Synthesis of m1G37-tRNA is catalyzed by TrmD in bacteria and by Trm5 in eukarya and archaea, using unrelated and dissimilar structural folds. The authors probe the mechanism of discrimination among functional groups of guanosine for synthesis of m1G37-tRNA by TrmD and Trm5. The data suggest a model in which both enzymes rapidly screen tRNA by direct recognition of G37 in order to monitor the global state of m1G37-tRNA synthesis.
Keywords: m1G37-tRNA, TrmD, Trm5, guanosine analogs, reconstitution of tRNA by ligation
Abstract
Guanosines are important for biological activities through their specific functional groups that are recognized for RNA or protein interactions. One example is recognition of N1 of G37 in tRNA by S-adenosyl-methionine (AdoMet)–dependent tRNA methyltransferases to synthesize m1G37-tRNA, which is essential for translational fidelity in all biological domains. Synthesis of m1G37-tRNA is catalyzed by TrmD in bacteria and by Trm5 in eukarya and archaea, using unrelated and dissimilar structural folds. This raises the question of how dissimilar proteins recognize the same guanosine. Here we probe the mechanism of discrimination among functional groups of guanosine by TrmD and Trm5. Guanosine analogs were systematically introduced into tRNA through a combination of chemical and enzymatic synthesis. Single turnover kinetic assays and thermodynamic analysis of the effect of each analog on m1G37-tRNA synthesis reveal that TrmD and Trm5 discriminate functional groups differently. While both recognize N1 and O6 of G37, TrmD places a much stronger emphasis on these functional groups than Trm5. While the exocyclic 2-amino group of G37 is important for TrmD, it is dispensable for Trm5. In addition, while an adjacent G36 is obligatory for TrmD, it is nonessential for Trm5. These results depict a more rigid requirement of guanosine functional groups for TrmD than for Trm5. However, the sensitivity of both enzymes to analog substitutions, together with an experimental revelation of their low cellular concentrations relative to tRNA substrates, suggests a model in which these enzymes rapidly screen tRNA by direct recognition of G37 in order to monitor the global state of m1G37-tRNA.
INTRODUCTION
Guanosines serve as building blocks for RNA structure and provide specific sites for RNA activity. This dual function is enabled by the diverse functional groups of the nucleoside, including the N1-imino group, N2-amino group, O6-carbonyl group, N7-nitrogen, C8-proton, and the 2′-OH of the ribose. These functional groups contribute to both short- and long-range interactions in RNA structure and offer recognition sites for protein interaction and activity. For example, a conserved G3-U70 base pair in the acceptor stem is a marker for aminoacylation of cytoplasmic tRNAAla (Hou and Schimmel 1988; McClain and Foss 1988; Francklyn and Schimmel 1989), where G3 forms a long-range interaction with U70, while exposing its exocyclic N2-amino group in the minor groove for recognition by alanyl-tRNA synthetase (Musier-Forsyth et al. 1991). Similarly, a G-U base pair is conserved as the defining feature for 5′ splice site cleavage within group I introns, where the ribozyme recognizes the G through its exocyclic N2-amino group during the cleavage reaction (Strobel and Cech 1995). Also, in the Escherichia coli peptidyl transferase center that catalyzes peptide bond formation in the large subunit of the ribosome, a conserved G2553 in the A loop interacts with conserved C75 in the CCA end of an aminoacyl-tRNA, while conserved G2251 and G2252 in the P loop interact, respectively, with C75 and C74 of the adjacent peptidyl tRNA (Yusupov et al. 2001; Valle et al. 2003). These stabilizing interactions in the ribosome–tRNA complexes are provided by precise Watson-Crick base-pairing interactions during peptide bond formation (Puglisi et al. 1997) and are involved in initiating translocation movements of ribosome complexes (Dorner et al. 2006).
While different RNA activities often rely on different guanosine functional groups, it is unknown how the same activity, catalyzed by different enzymes, recognizes guanosine. In nature, enzymes that catalyze the same activity, but having unrelated structures, are known as analogous enzymes (Galperin et al. 1998), which usually evolved from different phylogenetic lineages. While relatively rare as compared with enzymes sharing homologous origins, analogous enzymes are fundamentally intriguing, because they illustrate the existence of distinct structural solutions to the conversion of substrate to product. For pharmaceutical purposes, analogous enzymes are also highly attractive, because their unrelated structures permit structure-specific (and thus species-specific) drug targeting. For these reasons, an understanding of how analogous enzymes recognize the same substrate at the atomic resolution is of high importance.
The recognition of the N1 of the guanosine at position 37 of tRNA for synthesis of m1G37, a modified base 3′ adjacent to the anticodon, is essential for tRNA translational fidelity by preventing frameshift errors in all three domains of life (Bjork et al. 1989; Hagervall et al. 1993; Bjork et al. 2001). This recognition is required for growth both for bacteria—e.g., E. coli (Baba et al. 2006), Salmonella typhimurium (Bjork et al. 1989), Streptococcus pneumoniae (O'Dwyer et al. 2004)—and for eukaryotes—e.g., Saccharomyces cerevisiae (Bjork et al. 2001). Additionally, in yeast, the m1G37 base is necessary to prevent mischarging of tRNAAsp with arginine (Putz et al. 1994). In methanogenic archaea, the m1G37 base is important for tRNA aminoacylation by phosphoseryl-tRNA synthetase and by cysteinyl-tRNA synthetase (Hauenstein and Perona 2008; Hauenstein et al. 2008; Zhang et al. 2008). The synthesis of m1G37-tRNA is performed by the TrmD methyltransferase in bacteria but by a structurally unrelated Trm5 in eukaryotes and in archaea (Christian et al. 2004), providing an example of guanosine recognition by analogous enzymes. While both TrmD and Trm5 use S-adenosyl-methionine (AdoMet) as the methyl donor and recognize G37-tRNA as the substrate, they share no sequence or structural homology.
TrmD functions as a homodimer and binds AdoMet at the dimer interface in a deep-knotted trefoil-fold structure, where the methyl donor adopts an unusual bent conformation by folding the adenosine moiety toward the methionine moiety (Ahn et al. 2003; Elkins et al. 2003). In contrast, Trm5 functions as a monomer and binds AdoMet in a straight conformation within the open-sandwich structure of the Rossmann fold that is typically involved in binding nucleotides (Brule et al. 2004; Christian et al. 2004; Goto-Ito et al. 2008, 2009). Additionally, TrmD and Trm5 differ in the recognition of tRNA structure; while TrmD requires just an extended anticodon stem–loop structure, Trm5 requires the complete and intact tRNA L shape (Christian and Hou 2007). Furthermore, the two enzymes also differ in their kinetic mechanism; while TrmD is slow in the synthesis of m1G37-tRNA, Trm5 is slow in the release of the m1G37-tRNA product (Christian et al. 2010b). Finally, to discriminate among diverse tRNA sequences, TrmD requires the presence of a preceding G36 for synthesis of m1G37, whereas Trm5 does not require G36 (Redlak et al. 1997; Christian et al. 2004). However, despite the differences shown to date, the key question of how TrmD and Trm5 recognize G37 to perform the AdoMet-dependent methyl transfer remains unknown.
To address the key question directly, we use the bacterial E. coli TrmD (EcTrmD) and the archaeal Methanococcus jannaschii Trm5 (MjTrm5) as a pair of model enzymes. Both enzymes have been extensively characterized by biochemical analysis (Elkins et al. 2003; Christian et al. 2004, 2006, 2010a,b; Christian and Hou 2007) and by AdoMet-bound crystal structures (Ahn et al. 2003; Goto-Ito et al. 2008, 2009). Two aspects of guanosine recognition are of interest. First, is it feasible to probe the recognition by using analogs that contain position-specific substitutions of functional groups? While position-specific substitutions are typically achieved by chemical synthesis of RNA, this can be challenging for tRNAs with lengths in 70–90 nucleotides. Here we address the challenge by developing a method using both chemical synthesis and multipiece enzymatic ligation to replace G37 with a series of analogs. Second, is it feasible to identify guanosine functional groups specifically required in the rate-determining step of the methyl transfer reaction? The rate-determining step is the slowest step in the overall scheme of methyl transfer, affording an enzyme the highest potential to scrutinize its tRNA substrate. Here we show that, by using our previously developed single-turnover kinetic assays for both EcTrmD and MjTrm5 (Christian et al. 2006, 2010b), we can directly probe the rate-determining step. These kinetic assays are performed with an enzyme in molar excess of its tRNA, in the presence of saturating AdoMet, to ensure only one round of methyl transfer, so that the observed rate constant (kobs) corresponds to the slowest step of the reaction. In contrast, steady-state assays are performed with molar excess of tRNA over enzyme, allowing multiple rounds of turnover, such that the meaning of the observed rate constant is obscure and cannot be clearly defined.
