This article describes the role of AUX/LAX auxin influx carriers in plant development, revealing that the auxin influx carrier LAX2 regulates vascular patterning in cotyledons. Although the AUX1/LAX family members share auxin transport characteristics, these transport activities seem to be dependent on their unique cell- or tissue-type expression patterns.
Abstract
Auxin transport, which is mediated by specialized influx and efflux carriers, plays a major role in many aspects of plant growth and development. AUXIN1 (AUX1) has been demonstrated to encode a high-affinity auxin influx carrier. In Arabidopsis thaliana, AUX1 belongs to a small multigene family comprising four highly conserved genes (i.e., AUX1 and LIKE AUX1 [LAX] genes LAX1, LAX2, and LAX3). We report that all four members of this AUX/LAX family display auxin uptake functions. Despite the conservation of their biochemical function, AUX1, LAX1, and LAX3 have been described to regulate distinct auxin-dependent developmental processes. Here, we report that LAX2 regulates vascular patterning in cotyledons. We also describe how regulatory and coding sequences of AUX/LAX genes have undergone subfunctionalization based on their distinct patterns of spatial expression and the inability of LAX sequences to rescue aux1 mutant phenotypes, respectively. Despite their high sequence similarity at the protein level, transgenic studies reveal that LAX proteins are not correctly targeted in the AUX1 expression domain. Domain swapping studies suggest that the N-terminal half of AUX1 is essential for correct LAX localization. We conclude that Arabidopsis AUX/LAX genes encode a family of auxin influx transporters that perform distinct developmental functions and have evolved distinct regulatory mechanisms.
INTRODUCTION
The phytohormone auxin indole-3-acetic acid (IAA) is a versatile trigger for plant development (Vanneste and Friml, 2009). Auxin regulates embryogenesis, organogenesis, vascular tissue formation, and tropic responses in plants (Vieten et al., 2007; Petrásek and Friml, 2009). The polar transport of auxin from cell to cell is achieved through the coordinated process of efflux and influx transporters, encoded by PIN-FORMED (PIN) and P-GLYCOPROTEIN (PGP), respectively (Geisler et al., 2005; Petrásek et al., 2006; Cho et al., 2007) and AUXIN1/LIKE AUX1 (AUX/LAX) genes (Bennett et al., 1996; Swarup et al., 2008). The PIN efflux transporters have a polar plasma membrane (PM) localization that regulates the direction of auxin flow (Wisniewska et al., 2006). Their mode of action during plant development shows strong redundancy and auxin-dependent cross-regulation of their expression (Vieten et al., 2005). Localization of AUX1 has been described to be cell type–dependent and, together with PIN efflux transporters, it provides directionality of intercellular auxin flow (Swarup et al., 2001; Kleine-Vehn et al., 2006).
In Arabidopsis thaliana, the AUX/LAX family is represented by four highly conserved genes called AUX1, LAX1, LAX2, and LAX3 (see Supplemental Figure 1A and Supplemental Data Set 1 online), which encode multimembrane-spanning transmembrane proteins and share similarities with amino acid transporters. This protein family forms a plant-specific subclass within the amino acid/auxin permease super family (Young et al., 1999). Mutations in AUX1 or LAX3 result in auxin-related developmental defects. For example, aux1 mutants are agravitropic and have a decreased number of lateral roots. By comparison, a loss-of-function mutation in LAX3 results in delayed lateral root emergence, and together, LAX3 and AUX1 act concomitantly to regulate lateral root development by regulating the emergence (Swarup et al., 2008) and initiation (Marchant et al., 2002) steps, respectively. Auxin uptake experiments in heterologous expression systems have confirmed that AUX1 and LAX3 are high-affinity auxin transporters (Yang et al., 2006; Carrier et al., 2008; Swarup et al., 2008).
In contrast with AUX1 and LAX3, the functional roles of the other two members of the AUX/LAX family are not well understood. Experimental observations suggest that both may also function as auxin influx carriers (Bainbridge et al., 2008), because mutating multiple members of the AUX/LAX family affects phyllotactic patterning—a process that is known to be regulated by auxin. This is supported by the fact that AUX1 shares 82, 78, and 76% identity with LAX1, LAX2, and LAX3, respectively (see Supplemental Figure 1B online). Examination of their gene structure revealed well-conserved exon/intron boundaries for most of the sequence (see Supplemental Figure 1C online), indicating that all four members of the family have originated from a common ancestor through gene duplication. In this study, using a combination of genetic, molecular, and biochemical approaches, we provide experimental evidence that all members of the AUX/LAX family have auxin influx activity. Despite the conservation of biochemical function, we demonstrate that their regulatory and coding sequences have undergone subfunctionalization. We also show that the N-terminal domain of AUX1 provides information for correct localization of LAX proteins in the AUX1 expression domain.
