoring the transitions for precursor [M+H]+ –> m/z 102 (shown as a dashed line) and the transition from [M+H]+ –> [M+H-101]+ (shown as solid lines), and the IS peaks (dotted lines). The identified AHLs are labeled. In this experiment, it is clear that the integrated peak areas for the ion transitions of 3-oxo-AHLs are of nearly equally size (approximately 40:60) for the two transitions (to the acyl and lactone moieties, repectively). (B) Retention times of the transitions observed for the lactone and acyl moieties plotted versus the number of carbon atoms in the acyl chain of each AHL. The plot shows that all the AHLs with the same substitution lie on a straight line, which coincides with a line that was generated by using the reference standards. Deviation from the line of the reference standards indicates that the compound, which has been named in the data analysis is likely to be incorrect. (C) Bar graph representation of the AHLs purified from two independent experiments of LasI expressed in E. coli, with two slightly different growth and extraction conditions. Sample 1 and sample 2 differ in number of hours that LasI was grown as well as the addition of Amberlite XAD 16. In sample 1, the Amberlite XAD 16 resin was added immediately after induction and the culture was grown for only eight hours, whereas for sample 2 the Amberlite XAD 16 resin was added right before inoculation and the culture was grown for 24 hours. The height of the bars in the graph represents the percentage of each AHL and not the total amounts of AHL in the sample.
2. Materials
2.1 Synthesis of AHLs using carbodiimide chemistry
N-cyclohexylcarbodiimide-N’-propyloxymethyl polystyrene resin (PS-Carbodiimide resin; Argonaut Technologies).
Homoserine lactone starting material: α-amino-γ-butyrolactone hydrobromide (HSL-HBr) (Aldrich)
Acyl-chain starting materials for deuterium labeled mass spectrometry standards include D3-hexanoic acid or those with other lengths of acyl chain (Cambridge Isotope)
Acyl-chain starting materials for production of reference standards including unsubstituted and 3-hydroxy-AHLs: Fatty acids were purchased from Sigma and the 3-hydroxy-C6 fatty acid was purchased from Larodan Lipids (Sweden) and was supplied as 25 mg in a liquid.
Solvents: methylene chloride (CH2Cl2), N, N-dimethyl-formamide (DMF), triethylamine (TEA), methanol (Sigma) (Note 1).
Teflon fritted filters and glass reaction vials of appropriate scale. MUST USE ONLY ORGANIC-SAFE GLASSWARE (Note 2).
2.2 Production of AHLs from bacterial culture
Any media may be used; the selection of the media depends on the growth of the desired bacterial strain.
Amberlite XAD 16 (Sigma; (XAD16)) resins must be added to media after autoclaving, at 2 grams per liter.
Methanol (Fisher, optima 0.2 micron filtered (A454)) is used to initially re-suspend the resin and cells.
Syringe filters (non sterile filter, 0.2 um, Nalgene 191–2020) are used to filter the methanol re-suspended supernatant.
2.3 Solid phase extraction purification of AHLs
Isooctane (Fisher O296; HPLC grade, submicron filtered) and Diethyl ether anhydrous (Fisher E138) are mixed at a ratio of 1:1, and are used in the purification procedure. MUST USE ONLY GLASSWARE (Notes 1 and 2).
Ethyl acetate (Fisher E195 HPLC Grade) is used as an elution buffer. MUST USE ONLY GLASSWARE.
Sep-Pak plus, silica cartridges (Waters WAT020520) used to purify AHLs.
Supelco Visiprep vacuum purification system.
Fisher brand disposable culture tubes (14-961-27 Borosilicate Glass) to collect AHLs from Sep-Pak plus cartridges.
Glass pipettes (Fisher 10 mL CC65G and 9” disposable Pasteur pipettes 13-678-20D).
2.4 Liquid chromatography/ Mass Spectrometry/ Mass Spectrometry (LC/MS/MS)
-
1.