Using these two approaches, we show that TrmD and Trm5, while recognizing common features of G37, differ from each other by placing discernable emphasis on individual functional groups. This difference is attributable to their structural distinctions. We also show that the functional groups identified by kinetic analysis of MjTrm5 do not completely agree with those identified in the crystal structure of the MjTrm5–tRNA–AdoMet ternary complex (Goto-Ito et al. 2009). The discrepancy is attributable to the emphasis of the kinetic approach on probing the rate-limiting step, which may not be captured by the structural approach. By combining both approaches, we gain insight into a dynamic and time-dependent rearrangement of functional groups during the Trm5 synthesis of m1G37-tRNA. Finally, based on the observation that both TrmD and Trm5 recognize the target guanosine during ground-state binding of tRNA and that both exist in low cellular concentrations relative to their tRNA substrates, we suggest that they both use ground-state binding to screen tRNA for m1G37 synthesis.
RESULTS
Incorporation of guanosine analogs to tRNA
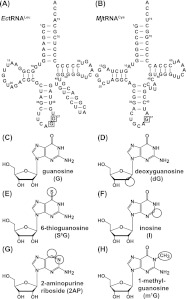
We designed a systematic approach to replace functional groups of G37 by guanosine analogs in EctRNALeu and MjtRNACys (Fig. 1A,B), the respective optimal substrates for EcTrmD and MjTrm5. These analogs included the following (Fig. 1C–G): deoxyG (dG) for evaluation of the importance of the ribose 2′-hydroxyl; 6-thioG (S6G) for evaluation of the O6 of guanosine; inosine (I) for evaluation of the exocyclic 2-NH2; and 2-aminopurine (2AP) for evaluation of the N1 and O6 of the guanine base. All but one (dG) of these analogs probed the Watson-Crick base-pairing interface of G37. In addition, because TrmD requires G36 for synthesis of m1G37, we evaluated the effect of a methylated m1G36 substitution (Fig. 1H).
FIGURE 1.
Guanosine analogs in tRNA. (A) Sequence and cloverleaf structure of E. coli tRNALeu (EctRNALeu), where G36 and G37 are boxed. (B) Sequence and cloverleaf structure of M. jannaschii tRNACys (MjtRNACys), where G37 is boxed. (C–H) Chemical structures of guanosine (G), deoxyguanosine (dG), 6-thioguanosine (S6G), inosine (I), 2-aminopurine (2AP) riboside, and 1-methyl guanosine (m1G). The altered atomic groups relative to guanosine are circled.
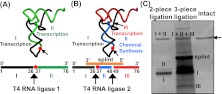
Each analog was introduced into tRNA by an enzyme-mediated ligation of shorter fragments that were generated by in vitro transcription or by chemical synthesis. Except for the intended analog, these fragments contained no other modification. In a two-piece ligation approach (Fig. 2A), which was used to synthesize dG37-tRNA, the sequence of a tRNA fragment encoding nucleotides 1–36 (fragment I) and the sequence of a second fragment encoding nucleotides 37–76 (fragment II) were transcribed in vitro by T7 RNA polymerase with standard NTPs. Two features were noted to reconstitute a full-length tRNA. First, transcription of fragment I was initiated with guanosine, whereas transcription of fragment II was initiated with dGMP, such that the two fragments could be joined in order by T4 RNA ligase 1, using the monophosphate of dGMP of fragment II as the primary acceptor. Driven by the facile annealing of the two fragments at their mutual complementary regions, the ligation yield was robust, as evidenced in a denaturing PAGE analysis (Fig. 2C), where the limiting fragment II was easily converted to the full-length tRNA. Similarly high yield of two-piece ligation to reconstitute tRNA was reported previously (Sherlin et al. 2001). For all other guanosine analogs, the incorporation into a full-length tRNA was achieved by a three-piece ligation approach (Fig. 2B). For example, fragments I and III (encoding nucleotides 1–36 and 49–76, respectively) were prepared by T7 transcription, while fragment II (encoding an analog at position 37 in the nucleotide sequence 37–48) was prepared by chemical synthesis. The three fragments were joined in order by T4 RNA ligase 2 with the assistance of a DNA splint template. While multiple side products were obvious in the three-piece ligation approach, the yield of the full-length tRNA was usually 30%–60% as estimated from the conversion of the limiting fragment II (Fig. 2C). Thus both ligation methods produced stoichiometric quantities of the full-length tRNA, which was purified from denaturing PAGE.
FIGURE 2.
Construction of modified tRNA. (A) Schematic illustration of two-piece ligation. The 5′ fragment (fragment I; black) and the 3′ fragment (fragment II; green) were transcribed by T7 RNA polymerase, using molar excess of dGMP as the initiation nucleotide for fragment II. These two fragments were ligated with T4 RNA ligase 1 to construct a full-length tRNA. (B) Schematic illustration of three-piece ligation. The 5′ fragment (fragment I; black) and the 3′ fragment (fragment III; red) were transcribed by T7 RNA polymerase. The middle piece (fragment II; blue) with the modification was chemically synthesized. These three fragments were ligated with T4 RNA ligase 2 with the assistance of a DNA splint oligonucleotide to construct a full-length tRNA. Note that the actual sites of ligation vary depending on the sequence. See Materials and Methods for details. The red circle in A and B indicates the modified position 37. (C) Denaturing gel analysis of the ligated product. Individual fragments are indicated, except for fragment II in the three-piece ligation, which is too small for the gel analysis. Both the two-piece and three-piece ligation mixtures yield a product that migrates to the same position as the intact tRNA.
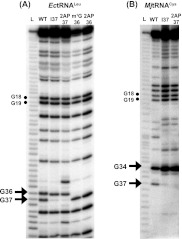
Each reconstituted full-length tRNA was verified for the incorporation of the intended analog. In one method, the analog was detected by its resistance to RNase T1, which cleaved the 3′-phosphodiester linkage of structurally unhindered guanosines (Hartmer et al. 2003). The T1 cleavage analysis was performed under conditions of limited digestion, such that each cleavage generated a fragment that migrated on denaturing PAGE according to the position of the cleaved-off G. Using a reconstituted EctRNALeu that was 3′-end-labeled with 32P as an example (Fig. 3A), denaturing PAGE analysis revealed that G36 and G37 were both readily cleaved in the wild-type (wt) tRNA, along with the cleavage markers at G18 and G19 in the D loop. However, while the cleavage markers remained the same in several reconstituted tRNAs, the cleavage at position 37 was greatly reduced in the reconstituted I37- and 2AP37-tRNA, and the cleavage at position 36 was abolished in the reconstituted m1G36- and 2AP36-tRNAs. These results were consistent with the resistance of the guanosine analog to RNase T1 cleave in each case (Heinemann and Saenger 1983). For MjtRNACys (Fig. 3B), G37 in the wt tRNA was readily cleaved by T1, along with the cleavage markers at G34 of the anticodon loop and G18 and G19 of the D loop. While the cleavage markers remained the same in the reconstituted I37-tRNA and 2AP37-tRNA, the cleavage at position 37 was greatly reduced, consistent with the resistance of the guanosine analog to cleavage in each case. Similarly, analysis of dG37-EctRNALeu and of dG37-MjtRNACys, the only substrates reconstituted by the two-piece ligation approach, confirmed the resistance of the dG residue in each tRNA to T1 cleavage (Supplemental Fig. S1; Hougland et al. 2008). In all cases, the cleavage pattern between G37-tRNA and analog-containing tRNAs outside the region of interest was preserved, providing a strong indication that all reconstituted tRNAs had retained the sequence and structural framework of G37-tRNA.
FIGURE 3.
RNase T1 analysis of modified tRNAs. (A) Denaturing PAGE analysis of T1 digestion of wild type (wt)-, I37-, 2AP37-, m1G36-, and 2AP36-modified EctRNALeu. A ladder of digested fragments (L) generated by heating the tRNA in 50 mM NaHCO3 for 5 min at 90°C is provided on the left. The cleavage fragments after G18 and G19 are marked by dots, while those after G36 and G37 are marked with arrows. (B) Denaturing PAGE analysis of T1 digestion of wt-, I37-, and 2AP37-modified MjtRNACys. A ladder of digested fragments (L) generated by heating the tRNA in 50 mM NaHCO3 for 5 min at 90°C is provided on the left. The cleavage fragments after G18 and G19 are marked by dots, while those after G34 and G37 are marked with arrows.