RESULTS
AUX/LAX Genes Exhibit Nonredundant and Complementary Expression Patterns in Roots
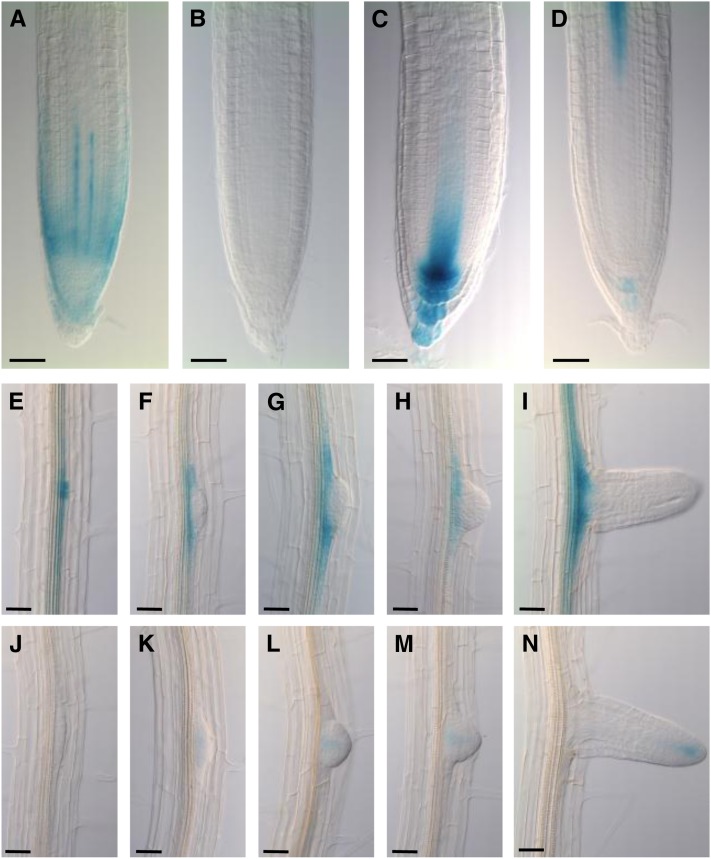
To provide insight into the roles of AUX/LAX family members in plant growth and development, their expression was analyzed in detail using in situ immunolocalization and/or promoter:β-glucuronidase (GUS) fusions and genomic yellow fluorescent protein (YFP)/VENUS translational fusions. These studies revealed that the expression patterns of AUX/LAX genes are mostly nonredundant and complementary in the primary root apex. Previous studies have shown that AUX1 is expressed in the columella, lateral root cap (LRC), epidermis, and stele tissues (Figure 1A; see Supplemental Figure 2A online) (Swarup et al., 2001; Swarup et al., 2005), whereas LAX3 is expressed in the columella and stele (Figure 1D; see Supplemental Figure 2D online) (Swarup et al., 2008).
Figure 1.
Promoter:GUS Studies Show That AUX/LAX Genes Exhibit Complementary Expression Patterns.
(A) to (D) Expression profile of AUX1 (A), LAX1 (B), LAX2 (C), and LAX3 (D) in the primary root apex.
(E) to (H) Expression profile of LAX1 ([E] to [I]) and LAX2 ([J] to [N]) during lateral root primordium development.
Bars in (A) to (D) = 35 μm; bars in (E) to (N) = 40 μm.
As part of this investigation, using two different approaches (promoter:GUS and genomic YFP/VENUS translational fusions), we report that LAX1 is expressed in the mature regions of primary root vascular tissues (Figures 1E to 1I; see Supplemental Figures 2E to 2I online). Weak LAX1 expression was also detected in the vascular tissues in the primary root apex in ProLAX1:LAX1-VENUS lines (see Supplemental Figure 2B online) but was not detectable in the ProLAX1:GUS lines (Figure 1B) even after prolonged GUS staining. This discrepancy is likely to be caused by the much larger genomic region used in ProLAX1-LAX1-VENUS lines.
LAX2 expression is detected in young vascular tissues, the quiescent center, and columella cells (Figures 1C, 6A, and 6B; see Supplemental Figure 2C online). LAX2 signal in the columella cells is most pronounced in the ProLAX2:GUS lines (Figure 1C), but is almost absent or very weak in the ProLAX2:LAX2-VENUS lines (see Supplemental Figure 2C online). Localization of LAX2 by in situ immunolocalization using anti-LAX2 antibody also showed a relatively weak expression of LAX2 in the columella cells (Figures 6A and 6B), suggesting that the stronger signal of LAX2 in ProLAX2:GUS lines is likely to be caused by the more stable nature of the GUS reporter.