Reagents for standard curve 10 × 32 culture tubes (Fisher 14-961-27 Borosilicate Glass), 3-oxo-C6-HSL (Aldrich K3007), C6-HSL (Fluka 09926), and C12-HSL (Sigma 17247) and 3-oxo-C12-HSL (Sigma 09139) and deuterium labeled HSL (see Materials section 2.1).
-
2.
High Pressure Liquid Chromatography (HPLC) Shimdzu SCL-10AVP controller and LC10ASVP system.
-
3.
1.0 × 150 mm 5µm C18 Columbus reversed-phase column, Phenomenex (00F-4108A0) at a flow rate of 0.050 µL/min.
-
4.
PAL autosampler (LEAP Technologies); vials used for autosampler: Supelco 10× 32 0.9 mL crimp vials.
-
5.
HPLC solvent A: 8.3 mM acetic acid (Fisher A38-500), buffered to pH 5.7 with Ammonium Hydroxide (Fisher A669-500). Solvent B: Methanol (Fisher A452-4).
-
6.
Mass spectrometer; Applied Biosystems MDS Sciex Q Trap LC/MS/MS system (Applied Biosystems, Foster City, CA).
-
8.
Areas of peaks are integrated using a quantitation software package Analyst 1.5 (Applied Biosystems).
3. Methods
3.1 Synthesis of 3-hydroxy, unsubstituted, and deuterium labeled AHLs using carbodiimide chemistry (Figure 1C)
4.2 mg (23 µmol) of HSL-HBr was weighed into a small glass reaction vial and was completely dissolved (5–10 min) by vortexing in 200 µL of methylene chloride (CH2Cl2) with 10% N, N-dimethyl-formamide (DMF) and 4.6 µmol of triethylamine (TEA) (6.4 µL).
For the deuterium labeled or other unsubstituted fatty acids that are supplied as neat liquids (25 mg; density ~0.93 g/mL), 4.4 µL (~35 µmol) was added to 400 µL of CH2Cl2 with 10% DMF in a separate glass reaction vial. To this reaction vial, 40 mg of PS-Carbodiimide was added, and stirred for 5 min at room temperature. (Go to step 4)
For the unlabeled unsubstituted or 3-hydroxy-fatty acids, which are supplied as liquid or wax, 25 mg was weighed out and resuspended in 100 µL of methanol, transferred to a clean glass vial, dried down by vacuum evaporation and then resuspended in 25 µL of methanol, and treated as 1 mg/µL. The 25 µL was added to 400 µL of CH2Cl2 with 10% DMF in a separate glass reaction vial. To this reaction vial, 40 mg of PS-Carbodiimide was added, and stirred for 5 min at room temperature. (Go to step 4)
After the fatty acid had been allowed to equilibrate with the resin, the 200 µL solution containing the HSL-HBr was added and stirred for 30 h at room temperature.
The reaction mixture was then diluted into 6 mL of isooctane: ethyl ether (1:1) and passed through the teflon frit of the glass tube directly onto the activated normal phase silica SPE cartridge for purification as described in section 3.4
3.2 Preparative scale production of AHLs from bacterial culture (Figure 2)
Figure 2.