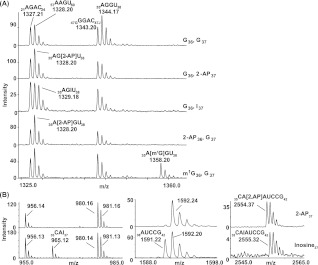
In a second method of verification, each reconstituted tRNA was digested to completion by RNase A or RNase T1 to generate site-specific fragments, followed by analysis of these fragments by MALDI-TOF mass spectrometry (MS) or by the combination of nucleases and alkaline phosphatase, followed by LC/tandem-MS for detection of the analog. In the MALDI-TOF MS analysis, G37-EctRNALeu was digested by RNase A, which cleaved after pyrimidines, to reveal the presence of a 35AGGU38 fragment at m/z 1344.17 (Fig. 4A). The reconstituted 2AP37-tRNA contained a fragment at m/z 1328.20, and the reconstituted I37-tRNA contained a fragment at m/z 1329.18, with concomitant loss of the 35AGGU38 fragment at m/z 1344.17. These results are consistent with the replacement of G37 with 2AP37 and I37, respectively. Similarly, the reconstituted 2AP36-tRNA contained a fragment at m/z 1328.20, and the reconstituted m1G36-tRNA contained a fragment of m/z 1358.20, with concomitant loss of the m/z 1344.17 fragment, consistent with the replacement of G36 with 2AP36 and m1G36, respectively. For MjtRNACys (Fig. 4B), digestion of the reconstituted 2AP37-tRNA by RNase T1 revealed the presence of a 35CA[2AP]AUCCG42 fragment at m/z 2554.37, consistent with the incorporation of 2AP at position 37. Digestion of the reconstituted I37-tRNA revealed the presence of two fragments, one corresponding to 35CAI37 at m/z 965.12 and the other corresponding to 35CAIAUCCG42 at m/z 2555.32, both of which were consistent with the incorporation of I37. The detection of two RNase T1 fragments originating from the region of positions 35–42 was consistent with the partial sensitivity of I to RNase T1 cleavage, as shown in denaturing PAGE (Fig. 3B). Separately, the detection of the site-specific guanosine analog of each tRNA was confirmed by LC/tandem mass spectrometry (electrospray ionization) (see Supplemental Fig. S2 for an example). The only guanosine analog that was not clearly detected by LC/MS was S6G, due to the lability of the thio substitution to chemical degradation during the analysis. The presence of the S6G analog in each reconstituted tRNA (S6G37-EctRNALeu and S6G37-MjtRNACys) was verified with the thio-specific absorption peak at 342 nm, which was absent from the corresponding G37-tRNA (Supplemental Fig. S3).
FIGURE 4.
MALDI-TOF-MS analysis of nuclease-digested tRNAs. (A) Analysis of EctRNALeu digested by RNase A. (G36, G37) Fragments of unmodified tRNA. The labeled signals at 1327.21, 1328.20, and 1343.20 indicate the monoisotopic species of RNase A digestion products from unmodified regions in the sample. Notice that digestion products are represented by multiple signals, generating partially overlapping clusters of peaks, which reflect the natural isotope distribution in the sample. (G36, 2AP37) Fragments of 2-AP37-modified tRNA, showing a decrease in intensity of m/z 1344.17 (the disappearance of 35AGGU38) and an increase in intensity of m/z 1328.20 corresponding to the appearance of 35AG[2-AP]U38. (G36, I37) Fragments of I37-modified tRNA, showing a decrease in intensity of m/z 1344.17 (the decrease of 35AGGU38) and an increase in intensity of m/z 1329.18 corresponding to the appearance of 35AG[I]U38. (2AP36, G37) Fragments of 2-AP36-modified tRNA, showing a decrease in intensity of m/z 1344.17 (the decrease of 35AGGU38) and an increase in intensity of m/z 1328.20 corresponding to the appearance of 35A[2-AP]GU38. (m1G36, G37) Fragments of m1G36-modified tRNA, showing a decrease in intensity of m/z 1344.17 (the decrease of 35AGGU38) and an appearance of m/z 1358.20 corresponding to 35A[m1G]GU38. Signals of unmodified digestion products are only assigned in the spectrum from the unmodified tRNA. (B) Analysis of MjtRNALeu digested by RNase T1. The labeled signals around m/z 955.1, 980.2, 981.2, and 1592.2 indicate the monoisotopic species of RNase T1 digestion products from unmodified regions in the sample. All digestion products are represented by multiple signals, which reflect the natural isotope distribution in the sample. Upper row of zooms (all from the same mass spectrum): fragments of 2AP37-modified tRNA, showing the composite oligonucleotide 35CA[2-AP]AUCCG42. Lower row of zooms (all from the same mass spectrum): fragments of I37-modified tRNA, showing low signal of the composite oligonucleotide 35CAIAUCCG42, with a simultaneous appearance of signals of 35CAI37 and 38AUCCG42, as a result of RNase T1 cleavage after I.
Together, the two complementary verification methods confirmed the presence of the intended analog in each reconstituted tRNA. Because the intended analog was introduced by chemical synthesis (except for dG), its incorporation into the full-length tRNA via ligation would be stoichiometric with the extent of ligation. The incorporation of dG was also quantitative, as determined from denaturing gel analysis of RNase T1 cleavage (Supplemental Fig. S1).
Kinetic analysis of methyl transfer to G37-substituted tRNA
The previously developed single-turnover assay (Christian et al. 2006, 2010b) was used to evaluate the effect of each guanosine analog on the EcTrmD- and MjTrm5-catalyzed reactions. The enzyme concentration was maintained in molar excess of each tRNA substrate, while 3H-AdoMet was saturating relative to the Kd of the methyl donor for each enzyme (Christian et al. 2010b). Reaction of methyl transfer was initiated upon rapid mixing of an enzyme with a mixture of tRNA and [3H-methyl]-AdoMet, and the kinetics of m1G37-tRNA synthesis was monitored over time by the incorporation of the radioactive label into acid-precipitable counts on filter pads. All time courses of EcTrmD were well fit to a single exponential equation, and all data of kobs as a function of enzyme concentration were well fit to a hyperbola (Supplemental Fig. S4a,b), validating the rapid equilibrium binding condition. The same criteria were met for kinetic analysis of MjTrm5 (Supplemental Fig. S4c,d). For each enzyme–tRNA complex, analysis of the hyperbola fit from the average values of at least two sets of independent experiments revealed the dissociation constant Kd of the complex and the maximum rate constant of methyl transfer (defined as kchem), the latter of which is a composite term of both the rate constant of the pre-transfer conformational adaptation of the enzyme–tRNA–AdoMet complex and the rate constant of methyl transfer. The kinetic parameters of methyl transfer are summarized in Table 1. The parameters of methyl transfer to G37-tRNA for EcTrmD [Kd (tRNA) = 0.7 ± 0.2 μM and kchem = 0.07 ± 0.01 sec−1] and for MjtRNATrm5 [Kd (tRNA) = 0.8 ± 0.1 μM and kchem = 0.14 ± 0.05 sec−1] were closely similar to values reported previously (Christian et al. 2010b).
TABLE 1.
Kinetic parameters of TrmD and Trm5 with respect to modifications of G37
The dissociation constant measured by single-turnover kinetics was an estimation of ground-state binding. To measure ground-state binding independently, a titration experiment to measure the equilibrium binding of tRNA to EcTrmD or to MjTrm5, based on the intrinsic enzyme tryptophan fluorescence, was performed (Supplemental Fig. S5). G37-tRNA and two derivatives (I37-tRNA and 2AP37-tRNA) were measured for each enzyme in the presence of a saturating concentration of sinefungin, a nonreactive AdoMet analog. In all cases, the addition of tRNA to each enzyme resulted in a quench of fluorescence; analysis of the quench as a function of tRNA concentration was fit to a hyperbolic equation to determine the equilibrium Kd. This analysis confirmed that the equilibrium Kd of G37-tRNA for EcTrmD (1.5 ± 0.7 μM) was closely similar to the kinetic Kd (0.7 ± 0.2 μM), and that the equilibrium Kd of G37-tRNA for MjTrm5 (1.3 ± 0.6 μM) was closely similar to the kinetic Kd (0.8 ± 0.1 μM) (Table 1).