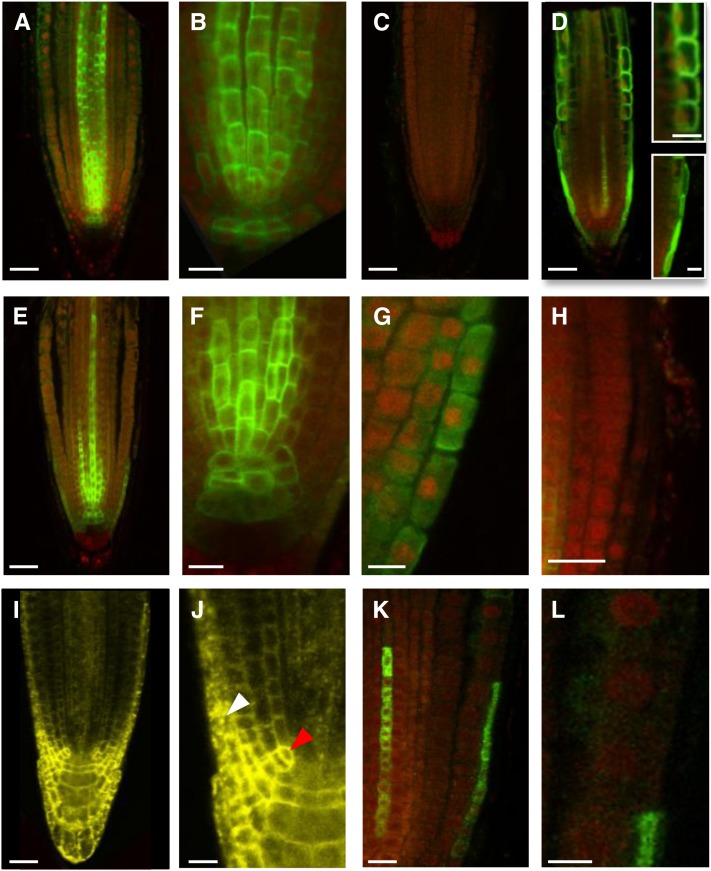
Figure 6.
LAX2 and LAX3 Cannot Be Correctly Targeted in AUX1-Expressing Cells.
(A) to (C) In situ immunodetection of LAX2 (green) in the wild type ([A] to [B]) or lax2 (C) primary roots counter stained with propidium iodide (red).
(D) In situ immunodetection of NHA-AUX1 in root apex. Inset: Close-up of epidermal (Top) and LRC (Bottom) cells.
(E) to (H) In situ immunodetection of LAX2 in aux1-22 ProAUX1:LAX2 roots showing targeting defect of LAX2 in AUX1-expressing cells, including LRC (G) and epidermal cells (H).
(I) and (J) Confocal imaging of seedlings expressing LAX2-YFP under the control of CaMV35S promoter showing correct targeting of LAX2 in LAX2-expressing cells (red arrowhead) but not in AUX1-expressing cells (white arrowhead).
(K) and (L) In situ immunodetection of LAX3-FLAG in aux1-22 ProAUX1>>LAX3 (Methods) roots, showing targeting defects of LAX3 in AUX1 expression domains including LRC and epidermal cells (L).
Bars in (A), (C) to (E), and (I) = 25 μm; bars in (B), (F) to (H), (J), and (K) = 10 μm; bars in (L) and insets = 5 μm.
The divergence in spatial expression patterns of AUX/LAX members is also clearly illustrated during lateral root development. As previously described, LAX3 is expressed outside the emerging lateral root primordia (Swarup et al., 2008), whereas AUX1 is localized within the lateral root primordia during all stages of development (Marchant et al., 2002). In comparison, LAX1 expression is first detected in stage I primordia and then mainly persists at the primordium base throughout lateral root formation (Figures 1E to 1I; see Supplemental Figures 2E to 2I online). By contrast, LAX2 expression is only detected in the central region of lateral root primordia (Figures 1J to 1N; see Supplemental Figures 2J to 2N online).
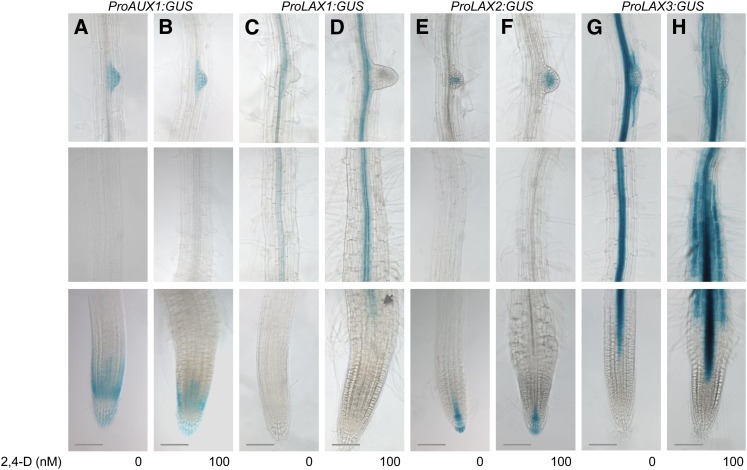
As previously reported, LAX3 expression is auxin inducible (Swarup et al., 2008) (Figures 2G and 2H; see Supplemental Figure 4G and 4H online). We then tested whether the expression of other AUX/LAX genes can be regulated by auxin. A bioinformatic search for auxin-related transcription factor binding sites and the presence of canonical auxin response elements in a 2-kb upstream sequence from ATG of the AUX/LAX promoters revealed that LAX3 and LAX1 have the highest number of transcription factor binding sites (see Supplemental Figure 3 online). To test this directly, 7-d-old seedlings were treated for 16 h with 100 nM 2,4 dichlorophenoxyacetic acid (2,4-D). Under these conditions, both LAX3-GUS (Figures 2G and 2H) and LAX3-YFP (see Supplemental Figures 4G and 4H online) expression was strongly induced by auxin. Our results also revealed that LAX1 transcript abundance was upregulated by auxin (Figures 2C and 2D; see Supplemental Figures 4C and 4D online). LAX1 expression seems stronger in the presence of auxin and is detected much closer to the root apex compared with untreated controls (arrow in Figure 2D). However, unlike LAX3, LAX1 is not induced in outer root tissues (compare Figure 2D with Figure 2H and Supplemental Figure 4D with Supplemental Figure 4H online). In contrast with LAX3 and LAX1, neither AUX1 (Figures 2A and 2B; see Supplemental Figures 4A and 4B online) nor LAX2 (Figures 2E and 2F; see Supplemental Figures 4E and 4F online) expression seem to be altered in the presence of auxin.