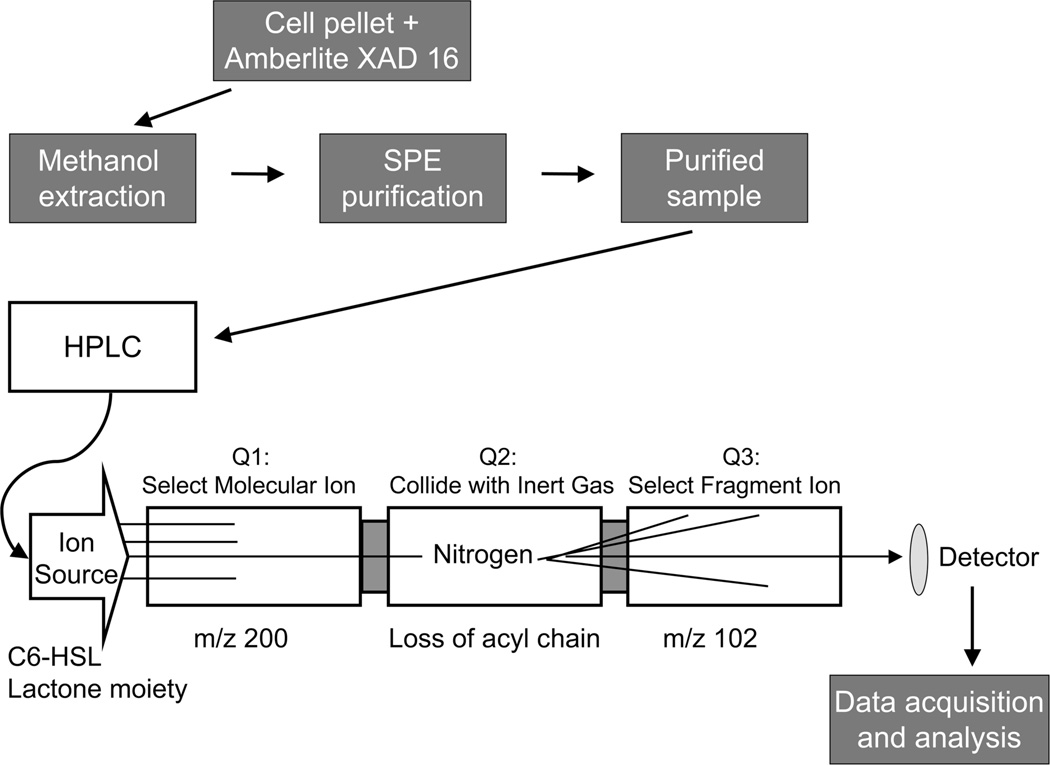
Flowchart of the methods described in this chapter. The cell cultures were grown in the presence of Amberlite XAD 16, which adsorbs AHLs. In order to extract the AHLs from the resin and cell debris, the harvested pellet from the bacterial culture and Amberlite XAD 16 was re-suspended in methanol and left overnight. The methanol solution was filtered and centrifuged, and then subjected to solid phase extraction to remove any remaining impurities and to concentrate the AHLs. The purified sample was then injected onto the reversed phase HPLC column to separate the AHLs based on their lipophilicity and introduce them into the mass spectrometer. In the multiple reaction monitoring (MRM) experiment, the sample was passed through the ion source into Quadrupole1 (Q1), then the selected ions proceeded into Quadrupole 2 (Q2) where they collide with inert gas (nitrogen). After fragmentation, selected ions move into Quadrupole 3 (Q3) and are visualized by the detector. Data were processed by applying standard isotope dilution curves to the analytical data (as in Figures 3 and 4).
Prepare desired media and autoclave. Add 2 gm of Amberlite XAD 16 resin per liter of media after media has cooled down to room temperature.
Grow desired strain as if growing bacteria for recombinant protein expression. Each strain grows according to specific medium, antibiotic, temperature, and IPTG induction concentration, as well as induction OD.
Harvest resin by centrifugation at 5,000 rpm for 10 min.
Store at −80°C until ready to extract AHLs.
Re-suspend in a 10-fold volume of methanol, and vortex and shake vigorously until homogenized. Then shake and vortex vigorously at 20 minute intervals, for two h. For example, 10 gm of harvested cell culture should be re-suspended in 100 mL of methanol.
Leave overnight at room temperature with a final vigorous shake.
After leaving overnight shake and vortex vigorously then centrifuge at 5,000 rpm for 10 min.
Syringe filter supernatant into test tubes and dry down each tube to a final volume of approximately 2 mL.
Collect all 2 mL samples together then centrifuge at 5,000 rpm for 10 min.
Collect supernatant into new glass culture tubes.
Store supernatant at −20°C, for a maximum of 16 h until SPE purification.