Comparison of fluorescence and kinetic Kd values showed that the I37-tRNA derivative for EcTrmD has a more favorable fluorescence Kd (4.9 ± 0.7 μM) than kinetic Kd (13 ± 2 μM), with the trend preserved for MjTrm5 (fluorescence Kd of 0.42 ± 0.06 μM and kinetic Kd of 1.6 ± 0.6 μM). In addition, the I37-tRNA derivative for MjTrm5 has a more favorable fluorescence Kd of 0.42 ± 0.06 μM as compared with G37-tRNA with the fluorescence Kd of 1.3 ± 0.6 μM. The discrepancy is likely a result of the use of sinefungin in fluorescence measurement versus AdoMet in kinetic measurement, indicating that kinetic measurement should provide more accurate information. Nonetheless, the difference between the two measurements for each tRNA was about twofold, indicating that the kinetic measurement was a valid approach to gain insight into equilibrium binding. In the case of 2AP37-tRNA derivatives, kinetic Kd measurement for both enzymes was not feasible, due to the lack of a detectable activity under the experimental conditions, whereas fluorescence measurement was not limited by the lack of activity and indeed revealed a defective Kd in both cases (4.2 ± 0.7 μM for EcTrmD and 3.3 ± 0.5 μM for MjTrm5).
Using the kinetic Kd value of each reaction as the equilibrium binding constant, the catalytic efficiency of methyl transfer by each enzyme–tRNA complex was expressed as kchem/Kd, and the effect of an analog on the transfer efficiency was expressed as the fold-decrease in kchem/Kd relative to the efficiency of the wt-tRNA. The fold-decrease was then converted to the increase in the free energy of activation during the transition state of methyl transfer.
For methyl transfer to G37-substituted tRNA, a stronger deleterious effect on EcTrmD was observed relative to MjTrm5. For example, substitution of G37 with dG reduced catalytic efficiency by about sevenfold for EcTrmD (an increase of ΔΔG by 1.2 kcal/mol) but virtually had no effect on MjTrm5. Substitution of G with I reduced catalytic efficiency by 420-fold for EcTrmD (an increase of ΔΔG by 3.7 kcal/mol) but virtually had no effect on MjTrm5. The one exception was the substitution with S6G, which had no effect on EcTrmD but roughly a sevenfold effect on MjTrm5 (an increase of ΔΔG by 1.2 kcal/mol). Notably, the 2AP37 substitution had the most profound effect on both enzymes, eliminating activity under the experimental condition.
The deleterious effect of I37 and 2AP37 substitutions prompted further analysis of the reaction product in each case. A reconstituted I37- or 2AP37-tRNA was used as a substrate for EcTrmD or MjTrm5, and the product of each reaction was analyzed by mass spectrometry. MALDI-TOF analysis detected methylation of m1I37-tRNA by both enzymes (Supplemental Fig. S6a,b). Additionally, MALDI tandem MS analysis also revealed that the methylation occurred at I37 in tRNA by MjTrm5 (Supplemental Fig. S6c); a similar analysis of the EcTrmD product was not possible due to overlap of RNase A digestion fragments (see Supplemental Fig. S6a, lower panel). For 2AP37-tRNA, mass spectrometry analysis detected no methylation for either reaction, consistent with the lack of activity in kinetic measurement. Indeed, the N1 of 2AP was saturated in valence and would be inaccessible to methylation.
Kinetic analysis of methyl transfer to G36-substituted tRNA
The parameters for methyl transfer on G36-substituted tRNA substrates are summarized in Table 2. Here the tRNA substrates were derived from EctRNALeu for both enzymes, due to its possession of G36 and G37, which are required elements for TrmD and are recognizable by Trm5 (Fig. 1A). With EctRNALeu as the substrate, MjTrm5 displayed similar kinetic parameters [Kd (tRNA) = 0.7 ± 0.1 μM, kchem = 0.07 ± 0.01 sec−1] as with MjtRNACys, indicating a lack of discrimination between the two sequences. Of interest was the m1G36 substitution—while the substitution severely reduced catalytic efficiency of EcTrmD by up to 90-fold (an increase of ΔΔG by 2.8 kcal/mol), it had no effect on MjTrm5. The sensitivity of EcTrmD to other atomic substitutions of G36 was also evident; the most deleterious substitution was 2AP (with a decrease in catalytic efficiency by ∼70-fold and an increase of ΔΔG by 2.6 kcal/mol), followed by substitution with I (a decrease by ∼10-fold and an increase of ΔΔG by 1.4 kcal/mol), and followed by substitution with dG (a decrease of twofold and an increase of ΔΔG by 0.4 kcal/mol).
TABLE 2.
Kinetic parameters of TrmD and Trm5 with respect to modifications of G36
G37 recognition in vivo
A key finding of the kinetic analysis is that both EcTrmD and MjTrm5 were sensitive to the ground-state binding parameter [Kd (tRNA)] of substrates that contain a deleterious guanosine analog (Tables 1 and 2). For example, when substitution of a functional group resulted in a negative effect, the effect was always associated with an increase in Kd (tRNA), whereas in a few cases, it was associated with both an increase in Kd (tRNA) and a decrease in kchem (e.g., substitution of G37 with I and 2AP for EcTrmD). The sensitivity of Kd (tRNA) to analog substitution illustrates the importance of ground-state binding as an initial step of discrimination, which suggested that an evaluation of the dynamics of the binding interaction in vivo would be of interest. The objective was to gain insight into how these enzymes locate and then catalyze m1G37 synthesis on a tRNA substrate. This was addressed by determination of the cellular concentration of enzyme versus tRNA, because the ratio would impact on the nature of the binding interaction. We included all tRNA species possessing G36 and G37 as substrates for TrmD and those possessing G37 as substrates for Trm5. These criteria are held with few exceptions across all three biological domains based on analysis of tRNA databases between gene sequences and tRNA sequences isolated from cells.
The cellular concentration of EcTrmD in an E. coli cell lysate was determined by the AdoMet-dependent synthesis of m1G37-tRNA. As a substrate, the transcript of EctRNAPro was chosen (Supplemental Fig. S7a), because m1G37 synthesis on the tRNA is essential for bacterial cell growth (Nasvall et al. 2009) and key features of the synthesis had been previously identified (Christian and Hou 2007). The cell lysate activity of m1G37-tRNA synthesis was corrected by two controls (Supplemental Material). First, its activity was compared with the linear profile of activity versus concentration of a purified recombinant EcTrmD to estimate the concentration of the enzyme in the cell lysate and corrected for the number of cells used and the fraction of cytosol in each cell. Second, its activity was subtracted from the activity of methyl transfer generated from a mutant tRNA substrate (e.g., G37C-tRNA) to eliminate methyl transfer to non-G37 positions due to the activity of other AdoMet-dependent methyltransferases in the cell lysate. These corrections led to estimation of the cellular concentration of EcTrmD of ∼100 nM. To determine the cellular concentration of Trm5, we used the yeast Saccharomyces cerevisiae enzyme as a model, which was characterized previously (Lee et al. 2007) and could be detected in cell lysates generated in large quantities in normal growth conditions rather than in the extreme conditions of M. jannaschii. The substrate for ScTrm5 was the transcript of SctRNAPhe (Supplemental Fig. S7b), which must be converted to m1G37-tRNA before maturation to W37-tRNA (where W is wybutosine) (Noma et al. 2006). A similar analysis of the activity of m1G37-tRNA synthesis by a cell lysate of S. cerevisiae, normalized by comparison to a purified recombinant ScTrm5 and subtracted from methyl transfer to positions other than G37 in the G37C-mutant of SctRNAPhe, revealed a cellular concentration of ∼700 nM for ScTrm5.
The cellular concentration of tRNA substrates for EcTrmD was determined in two steps (Supplemental Material). First, total tRNA was isolated from E. coli, and, based on the number of cells and the volume of cytosol in each cell, the cellular tRNA concentration was calculated. Second, the cellular tRNA concentration was corrected for the fraction of tRNA gene sequences in the E. coli genome that encode the G36-G37 element (at 11%). This correction was based on the observation that the number of gene copies is proportional to the level of the tRNA (Percudani et al. 1997). These corrections led to a value of 60 μM for the total tRNA cellular concentration recognizable by EcTrmD. A similar analysis of tRNA in S. cerevisiae and correction for the fraction that contained G37 (at 28%) returned a value of 60 μM for tRNA recognizable by ScTrm5, identical to the tRNA concentration for EcTrmD. For both enzymes, the low concentration (100–700 nM) relative to tRNA (60 μM) suggests the need to rapidly screen and identify substrates from among the entire tRNA population.