Figure 2.
LAX1 and LAX3 Genes Are Induced by Auxin.
Expression profile of AUX1 ([A] and [B]), LAX1 ([C] and [D]), LAX2 ([E] and [F]), and LAX3 ([G] and [H]) in absence and presence of 100 nM 2,4-D. Note LAX1 expression in the presence of auxin is detected much closer to the root apex (arrow in [D]).
Bars = 50 μm.
These results indicate that the regulation of AUX/LAX gene expression has diverged during the course of evolution, suggesting that they have acquired distinct roles in different developmental/physiological processes, an evolutionary mechanism described as subfunctionalization.
Members of the AUX/LAX Family Facilitate Distinct Auxin-Regulated Developmental Programs
To probe whether the AUX/LAX family of proteins exhibit subfunctionalization, a genetic approach was used to test the roles of these genes during Arabidopsis growth and development. AUX1 has previously been reported to play an important role during the root gravitropic response (Swarup et al., 2001; Swarup et al., 2004; Swarup et al., 2005) as well as lateral root initiation (Marchant et al., 2002), whereas LAX3 has recently been shown to be involved in lateral root emergence (Swarup et al., 2008). As part of this study, lax1 and lax2 mutants were analyzed for auxin-regulated developmental phenotypes. No root growth–related defects were obvious in either lax1 or lax2 mutants (see Supplemental Figures 5 to 7 online). Unlike aux1, mutations in lax1 or lax2 did not affect their root gravitropic responses (see Supplemental Figures 5A and 5B online) or sensitivity to synthetic auxin 2,4-D (see Supplemental Figure 6 online). Similarly, unlike aux1 and lax3, no lateral root–related defects were observed for either lax1 or lax2 mutant alleles (see Supplemental Figure 7 online).
To test the possibility of genetic redundancy between AUX1, LAX1, and LAX2, growth responses to synthetic auxin 2,4-D and lateral root development were investigated in double and triple mutants. The growth responses of double and triple mutant combinations to synthetic auxin 2,4-D were similar to aux1, suggesting that loss of lax1 and/or lax2 did not enhance the aux1 phenotype (see Supplemental Figure 8A online). Similarly, the lateral root phenotypes of aux1 lax1 and aux1 lax2 double mutants or aux1 lax1 lax2 triple mutants were not significantly different from single aux1 mutants (see Supplemental Figures 8B and 8C online). Under the same conditions, the aux1 lax3 double mutant showed a severe reduction in emerged lateral roots, in agreement with Swarup et al. (2008).
These results suggest that during the course of evolution, at least two members of the AUX/LAX family, AUX1 and LAX3, have subfunctionalized, whereas LAX1 and LAX2 gene products do not seem to influence root system architecture. However, it cannot be ruled out that LAX1 and LAX2 perform more subtle patterning functions, and specific conditions are required to uncover a root-related mutant defect. Alternatively, these genes may have acquired novel functions (neofunctionalization—no longer auxin influx carriers) or new roles (subfunctionalization—still auxin influx carriers) in other plant organs. The latter view is supported by the discovery that mutating all four members of the AUX/LAX family affects phyllotactic patterning (Bainbridge et al., 2008), and both AUX1 and LAX3 have also been implicated in apical hook development (Vandenbussche et al., 2010), processes that are known to be regulated by auxin.