3.3 Analytical scale identification of AHL products (Figure 2)
Grow desired strain as if growing bacteria for recombinant protein expression. Each strain grows according to specific medium, antibiotic, temperature, and IPTG induction concentration, as well as induction OD. The only difference is that 2 gm of Amberlite XAD 16 resin per liter of growth medium is added immediately before inoculation.
Harvest the cells and resin by centrifugation at 5,000 rpm for 10 min.
Store at −80°C until ready to extract AHLs.
Re-suspend the samples in a 10-fold volume of methanol. Vortex and shake vigorously until homogenized and shake and vortex vigorously at 20 minute intervals, for two h.
Add 2 nmol of the internal standards (D3-C6-HSL and D3-C12-HSL).
Leave overnight at room temperature with a final vigorous shake.
Shake and vortex vigorously and centrifuge at 5,000 rpm for 10 min.
Filter the supernatant using a syringe into test tubes and dry down each tube to a final volume of approximately 2 mL.
Collect all 2 mL samples and centrifuge at 5,000 rpm for 10 min.
Store at −20°C for a maximum of 16 h until SPE purification.
3.4 Solid phase extraction purification of AHLs (Figure 2)
Wash all equipment with 1:1 Isooctane / Ethyl ether solution.
-
Equilibrate Sep-Pak Cartridges (WAT020520) plus Silica, with 8 mL of;
1:1 Isooctane : Ethyl ether
Ethyl Acetate
1:1 Isooctane : Ethyl ether
Take 0.2 mL of sample and bring up to 8 mL using 1:1 Isooctane: Ethyl ether then load on Sep-Pak Cartridges (WAT020520) plus Silica.
Wash twice with 8 mL of 1:1 Isooctane : Ethyl ether
Elute with 8 mL Ethyl acetate.
Dry down using Speed Vac concentrator under vacuum, until completely dry.
3.5 Liquid chromatography/Mass Spectrometry/Mass Spectrometry (LC/MS/MS) (Figures 3 and 4)
Figure 3.
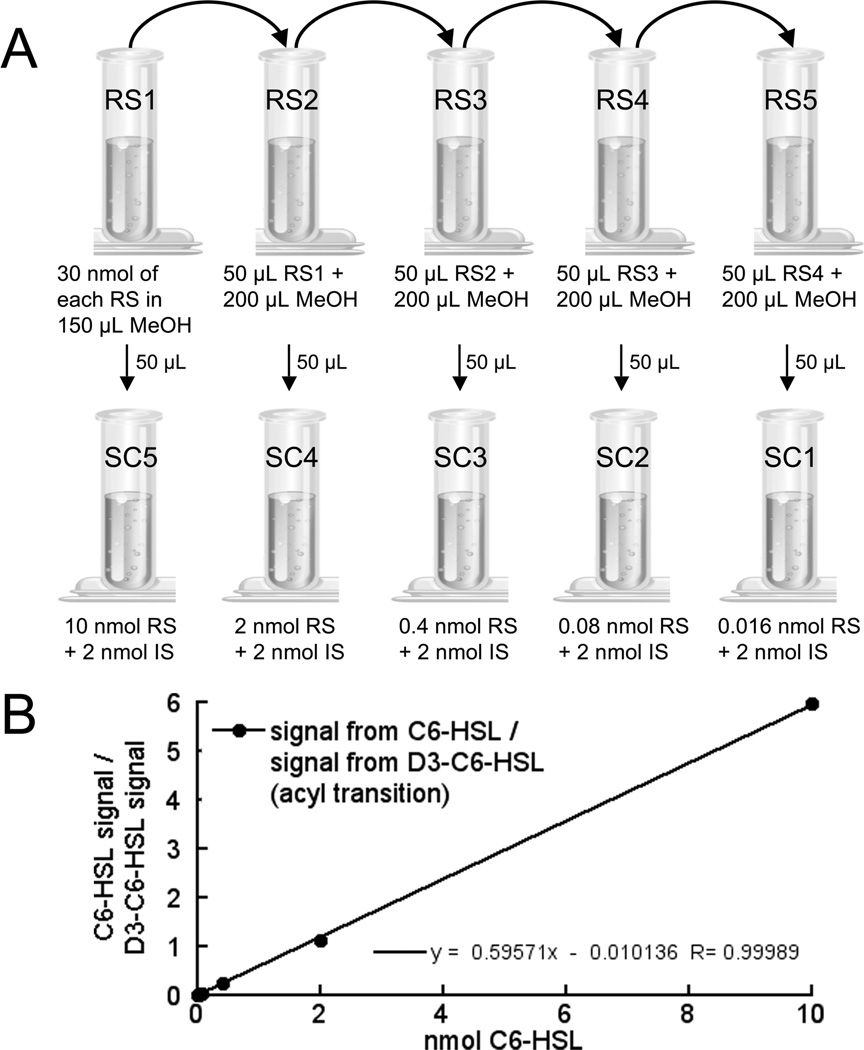
Preparation of a standard curve for mass spectrometry of AHLs. (A) Step-by-step preparation of internal standard and reference standard mixtures for mass spectrometry. First a series of serial dilutions (1:5) are prepared from a stock solution (vial 1A) containing 30 nmol each of reference standard (RS) in 150 µL methanol (Table 3). Here the four reference standards being used are 3-oxo-C6-HSL, 3-oxo-C12-HSL, C6-HSL, and C12-HSL. The samples in vials 2A, 3A, 4A, and 5A are