DISCUSSION
How TrmD and Trm5 recognize guanosine functional groups for synthesis of m1G37-tRNA is an important question that probes the consequence of their distinct structures. While the two enzymes lack structural similarity, this does not necessarily guide them to read different guanosine functional groups. Indeed, we recently showed that, while the two enzymes recognize the same functional groups of AdoMet, they exhibit substantial differences in the energetic costs of recognition of each (Lahoud et al. 2011). Such differences are attributable to their distinct structures. Here we introduced guanosine analogs to replace specific functional groups in the G36-G37 motif of tRNA as a way to probe how TrmD and Trm5 recognize the target G37. Using EcTrmD and MjTrm5 as the model enzymes, each of which exhibits high sequence homology with members of the respective families (∼80%), we have identified subtle but important differences between the two enzymes. These differences offer new insight into the impact of their structural differences on recognition of the target base and provide a basis for enzyme-specific drug development. Recently, TrmD has been identified as a leading candidate among several highly promising new anti-bacterial targets, based on an extensive genomewise analysis of currently clinical challenging microbial pathogens (White and Kell 2004). The difference in target recognition between TrmD and Trm5, together with the differences identified previously, should provide an important foundation upon which novel and highly enzyme-specific drugs against TrmD can be developed.
Recognition of guanosine for m1G37 synthesis
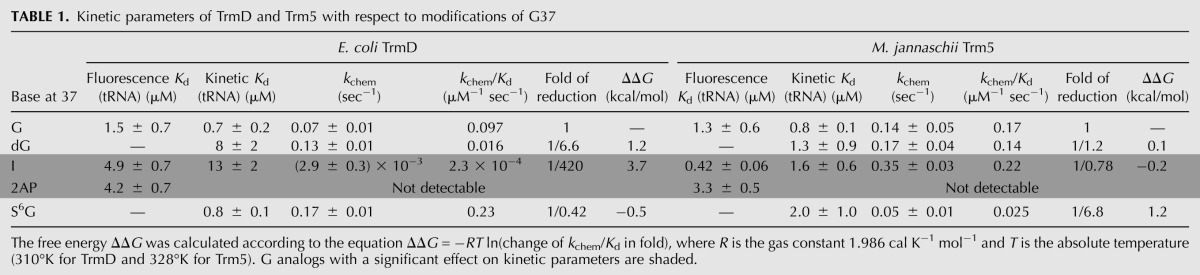
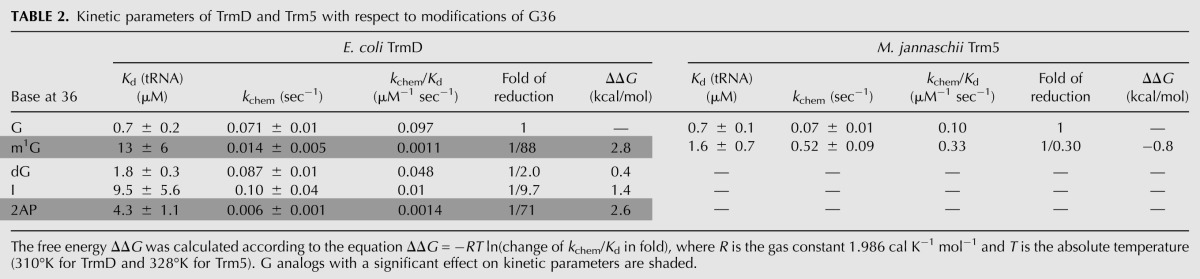
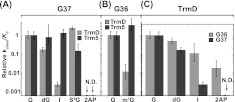
A logarithmic comparison of the sensitivity of TrmD versus Trm5 to guanosine substitutions has identified three important features (Fig. 5). First, at position 37, while TrmD is sensitive to both 2AP and I substitutions, Trm5 is sensitive to only 2AP substitution (Fig. 5A). Second, at position 36, while TrmD discriminates m1G substitution by two orders of magnitude, Trm5 exhibits no discrimination (Fig. 5B). Third, while TrmD most strongly discriminates against 2AP substitution at both G36 and G37, the discrimination at G37, the site of methyl transfer, is significantly stronger (Fig. 5C). All three features have implications for the structural differences between the two enzymes.
FIGURE 5.
Effect of analog relative to guanosine on synthesis of m1G37-tRNA. (A) Activity in log(kchem/Kd) of TrmD and Trm5 with each analog modification of G37 relative to the respective G37-tRNA. (B) Activity in log(kchem/Kd) of TrmD and Trm5 with each analog modification of G36 relative to the respective normal G37-tRNA. (C) Activity in log(kchem/Kd) of TrmD with each analog modification of G36 and of G37 relative to the normal G37-tRNA. Data are obtained from Tables 1 and 2. (N.D.) Not detectable.
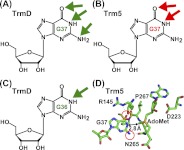
In the recognition of G37, the common sensitivity between TrmD and Trm5 to 2AP substitution indicates a shared emphasis on two functional groups (the N1-proton and the O6-carbonyl of guanosine) (Fig. 6A,B). However, the additional sensitivity of TrmD to I substitution reveals an extra emphasis placed on the N2-amino group of G37 (Fig. 6A). The further sensitivity of TrmD to substitutions at G36 indicates yet another layer of structural constraint that is not observed for Trm5. We note that at G36, 1-methyl or 2AP substitution reduces the TrmD activity by ∼80-fold (Table 2). Because the N1-proton of G36 is lacking in both substitutions, whereas the O6-carbonyl of G36 is lacking only in the 2AP substitution, the similar catalytic defect of the two substitutions suggests that it is the N1-proton of G36 that is a major determinant (Fig. 6C). Importantly, while TrmD recognizes the N1-proton of both G36 and G37, quantitative analysis shows that the N1-proton of G37 is far more critical.
FIGURE 6.
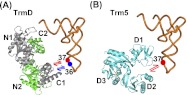
Recognition of guanosine by TrmD and Trm5 determined by analog substitutions. (A) Key determinants for recognition of G37 by TrmD. All three positions of the Watson-Crick base-pairing interface (N1-proton, N2-amino, and O6-carbonyl; each indicated by a green arrow) are critical for TrmD. (B) Key determinants for recognition of G37 by Trm5. Two of the three Watson-Crick base-pairing interfaces (N1-proton and O6-carbonyl, each indicated by a red arrow) are critical for Trm5. (C) The key determinant for recognition of G36 by TrmD, indicated by the green arrow at the N1-proton. (D) The active-site structure of MjTrm5 (PDBID: 2ZZN), showing enzyme stabilization of the O6-carbonyl by R145 and stabilization of the N2-amino by N265 (circled in red), and positioning the methyl group of AdoMet directly opposite from the N1-proton by a distance of 2.8 Å (indicated by arrows).
One interpretation for the requirement of extensive guanosine functional groups for TrmD is that this enzyme operates within a more rigid structure for catalysis, whereas Trm5 operates with more flexibility. Notably, the active site of TrmD is built at the bottom of a deep trefoil-knot structure that is stabilized by a large dimer interface formed by interdigitation between the two monomers of the enzyme. Such a deep-knotted structure is topologically constrained and is predicted by biophysical analysis to possess limited conformational space with significantly slower folding kinetics relative to an unknotted structure (King et al. 2010). Indeed, the rigidity of TrmD is supported by the lack of noticeable movements of the enzyme in complex with the adenosine fragment of AdoMet relative to the structure in complex with AdoMet, whereas the flexibility of Trm5, derived from the more fluid Rossmann fold, is evident from the marked structural shifts upon binding adenosine relative to AdoMet (Lahoud et al. 2011). Substantial flexibility of the Trm5 active site is also implied from the significant physical separation of the proposed general base (E185 in MjTrm5) from the N1-proton of G37 (by >5 Å), as observed in the ternary crystal structure of the enzyme complex with tRNA and AdoMet (Goto-Ito et al. 2009; Christian et al. 2010a).
Comparison with crystal structure
Kinetic analysis probes the rate-determining step of methyl transfer, revealing insight that is highly valuable for comparison with structural analysis. A ternary complex of MjTrm5-tRNACys-AdoMet is available (Goto-Ito et al. 2009), although a tRNA-bound ternary complex of TrmD is lacking. In the Trm5 ternary complex, the enzyme places the methyl group of AdoMet directly opposite from the N1 of G37 within a short H-bonding distance (2.8 Å), while stabilizing the O6-carbonyl and the N2-amino groups with the side chains of R145 and N265, respectively (Fig. 6D). The stabilization by R145 (a strictly conserved residue in the Trm5 family) is suggested to neutralize the electron-rich O6-carbonyl after proton abstraction from the N1 of G37 (Christian et al. 2006) (see a proposed model in Supplemental Fig. S8), while the stabilization by N265 (in the signature NLP loop of Trm5) reminiscent of the asparagine in the NPPY motif of other Rossmann-fold methyltransferases (Gong et al. 1997; Goedecke et al. 2001; Schubert et al. 2003) is to orient the N1 of G37 for nucleophilic attack on the methyl group of AdoMet. Importantly, the short distance between N1 of G37 and the methyl of AdoMet excludes the presence of an N1-proton, suggesting that proton abstraction has already taken place and that the structure is captured at a state that is poised for methyl transfer.