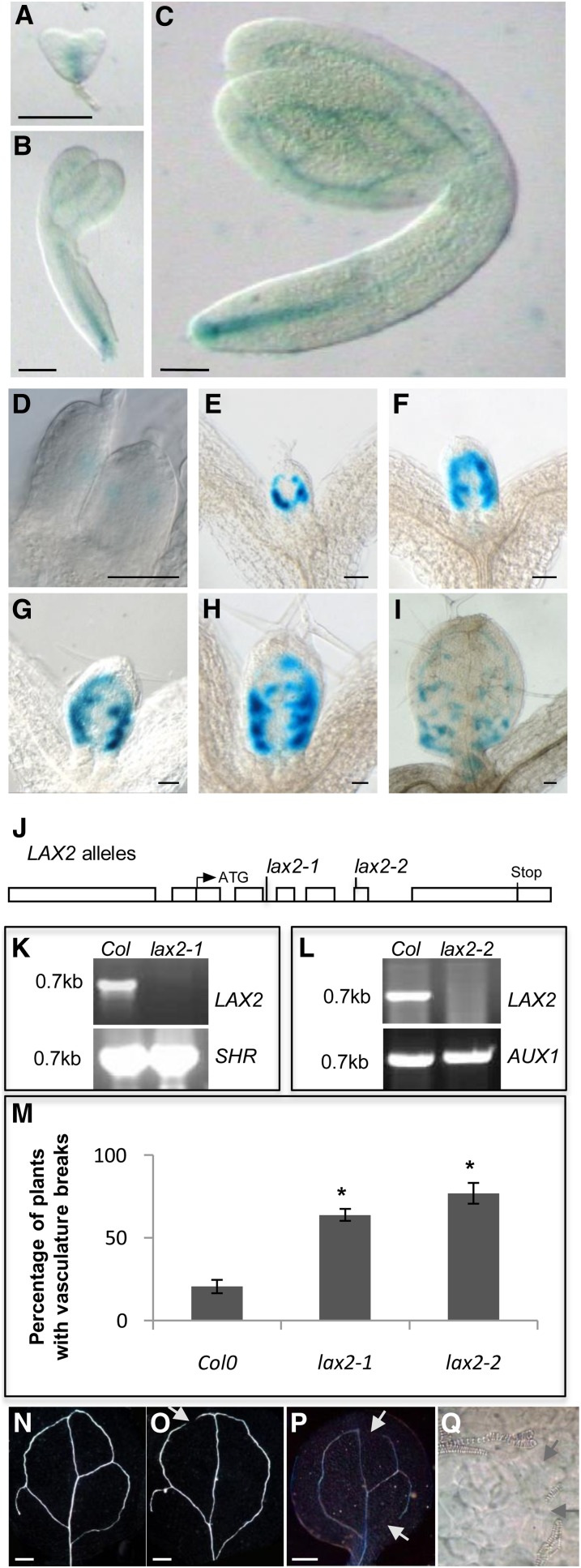
Auxin is also known to regulate vascular development, and many auxin transport and response mutants have defects in vascular development (Reinhardt, 2003; Petrásek and Friml, 2009). LAX2 promoter:GUS studies show that ProLAX2:GUS expression is associated with procambial and vascular tissues during embryogenesis (Figures 3A to 3C). In developing leaves, ProLAX2:GUS expression is detected very early at the sites of initiating veins, and starting from day 5, LAX2 is expressed along the secondary loops, starting with the first loop followed by the second, third, and fourth (Figures 3D to 3I). By days 7 to 8, LAX2 expression is also detected near the position of tertiary veins. Interestingly, LAX2 is not expressed along the midvein (Figures 3D to 3I).
Figure 3.
The lax2 Mutant Exhibits Vascular Patterning Defects in the Cotyledons.
(A) to (C) Promoter:GUS analysis of LAX2 expression in heart stage (A), torpedo (B), and mature (C) embryos.
(D) to (I) Promoter:GUS analysis of expression of LAX2 in developing leaf primordia.
(J) Structure of the LAX2 with the positions of the lax2 mutant alleles indicated. Boxes represent promoter, 5′, and 3′ untranslated regions and exons; lines represent introns.
(K) and (L) RT-PCR analysis of lax2-1 (K) and lax2-2 (L) alleles showing that LAX2 cDNA is detectable in the wild type (Col-0) but not in lax2-1 (K) and lax2-2 (L). Positive controls SHR (K) and AUX1 (L) are detected both in wild-type (Col-0) and lax2 alleles (n = 2).
(M) Graph showing the frequency of vascular breaks in cotyledons of lax2 mutant alleles compared with the wild type (Col-0). Error bars represent se. * indicates statistically significant difference compared with the wild type (Col-0); n = 30; Student’s t test, P < 0.01.
(N) to (P) Differential interference contrast images of wild-type (N), lax2-1 (O), and lax2-2 (P) cotyledons showing the vascular defect in lax2.
(Q) High-magnification differential interference contrast image pinpointing vascular break in a lax2 cotyledon.
Bars in (A) to (C) = 40 μm; bars in (D) to (I) = 100 μm; bars in (N) to (P) = 200 μm.
To assess the role of LAX2 during vascular development, two different alleles of LAX2 were analyzed (Figure 3J). The lax2-1 allele represents an En element inserted into intron 2 (position 452 from ATG), whereas lax2-2 has a T-DNA insertion in exon 6 (position 1239 from ATG). Both these alleles seem to be null alleles, because no LAX2 cDNA is detected by RT-PCR (Figures 3K and 3L). Examination of vascular development in lax2-1 and lax2-2 cotyledons revealed that both alleles exhibit a significantly higher propensity of discontinuity in vascular strands, with almost 64% of lax2-1 and 77% of lax2-2 seedlings showing vascular breaks in their cotyledons (Figures 3M to 3Q) compared with only 20% of control seedlings. In contrast with cotyledons, no defect in vascular patterning was apparent in lax2 leaves. This auxin-related developmental phenotype for lax2 provides indirect evidence for a role for LAX2 in facilitating auxin transport.