then made by serial dilution of 50 µL from the preceding vial into 200 µL of methanol. To produce the standard curve samples (SC1–5), 50 µL of each samples 1A–5A is transferred to a new vial and a fixed amount of the stable isotope labeled internal standard (IS) is added to each vial, as well as an empty control vial (SC0, not shown in the figure). Here the IS was D3-C6-HSL and 2 nmol was added to each of the SC vials. The samples are then dried down before use. (B) Standard curve of C6-HSL. The ratio of the integrated ion transition peak area of a particular transition for the RS (signal of C6-HSL [M+H]+ –> [M+H-101]+) to the integrated ion transition peak area of the same transition of the IS (signal of D3-C6-HSL [M+H]+ –> [M+H-101]+) was plotted and fitted with a linear equation (black line). This standard curve was then applied to analytical data.
The standard curve is prepared by making a 1 mg/mL stock solution of each of the following reference standards (RS) AHLs in methanol: 3-oxo-C6-HSL, C6-HSL, C12-HSL and 3-oxo-C12-HSL (Figure 3A).
The stock standard solutions were serially diluted to yield concentrations of 10, 2, 0.4, 0.08, 0.016 and 0 nmol (Table 3).
For each standard curve point (SC1–5) and sample, 2 nmol of D3-C6-HSL internal standard in methanol is added to each vial.
Standards and samples are dried down under nitrogen and resuspended in crimp vials with 60 µL of HPLC solvent A and 10 µL of HPLC solvent B.
30 µL of the sample is injected onto a 1.0 × 150 mm 5 µm C18 reversed-phase column.
The high pressure liquid chromatography (HPLC) flow rate is 50 µL/min with initial conditions at time 0 of 5% B. A typical HPLC program is shown in Table 1, but briefly the gradient is isocratic for 5 min, ramps up to 95% B in 30 min, is isocratic at 95% B for 20 min., and at 56 min, the concentration of B goes to 5% and the system re-equilibrates for 4 min with the effluent flowing directly into the mass spectrometer.
Once the sample has passed into the mass spectrometer, a Multiple Reaction Monitoring (MRM) experiment is used to collect data; the parameters are; ion spray voltage of 4,200 V, focusing potential of 200 V and collision energy of 25 V. The collision gas thickness in the collision cell is 1.6×1015 molecules/cm2; nitrogen is used as the collision gas.