While the ternary crystal structure of Trm5 reveals enzyme interaction with three functional groups of G37 (N1, O6-carbonyl, and N2-amino), kinetic analysis reveals interaction with only two functional groups (N1, O6-carbonyl) (Fig. 6B,D). The absence of the interaction with the N2-amino group in the kinetic analysis suggests that this interaction is not critical during the rate-determining step of methyl transfer. A recent activity-pH analysis of Trm5 has suggested that the rate-determining step involves the proton abstraction from the N1 of G37 (Christian et al. 2010a). Thus, a combination of kinetic and structural analysis provides unique insight into the temporal order of the Trm5 reaction, in which the enzyme interaction with the N1, O6 groups of G37 occurs earlier during proton extraction, while the additional interaction with the N2-amino group occurs later when the enzyme is ready to perform methyl transfer. Such insight is significant for the understanding of reaction mechanism.
A model of G37 recognition in vivo
While we have shown that TrmD and Trm5 occupy distinct biological domains and adopt unrelated structures with dissimilar recognition of the AdoMet substrate, this work further elaborates on their differences by showing their distinguishable recognition of the G37 substrate. However, despite these differences, an estimation based on enzyme activity and tRNA fraction in E. coli for TrmD and in yeast for Trm5 suggests that that both enzymes exist in relatively low concentration (100–700 nM) as compared with the concentration (∼60 μM) of their tRNA substrates in the respective cellular environment. Although the estimates cannot precisely determine the concentrations of these cellular components, the more than 100-fold molar excess of tRNA over enzyme in a cellular context is significant. Considering that each enzyme must screen among the total population of tRNA molecules to identify its substrates (∼11% for EcTrmD and ∼28% for ScTrm5), this difference can be as large as 5000-fold for TrmD and 300-fold for Trm5. Because the synthesis of m1G37-tRNA is essential for growth in bacteria and in yeast (Bjork et al. 1989, 2001; O'Dwyer et al. 2004; Baba et al. 2006), the large excess of tRNAs demands that TrmD and Trm5 must rapidly screen these molecules and perform catalysis on correct ones to support growth.
Our kinetic analysis reveals that the parameter Kd (tRNA) is consistently affected by various guanosine analogs (Tables 1 and 2), indicating that the recognition of appropriate guanosine functional groups is a major determinant in the ground-state binding of tRNA in the formation of the enzyme–AdoMet-tRNA encounter complex. This suggests a model in which the binding discrimination occurs directly at the G36-G37 site for TrmD and at the G37 site for Trm5 (Fig. 7). The direct discrimination at the site of interest upon binding excludes an alternative model, in which these enzymes bind to a distal site and slide around the tRNA molecule to reach the site of interest. Thus, we propose that each enzyme would use the search engine for the distinguishable functional groups of guanosine(s) to identify tRNA substrates, while rejecting molecules that lack these groups. Upon recognition of a substrate and synthesis of m1G37-tRNA, each enzyme would release the product and continue the search for the next substrate. By performing rapid binding and release and by hopping from one tRNA to the next, TrmD and Trm5 would have the ability to rapidly assess the global state of m1G37-tRNA to support cell growth.
FIGURE 7.
A model of tRNA recognition by direct recognition of G37. (A) Direct recognition of G36 and G37 by TrmD. (B) Direct recognition of G37 by Trm5. Both enzymes screen for tRNA substrate by rapid binding and discrimination of the site of interest.
MATERIALS AND METHODS
Materials
EcTrmD with an N-terminal His tag and MjTrm5 with a C-terminal His tag were expressed in E. coli and purified by the metal affinity Ni2+ resin (Redlak et al. 1997; Christian et al. 2004). Unmodified fragments of EctRNALeu and MjtRNACys were prepared by in vitro transcription based on DNA templates constructed from overlapping oligonucleotides and were gel-purified (Zhang et al. 2008). RNA fragments containing a guanosine analog were purchased from Dharmacon, while the fragment containing m1G36 in EctRNALeu was chemically synthesized on an Applied Biosystem (Broom et al. 1964; Agris et al. 1995). [3H-methyl]-AdoMet was purchased from PerkinElmer, while unlabeled AdoMet was purchased from Sigma-Aldrich.
Synthesis of m1G36-containing fragment
Several versions of m1G phosphoramidite were prepared in which 2′-hydroxyl groups were protected by TOM or TBDMS (Hoebartner et al. 2003; Porcher and Pitsch 2005). However, because introduction of TOM or TBDMS as 2′-hydroxyl protecting group always resulted in formation of significant amount of 3′-hydroxyl isomer byproduct, we developed an alternative scheme (see Supplemental Fig. S8) by selectively protecting the 3′ and 5′-hydroxyls first with a cyclic di-tert-butylsilyl group, followed by protecting the 2′-hydroxyl with TBDMS. Our synthesis started from guanosine free nucleoside (compound 1 in Supplemental Fig. S9). The N1-H of guanosine was first methylated regioselectively by treatment with methyl iodide in DMSO in the presence of potassium carbonate to give a crude product. Owing to the high polarity and poor solubility of N1-methyl guanosine free nucleoside, the crude product was treated with acetic anhydride in pyridine to give a 2′,3′,5′-acetyl protected analog (compound 2) in 64% yield for two steps of reaction. The exocyclic amine was kept intact under the acetylation conditions. Treatment of 2 with isobutyryl chloride in pyridine led to the formation of a doubly protected di-isobutyramide intermediate (compound 3) in 87% yield. Treatment of 3 with sodium hydroxide in methanol removed all of the three acetyl groups and one isobutyryl group to give compound 4 in 90% yield. Selective protection of the 3′,5′-hydroxyls by treatment of 4 with di-tert-butylsilyl-bis(trifluoromethanesulfonate) in DMF followed by protection of the 2′-hydroxyl by treatment with TBDMS-Cl and imidazole gave the intermediate compound 5 in 88% yield. Treatment of 5 with hydrogen fluoride-pyridine in THF removed the di-tert-butylsilyl protecting group selectively provided compound 6 in 92% yield. Selective protection of the 5′-OH of 6 with DMTr gave 7 (82%) and subsequent phosphitylation of the 3′-OH of 7 by standard procedure gave phosphoramidite 8 in 78% yield.
The m1G phosphoramidite 8 was then incorporated into a short 9-mer RNA sequence 5′-CAXGUGUUA-3′ (X = m1G) using double coupling. Wild-type phosphoramidites for A, C, G, U, and CPG carriers were purchased from Glen Research. The terminal DMTr protecting group was removed from the oligonucleotides by using the DMTr off mode. After each synthesis, the resin containing the oligonucleotide was transferred to a 2-mL vial and treated with 2.0 mL of 3:1 concentrated NH4OH:EtOH (ethanol) overnight at 55°C. The supernatant was collected and concentrated to dryness. The residue was then treated with a mixture of N-methyl pyrrolidinone, triethylamine, and triethylamine trihydrofluoride (0.3 mL, 6:3:4 [v/v/v]) for 90 min at 65°C. After it was cooled down for 10 min, NaOAc (3 M, 30 μL) was added followed by n-BuOH (1 mL) and the mixture cooled to −70°C for 1 h and centrifuged at 4°C (10,000g) for 20 min. The pellet was decanted and washed with 70% EtOH. The pellet was then dissolved in deionized water (1 mL) and purified by C18 reverse-phase HPLC with a gradient of 0%–20% acetonitrile in 0.1 M TEAA. The purified 9-mer RNA was further characterized by MALDI-TOF MS with the [MH]+ = 2870.