AUX1, LAX1, and LAX3 Encode Functional Auxin Influx Carriers
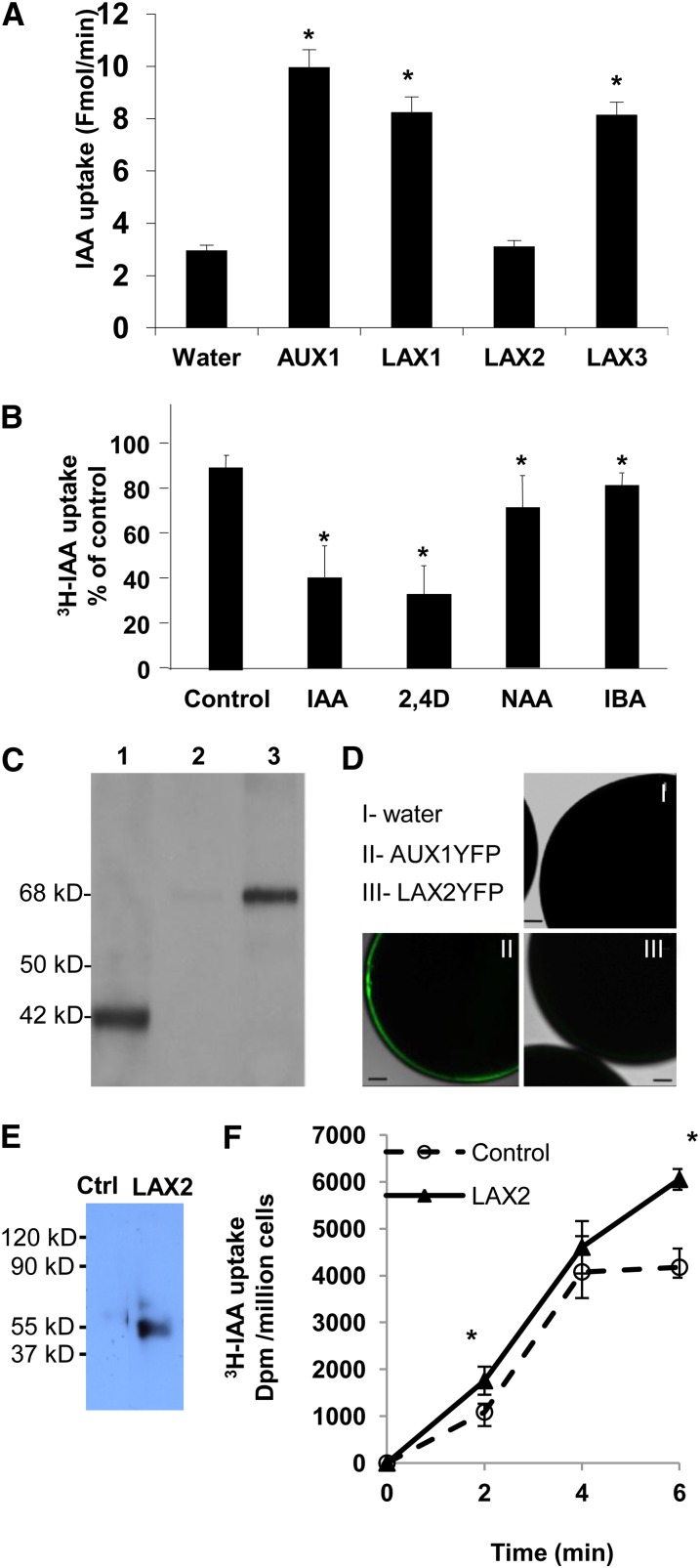
To directly test whether every AUX/LAX protein has auxin transport activity, experiments were performed in heterologous expression systems. Using an oocyte expression system, both AUX1 (Yang et al., 2006) and LAX3 (Swarup et al., 2008) were previously shown to be high-affinity auxin transporters. Similar experiments were performed for LAX1 and LAX2. These experiments revealed that LAX1 exhibited auxin uptake activity in oocytes (Figure 4A). Competition experiments with cold 2,4-D or IAA significantly reduced the uptake of radiolabeled IAA by oocytes injected with LAX1 complementary RNA (cRNA), suggesting a carrier-mediated uptake (Figure 4B). By contrast, there was only a small reduction in tritium-labeled IAA ([3H]IAA) uptake in the presence of the lipophilic auxin analog 1-naphthalene acetic acid or indole butyric acid (Figure 4B). Surprisingly, no auxin uptake activity was seen in LAX2-expressing oocytes (Figure 4A). Immunoblot experiments using specific anti-LAX2 antibodies revealed that the protein was correctly expressed in these oocytes (Figure 4C, lane 1), thus ruling out defects in its translation. To test whether LAX2 is correctly targeted to the PM in oocytes, a YFP-tagged version of LAX2 (LAX2-YFP) was expressed. Immunodetection again showed that LAX2-YFP was correctly expressed in these oocytes and was detected in the membrane and not the cytosolic fractions (Figure 4C, lanes 2 and 3). However, confocal analysis revealed no detectable LAX2-YFP on the PM (Figure 4D, panel III). In comparison, YFP fluorescence was clearly seen on the PM of oocytes expressing AUX1-YFP (Figure 4D, panel II). These results show that LAX2-YFP, unlike AUX1-YFP, is not properly targeted to the PM in Xenopus laevis oocytes and may suggest a requirement for some plant-specific accessory proteins/factors for its correct targeting that are lacking in X. laevis. As an alternative approach, LAX2 transport activity was also assayed using a yeast-based heterologous expression system (Yang and Murphy, 2009) to determine the role of LAX2 in IAA uptake. In this system, LAX2-expressing yeast cells displayed a weak but consistent IAA uptake activity compared with control cells (Figures 4E and 4F).