The ions monitored in Q1 include the parent AHL [M + H]+. In Q3, both the acyl moiety [M + H-101]+ and the lactone moiety at m/z 102 are monitored and appear at characteristic retention times (Figure 4A). The retention times and masses monitored in Q1 and Q3 are illustrated for a selection of AHLs in Table 2 that are used in the example presented in this chapter (Figure 4B and Note 3).
- Quantitation is accomplished by applying the standard curves to the analytical data. The amount of the unknown in each sample is determined by direct application of the slope and intercept of the standard curve to the ratios determined for the signals (from both transitions) of unknowns to internal standard, according to the following equation:
- Amount of unknown = ((Area counts of unknown/Area counts of internal standard) × (slope of standard curve) + intercept of standard curve)
Typically the ratios are calculated directly from the quantitative analysis routines included with mass spectrometer software, such as Analyst Software. Standard curves can be applied to the data using spreadsheet software, such as Microsoft Excel. For example, the standard curve for C6-HSL was generated by plotting the ratio of the integrated signals for the reference standard for C6-HSL to that of the labeled internal standard versus the amount of the reference standard for the transition [M+H]+ –> [M+H-101]+, as shown in Figure 3B and Table 2. A similar curve was also generated and used for the [M+H]+ –> m/z 102 transition (lactone moiety; not shown).
Finally the results can be presented in a graphical format. Figure 4C compares the 3-oxo-AHLs produced by LasI expressed recombinantly in E. coli from two independent experiments. It plots the type of AHL identified, based of the fragmentation pattern (Figure 4A), retention time (Figure 4B) and comparison to the reference standards (Table 2 and Note 4). The amount of AHL plotted is the sum of the amount determined for both of the monitored transitions [M+H]+ –> m/z 102 (lactone moiety) and [M+H]+ –> [M+H-101]+ (acyl moiety) as a percentage of the total AHL in the sample. This graph illustrates the utility of the method and gives an indication of the reproducibility achieved in detection of AHLs from similar samples.
Table 3.
Sample standard curve for C6-HSL
| nmol of C6-HSL | area of C6-HSL/area of D3-C6-HSL |
|---|---|
| 0 | 0 |
| 0.016 | 0.00999 |
| 0.08 | 0.0463 |
| 0.04 | 0.257 |
| 2 | 1.11 |
| 10 | 5.96 |
Table 1.
Chromatography profile used in LC/MS experiments
| Step | Time (min) | Flow rate (µL/min) |
A% | B% |
|---|---|---|---|---|
| 0 | 0 | 50 | 95 | 5 |
| 1 | 5 | 50 | 95 | 5 |
| 2 | 35 | 50 | 5 | 95 |
| 3 | 55 | 50 | 5 | 95 |
| 4 | 56 | 50 | 95 | 5 |
| 5 | 60 | 50 | 95 | 5 |
Table 2.
Subset of transitions monitored in MRM experiments used in Figure 4
| Transition to lactone moeity | Transition to acyl moeity | |||||
|---|---|---|---|---|---|---|
| Standards: | Q1 m/z |
Q3 m/z |
Retention Time (min) |
Q1 m/z |
Q3 m/z |
Retention Time (min) |
| D3-C6-HSL | 203.3 | 102.1 | 26.2 | 203.3 | 102.2 | 26.2 |
| D3-C12-HSL | 287.3 | 102.1 | 38.17 | 287.3 | 186.2 | 38.17 |
| C6-HSL | 200.3 | 102.1 | 26.14 | 200.3 | 99.2 | 26.15 |
| C12-HSL | 284.4 | 102.1 | 37.75 | 284.3 | 183.2 | 37.75 |
| 3-oxo-C6-HSL | 214.3 | 102.1 | 19.24 | 214.3 | 113.2 | 19.24 |
| 3-oxo-C12-HSL | 298.3 | 102.1 | 35.66 | 298.3 | 197.2 | 35.67 |
| Sample: | ||||||
| 3-oxo-C6-HSL | 214.3 | 102.1 | 19.27 | 214.3 | 113.2 | 19.30 |