Preparation of modified tRNA
Each reconstituted tRNA with a guanosine analog was constructed by enzymatic joining of RNA fragments as follows. To reconstitute dG37-EctRNALeu and dG37-MjtRNACys, fragment I encoding nucleotides 1–36 was transcribed by T7 RNA polymerase with natural NTPs (final 4 mM each) and with guanosine (final 16 mM) as the initiator nucleotide, while fragment II encoding nucleotides 37–76 was transcribed similarly, except with dGMP (final 16 mM) as the initiator nucleotide. The two fragments (80 μM of fragment I and 10 μM of fragment II) were annealed by heating for 3 min at 80°C, followed by slow cooling to 37°C over 20 min, and were subsequently joined by T4 RNA ligase 1 (prepared from an overproducer clone [from S. Strobel], 5 μM) in 50 mM Tris-HCl (pH 8.0), 10 mM DTT, 10 mM MgCl2, and 1 mM ATP for 2 h in 37°C. The reconstituted full-length tRNA was confirmed by analysis of denaturing PAGE (12% with 7 M urea) and was excised from the gel and purified by extraction and ethanol precipitation. To reconstitute other guanosine-modified MjtRNACys, fragment I was made by in vitro transcription to encode nucleotides 1–36 with guanosine as the initiator nucleotide, fragment II was made by chemical synthesis to encode nucleotides 37–48, where a guanosine analog was placed at position 37, and fragment III was made by transcription to encode nucleotides 49–76. All three fragments (80, 10, 80 μM, respectively) were heat-cooled, aligned by a DNA splint (30 μM, encoding nucleotides complementary to positions 17–68 of the tRNA), and joined by T4 RNA ligase 2 (prepared from an overproducer clone [Nandakumar et al. 2004], 5 μM). The ligation product was confirmed and purified as described above. To reconstitute other guanosine-modified EctRNALeu, fragment I was made by in vitro transcription to encode nucleotides 1–35 with guanosine as the initiator nucleotide, fragment II was made by chemical synthesis to encode nucleotides 36–42, where a guanosine analog was placed at position 36 or 37, and fragment III was made by transcription to encode nucleotides 43–76. All three fragments (40, 10, 40 μM, respectively) were heat-cooled, aligned by a DNA splint (30 μM, encoding nucleotides complementary to positions 16–62 of the tRNA), and joined by T4 RNA ligase 2 (5 μM) as described above. To reconstitute m1G36-EctRNALeu, fragment I was made by transcription to encode nucleotides 1–33 with guanosine as the initiator nucleotide, fragment II was made by chemical synthesis to encode m1G36 in nucleotides 34–42, and fragment III was made by transcription to encode nucleotides 43–76. All three fragments (40, 10, 40 μM, respectively) were joined with the assistance of a DNA oligonucleotide (30 μM) by T4 RNA ligase 2 as described above.
RNase T1 digestion
Each tRNA was labeled at the 3′ end by the Bacillus stearothermophilus CCA-adding enzyme (Cho et al. 2003) using [α-32P]ATP, and digested by RNase T1 (Roche; 109193) in 20 mM sodium citrate (pH 5.5), 1 mM EDTA, for 10 min at 50°C. The generated fragments were separated by denaturing 7 M urea/12% PAGE and analyzed by phosphorimaging.
Mass spectrometry
The procedures were based on previously published work (Crain 1990; Douthwaite and Kirpekar 2007; Giessing et al. 2011). For MALDI-TOF mass spectrometry, each tRNA (1–2 pmol) was digested by RNase A (for EctRNALeu transcripts) or by RNase T1 (for MjtRNACys transcripts) to completion, and the generated fragments were analyzed directly using 3-hydroxypicolinic acid as the matrix. Mass spectra were recorded in positive ion mode with a reflectron Time-of-Flight mass analyzer on a PerSeptive Voyager-DE STR instrument (Applied Biosystems) (Douthwaite and Kirpekar 2007). LC-tandem mass spectrometry of tRNA nucleosides was performed according to a recently published procedure (Giessing et al. 2011). After complete hydrolysis of the phosphodiester backbone (Crain 1990), tRNA nucleosides were separated on a porous graphitized carbon column with direct analysis on an ion trap mass spectrometer (Agilent XCT Ultra 6340) operated in positive ion mode. Nucleosides were identified based on their chromatographic retention and their mass spectrometric fragmentation pattern. Comparison with standard compounds was performed when relevant.
Kinetic assays
Single-turnover assays for synthesis of m1G37-tRNA was performed according to a previously published procedure (Christian et al. 2006, 2010b). Each tRNA was heat-denatured for 3 min at 85°C and annealed for 15 min at 37°C before use. The annealed tRNA (0.5 μM or 0.25 μM, depending on the enzyme concentration to maintain enzyme-excess condition) was then mixed with saturating [3H-methyl] of AdoMet (25 μM, specific activity 688–1050 cpm/pmol) in a reaction buffer and rapidly mixed with a specific enzyme in molar excess of the tRNA on the RQF-3 rapid chemical quench instrument. The reaction conditions for TrmD consisted of 100 mM Tris-HCl (pH 8.0), 24 mM NH4Cl, 4 mM DTT, 0.1 mM EDTA, 6 mM MgCl2, and 0.024 mg/mL BSA at 37°C, while the conditions for MjTrm5 consisted of 100 mM Tris-HCl (pH 8.0), 100 mM KCl, 4 mM DTT, 0.1 mM, EDTA, and 6 mM MgCl2 at 55°C. Methyl transfer was monitored by the incorporation of [3H-methyl] onto tRNA as acid-precipitable counts on filter pads. After correction for the filter quenching effect, the counts were calculated to determine the amount of methyl transfer. The time courses of methylated tRNA production were fit to the single exponential equation {y = m1 × [1 − exp(−m2 × m0)], where m0 is the time, m1 is the plateau level of the product, and m2 is the kobs} to determine kobs. The data of kobs as a function of enzyme concentration were fit to a hyperbola equation [y = m1 × m0/(m2 + m0), where m0 is the enzyme concentration, m1 is the kchem, and m2 is the Kd] to determine the parameters Kd (tRNA) and the maximum rate constant kchem. The catalytic efficiency of each tRNA substrate was calculated from kchem/Kd.
Determination of tRNA binding by fluorescence titration
The Kd of TrmD or Trm5 binding to tRNA substrates was determined by monitoring the quenching of intrinsic tryptophan fluorescence at room temperature. Each sample was excited at 280 nm, and the emission was monitored at 300–400 nm at room temperature in 100 mM Tris-HCl (pH 8.0), 24 mM NH4Cl, 0.1 mM EDTA, 6 mM MgCl2, 4 mM DTT, and 0.024 mg/mL BSA for EcTrmD and 100 mM Tris-HCl (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 6 mM MgCl2, and 4 mM DTT for MjTrm5. A mixture of enzyme (0.5 μM) and sinefungin (50 μM) was titrated with a series of tRNA transcript concentration (0.2–8.7 μM). The intensity at 330 nm was normalized (setting the initial intensity as 1), and the data were fit to the following equation using Kaleidagraph to obtain Kd:
where F is the normalized fluorescence signal, Fmax is the normalized fluorescence signal at saturation, and S is the concentration of tRNA.
Purification of methylated tRNA for mass spectrometry analysis
Methylated tRNAs were separated from unmethylated tRNAs using RNase H by the method previously described (Hou et al. 2006). Briefly, the tRNA sample after a methyl transfer reaction was subjected to RNase H cleavage in the presence of an oligonucleotide that only hybridized to the unmethylated fraction, rendering cleavage. The methylated fraction was resistant to hybridization or cleavage and was subsequently purified by denaturing PAGE. The sequence of the oligonucleotide specific for EctRNALeu was 5′-mAmCmAmCmUmAmAmCACCTmGmAmAG-3′, while that for MjtRNACys was 5′-mAmAmGmGmCmGmGmATCTGmCmAmGmU-3′, where “m” indicates 2′-O-methyl backbone modification. These oligonucleotides were synthesized by IDT (Integrated DNA Technologies) and used directly without further purification. To perform the RNase H cleavage, a tRNA and its specific oligonucleotide (final 4 μM each) were mixed in TE buffer (10 mM Tris-HCl at pH 8.0 and 1 mM EDTA), and heated for 3 min to 80°C, followed by addition of the RNase H buffer for annealing for 15 min at 37°C. The cleavage reaction was started by addition of RNase H (final 0.4 μM) and continued for 20 min at 37°C, followed by analysis on a 12% denaturing PAGE on a Bio-Rad Mini Protein Gel system. The band that was protected from RNase H cleavage was extracted from the gel for MS analysis.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Anette Rasmussen for technical support, Dr. Lincoln Scott (CASSIA, San Diego, CA) for providing the dG-modified nucleotide monophosphates, Dr. Scott Strobel (Yale University) for providing the expression clone of T4 RNA ligase 1, and Dr. Edward Winter (Thomas Jefferson University) for providing the S. cerevisiae strain. This work is supported by NIH grants GM081601 to Y.M.H., and GM088656 and AI081987 to J.P., and an FP7-REGPOT-2009-1 project Mobili to Z.L. and S.K., and grant VP1-3.1-SMM-07-K-01-105 to S.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.032029.111.