Figure 4.
AUX/LAX Proteins Are Functional Auxin Influx Transporters.
(A) Uptake of [3H]IAA into X. laevis oocytes injected with water or AUX1, LAX1, LAX2, and LAX3 cRNAs at pH 6.4. Oocytes injected with AUX1, LAX1, and LAX3 cRNAs showed increased [3H]IAA uptake when compared with the water-injected control (n = 8).
(B) Uptake of [3H]IAA into oocytes injected with LAX1 cRNA was examined in the presence of excess unlabeled IAA, the auxin analogs 2,4-D and 1-naphthalene acetic acid (NAA), and the naturally occurring auxin form indole butyric acid (IBA) (n = 5).
(C) Immunoblot analysis of oocytes injected with LAX2 (lane 1) or LAX2-YFP (lanes 2 and 3) cRNAs. Total oocyte extract expressing LAX2 (lane 1) or LAX2-YFP (cytosolic fraction, lane 2; microsomal fraction, lane 3) were separated by SDS-PAGE and immunodetected using anti-LAX2 antibodies (dilution 1/1000). Note the size difference between native LAX2 (42 kD) and LAX2-YFP (68 kD).
(D) Laser scanning confocal images of oocytes injected with water (I), AUX1-YFP cRNA (II), or LAX2-YFP cRNA (III).
(E) Immunoblot analysis of empty vector control or LAX2 expressing S. pombe cells. Proteins were separated by SDS-PAGE and immunodetected using anti-LAX2 antibodies (dilution 1/1000).
(F) Uptake of [3H]IAA into empty vector control (dashed line) versus LAX2-expressing (solid line) S. pombe cells compared with zero time point.
Error bars represent sd. * indicates statistically significant difference. Student’s t test P < 0.05.
Bar in (D) = 100 μm.
[See online article for color version of this figure.]
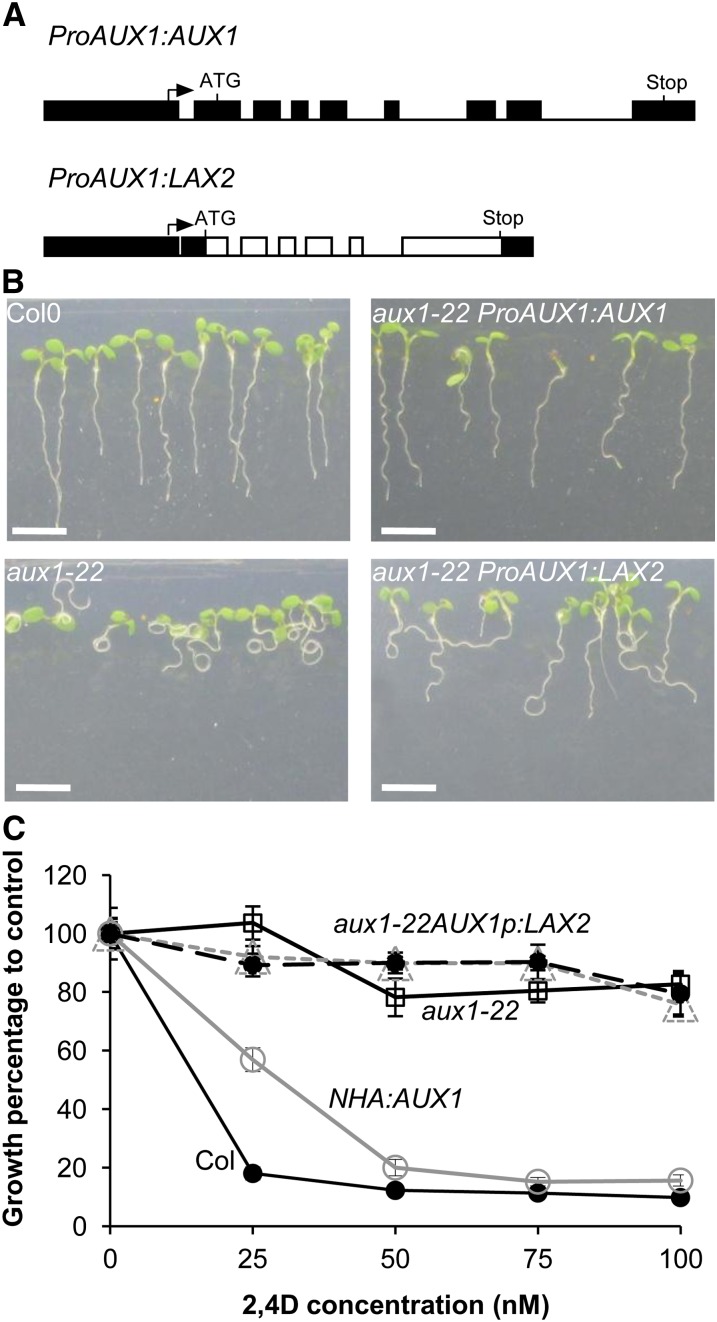
To further probe whether LAX2 encodes an auxin influx transporter, we also used a genetic assay. We reasoned that if LAX2 encodes a functional auxin transporter, an AUX1 promoter-driven LAX2 sequence would be expected to complement aux1 mutants. The aux1 mutant shows reduced sensitivity to auxins 2,4-D and IAA and has a strong agravitropic root phenotype (Bennett et al., 1996; Swarup et al., 2001, 2005) and a defect in lateral root initiation (Marchant et al., 2002). To test the ability of LAX2 to complement the aux1 mutant, a ProAUX1:LAX2 construct was created to express LAX2 under the control of the 1.7-kb AUX1 promoter and was then introduced into an aux1 mutant allele, aux1-22 (Figure 5A). Homozygous T3 seedlings were then tested for the restoration of the aux1 mutant phenotype (root gravitropic response and sensitivity to 2,4-D). As expected, ProAUX1:AUX1 lines (AUX1 promoter driving AUX1 that was used as a positive control) fully rescued the 2,4-D–resistant root growth and agravitropic phenotypes of aux1 seedlings (Figures 5B and 5C). By contrast, ProAUX1:LAX2 lines failed to rescue the root agravitropic phenotypes of aux1 seedlings (Figure 5B) as well as 2,4-D–resistant root growth (Figure 5C).