| 3-oxo-C8-HSL | 242.3 | 102.1 | 27.88 | 242.3 | 141.2 | 27.87 |
| 3-oxo-C9-HSL | 256.3 | 102.1 | 30.73 | 256.3 | 155.2 | 30.69 |
| 3-oxo-C10-HSL | 270.3 | 102.1 | 32.87 | 270.3 | 169.2 | 32.86 |
| 3-oxo-C11-HSL | 284.3 | 102.1 | 34.64 | 284.3 | 183.2 | 34.64 |
| 3-oxo-C12-HSL | 298.3 | 102.1 | 36.09 | 298.3 | 197.2 | 36.07 |
| 3-oxo-C13-HSL | 312.3 | 102.1 | 37.44 | 312.3 | 211.2 | 37.42 |
| 3-oxo-C14-HSL | 326.3 | 102.1 | 38.64 | 326.3 | 225.2 | 38.64 |
| 3-oxo-C15-HSL | 340.3 | 102.1 | 39.69 | 340.3 | 239.2 | 39.68 |
Acknowledgments
The authors would like to thank Drs. Ty Gould and Robert C. Murphy for their contributions in initially developing this method, Dr. Jake Herman for help in refining the method, and Dr. Joseph Hankin, Chris Johnson, and Wesley Martin for their assistance with the mass spectrometry. We also thank Dr. Robert C. Murphy for helpful suggestions and for use of the mass spectrometry equipment, which is supported by the Lipid Maps Large Scale Collaborative Grant (NIH GM069338 to R.C.M.). This work was supported by Grants from the National Science Foundation #0703467 and #0821220.
Footnotes
All organic solvents should be tested for the presence of AHLs. AHL contaminants have been found in commercially purchased solvents.
Use glass when working with organic solvents.
It is important to use a sufficient number of reference standards to define the retention times for a given series of AHLs.
True quantitative analysis can only be obtained with an identical isotope–labeled internal standard. The values obtained from this analysis are semi-quantitative.
References
- 1.Fuqua C, Eberhard A. Signal generation in autoinduction systems: synthesis of acylated homoserine lactones by LuxI-type proteins. In: Dunny G, Winans SC, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 211–230. [Google Scholar]
- 2.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Churchill MEA, Herman JP. Acyl-homoserinelactone Biosynthesis: Structure and Mechanism." Chapter 17. In: Winans Stephen, Bassler Bonnie., editors. Chemical Communication Among Bacteria. Washington, D.C.: ASM Press; 2008. [Google Scholar]
- 4.Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Rinehart KL, Farrand SK. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci U.S.A. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould TA, Herman JP, Krank J, Murphy RC, Churchill MEA. Specificity of acyl-homoserine lactone syntheses examined by mass spectrometry. J Bacteriol. 2006;188:773–783. doi: 10.1128/JB.188.2.773-783.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson WT, Minogue TD, Val DL, Beck von Bodman S, Churchill MEA. Structural Basis and Specificity of Acyl-homoserine lactone Signal Production in Bacterial Quorum Sensing. Mol Cell. 2002;9:685–694. doi: 10.1016/s1097-2765(02)00480-x. [DOI] [PubMed] [Google Scholar]
- 7.Khan SR, Herman JP, Krank K, Serkova NJ, Churchill MEA, Suga H, Farrand SK. N-(3-Hydroxy-Hexanoyl)-L-Homoserine Lactone Is the Biologically Relevant Quormone that Regulates the phz Operon of Pseudomonas chlororaphis (aureofaciens) Strain 30–84. Appl. Env. Microbiol. 2007;73:7443–7455. doi: 10.1128/AEM.01354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duerkop BA, Herman JP, Ulrich RL, Churchill MEA, Greenberg EP. The Burkholderia mallei BmaR3-BmaI3 Quorum-Sensing System Produces and Responds to N-3-Hydroxy-Octanoyl Homoserine Lactone. J. Bacteriol. 2008;190:5137–5141. doi: 10.1128/JB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]