REFERENCES
- Agris PF, Malkiewicz A, Kraszewski A, Everett K, Nawrot B, Sochacka E, Jankowska J, Guenther R 1995. Site-selected introduction of modified purine and pyrimidine ribonucleosides into RNA by automated phosphoramidite chemistry. Biochimie 77: 125–134 [DOI] [PubMed] [Google Scholar]
- Ahn HJ, Kim HW, Yoon HJ, Lee BI, Suh SW, Yang JK 2003. Crystal structure of tRNA(m1G37) methyltransferase: Insights into tRNA recognition. EMBO J 22: 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2: 2006.0008 doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR, Wikstrom PM, Bystrom AS 1989. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244: 986–989 [DOI] [PubMed] [Google Scholar]
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broom AD, Townsend LB, Jones JW, Robins RK 1964. Purine nucleosides. VI. Further methylation studies of naturally occurring purine nucleosides. Biochemistry 3: 494–500 [DOI] [PubMed] [Google Scholar]
- Brule H, Elliott M, Redlak M, Zehner ZE, Holmes WM 2004. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43: 9243–9255 [DOI] [PubMed] [Google Scholar]
- Cho HD, Oyelere AK, Strobel SA, Weiner AM 2003. Use of nucleotide analogs by class I and class II CCA-adding enzymes (tRNA nucleotidyltransferase): Deciphering the basis for nucleotide selection. RNA 9: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian T, Hou YM 2007. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol 373: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian T, Evilia C, Williams S, Hou YM 2004. Distinct origins of tRNA(m1G37) methyltransferase. J Mol Biol 339: 707–719 [DOI] [PubMed] [Google Scholar]
- Christian T, Evilia C, Hou YM 2006. Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline. Biochemistry 45: 7463–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian T, Lahoud G, Liu C, Hoffmann K, Perona JJ, Hou YM 2010a. Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA 16: 2484–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian T, Lahoud G, Liu C, Hou YM 2010b. Control of catalytic cycle by a pair of analogous tRNA modification enzymes. J Mol Biol 400: 204–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain PF 1990. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol 193: 782–790 [DOI] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R 2006. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol 13: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S, Kirpekar F 2007. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol 425: 1–20 [DOI] [PubMed] [Google Scholar]
- Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM 2003. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol 333: 931–949 [DOI] [PubMed] [Google Scholar]
- Francklyn C, Schimmel P 1989. Aminoacylation of RNA minihelices with alanine. Nature 337: 478–481 [DOI] [PubMed] [Google Scholar]
- Galperin MY, Walker DR, Koonin EV 1998. Analogous enzymes: Independent inventions in enzyme evolution. Genome Res 8: 779–790 [DOI] [PubMed] [Google Scholar]
- Giessing AM, Scott LG, Kirpekar F 2011. A nano-chip-LC/MSn based strategy for characterization of modified nucleosides using reduced porous graphitic carbon as a stationary phase. J Am Soc Mass Spectrom 22: 1242–1251 [DOI] [PubMed] [Google Scholar]
- Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E 2001. Structure of the N6-adenine DNA methyltransferase M•TaqI in complex with DNA and a cofactor analog. Nat Struct Biol 8: 121–125 [DOI] [PubMed] [Google Scholar]
- Gong W, O'Gara M, Blumenthal RM, Cheng X 1997. Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res 25: 2702–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto-Ito S, Ito T, Ishii R, Muto Y, Bessho Y, Yokoyama S 2008. Crystal structure of archaeal tRNA(m1G37) methyltransferase aTrm5. Proteins 72: 1274–1289 [DOI] [PubMed] [Google Scholar]
- Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S 2009. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol 16: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Hagervall TG, Tuohy TM, Atkins JF, Bjork GR 1993. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J Mol Biol 232: 756–765 [DOI] [PubMed] [Google Scholar]
- Hartmer R, Storm N, Boecker S, Rodi CP, Hillenkamp F, Jurinke C, van den Boom D 2003. RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis. Nucleic Acids Res 31: e47 doi: 10.1093/nar/gng047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenstein SI, Perona JJ 2008. Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J Biol Chem 283: 22007–22017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenstein SI, Hou YM, Perona JJ 2008. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J Biol Chem 283: 21997–22006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U, Saenger W 1983. Crystallographic study of mechanism of ribonuclease T1–catalysed specific RNA hydrolysis. J Biomol Struct Dyn 1: 523–538 [DOI] [PubMed] [Google Scholar]
- Hoebartner C, Kreutz C, Flecker E, Ottenschlaeger E, Pils W, Grubmayr K, Micura R 2003. The synthesis of 2′-O-[(tri-isopropylsilyl)oxy]methyl (TOM) phosphoramidites of methylated ribonucleosides (m1G, m2G, m22G, m1I, m3U, m4C, m6A, m62A) for use in automated RNA solid-phase synthesis. Monatsh Chem 134: 851–873 [Google Scholar]
- Hou YM, Schimmel P 1988. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333: 140–145 [DOI] [PubMed] [Google Scholar]
- Hou YM, Li Z, Gamper H 2006. Isolation of a site-specifically modified RNA from an unmodified transcript. Nucleic Acids Res 34: e21 doi: 10.1093/nar/gnj018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland JL, Sengupta RN, Dai Q, Deb SK, Piccirilli JA 2008. The 2′-hydroxyl group of the guanosine nucleophile donates a functionally important hydrogen bond in the Tetrahymena ribozyme reaction. Biochemistry 47: 7684–7694 [DOI] [PubMed] [Google Scholar]
- King NP, Jacobitz AW, Sawaya MR, Goldschmidt L, Yeates TO 2010. Structure and folding of a designed knotted protein. Proc Natl Acad Sci 107: 20732–20737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahoud G, Goto-Ito S, Yoshida K, Ito T, Yokoyama S, Hou YM 2011. Differentiating analogous tRNA methyltransferases by fragments of the methyl donor. RNA 17: 1236–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Kramer G, Graham DE, Appling DR 2007. Yeast mitochondrial initiator tRNA is methylated at guanosine 37 by the Trm5-encoded tRNA (guanine-N1-)-methyltransferase. J Biol Chem 282: 27744–27753 [DOI] [PubMed] [Google Scholar]
- McClain WH, Foss K 1988. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240: 793–796 [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K, Usman N, Scaringe S, Doudna J, Green R, Schimmel P 1991. Specificity for aminoacylation of an RNA helix: An unpaired, exocyclic amino group in the minor groove. Science 253: 784–786 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Ho CK, Lima CD, Shuman S 2004. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J Biol Chem 279: 31337–31347 [DOI] [PubMed] [Google Scholar]
- Nasvall SJ, Nilsson K, Bjork GR 2009. The ribosomal grip of the peptidyl-tRNA is critical for reading frame maintenance. J Mol Biol 385: 350–367 [DOI] [PubMed] [Google Scholar]
- Noma A, Kirino Y, Ikeuchi Y, Suzuki T 2006. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J 25: 2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dwyer K, Watts JM, Biswas S, Ambrad J, Barber M, Brule H, Petit C, Holmes DJ, Zalacain M, Holmes WM 2004. Characterization of Streptococcus pneumoniae TrmD, a tRNA methyltransferase essential for growth. J Bacteriol 186: 2346–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Pavesi A, Ottonello S 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol 268: 322–330 [DOI] [PubMed] [Google Scholar]
- Porcher S, Pitsch S 2005. Synthesis of 2′-O-[(triisopropylsilyl)oxy]methyl (=tom)-protected ribonucleoside phosphoramidites containing various nucleobase analogs. Helv Chim Acta 88: 2683–2704 [Google Scholar]
- Puglisi EV, Green R, Noller HF, Puglisi JD 1997. Structure of a conserved RNA component of the peptidyl transferase centre. Nat Struct Biol 4: 775–778 [DOI] [PubMed] [Google Scholar]
- Putz J, Florentz C, Benseler F, Giege R 1994. A single methyl group prevents the mischarging of a tRNA. Nat Struct Biol 1: 580–582 [DOI] [PubMed] [Google Scholar]
- Redlak M, Andraos-Selim C, Giege R, Florentz C, Holmes WM 1997. Interaction of tRNA with tRNA (guanosine-1)methyltransferase: Binding specificity determinants involve the dinucleotide G36pG37 and tertiary structure. Biochemistry 36: 8699–8709 [DOI] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X 2003. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem Sci 28: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlin LD, Bullock TL, Nissan TA, Perona JJ, Lariviere FJ, Uhlenbeck OC, Scaringe SA 2001. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA 7: 1671–1678 [PMC free article] [PubMed] [Google Scholar]
- Strobel SA, Cech TR 1995. Minor groove recognition of the conserved G•U pair at the Tetrahymena ribozyme reaction site. Science 267: 675–679 [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J 2003. Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- White TA, Kell DB 2004. Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs. Comp Funct Genomics 5: 304–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zhang CM, Liu C, Slater S, Hou YM 2008. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNACys. Nat Struct Mol Biol 15: 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]