Figure 5.
AUX/LAX Genes Are Not Fully Functionally Interchangeable.
(A) Gene constructs used for genetic complementation assays. AUX1 (control) or LAX2 genomic sequences were cloned between the AUX1 promoter and terminator to create ProAUX1:AUX1 and ProAUX1:LAX2 (boxes represent promoter, 5′, and 3′ untranslated regions and exons; lines represent introns).
(B) Root gravitropic phenotypes of the wild type (Col-0), aux1-22, and aux1-22 complemented by either ProAUX1:AUX1 (control) or ProAUX1:LAX2 transgenes (n = 40).
(C) Growth responses of the wild type (Col-0), aux1-22, and aux1-22 complemented by either ProAUX1:AUX1 (control) or ProAUX1:LAX2 transgenes grown at various concentrations of 2,4-D and root growth expressed as percentage of zero control (n = 40). Error bars represent se.
Bar in (B) = 5 cm.
[See online article for color version of this figure.]
LAX2 Is Mistargeted in AUX1-Expressing Cells
To determine why LAX2 did not rescue the aux1 phenotypes, a quantitative RT-PCR experiment was initially used to measure transgene expression levels of ProAUX1:AUX1, ProAUX1:LAX2, and ProAUX1:N-terminal HA epitope–tagged AUX1 (NHA-AUX1) (Swarup et al., 2001) lines compared with wild-type AUX1 levels (see Methods). Quantitative RT-PCR revealed that LAX2 transgene was consistently expressed at equivalent levels to those of ProAUX1:AUX1 and ProAUX1:NHA-AUX1 (see Supplemental Figure 9 online). Hence, transgene expression was not the basis for the lack of rescue of the aux1 phenotypes. Next, we tested whether LAX2 was either incorrectly translated or trafficked in AUX1-expressing cells in these lines by in situ immunolocalization using anti-LAX2 antibodies. Because of the high similarity between all AUX/LAX family members, the specificity of the antipeptide antibody was tested. In wild-type seedling roots, a strong signal was seen in vascular tissues, the S1 columella layer, and the quiescent center (Figures 6A and 6B), but no signal was detected in equivalent tissues of lax2 seedlings (Figure 6C), confirming the high specificity of this antibody for LAX2. Furthermore, immunolocalization of LAX2 exhibited a broadly similar spatial expression pattern to that obtained using ProLAX2:GUS (Figure 1C) and ProLAX2:LAX2-VENUS lines (see Supplemental Figure 2C online), suggesting the absence of posttranscriptional control of LAX2 in LAX2-expressing cells (Figures 1C, 6A, and 6B; see Supplemental Figure 2C online). Slight differences in expression of ProLAX2:GUS, particularly in the columella cells, may be caused by differences in stability of GUS and LAX2 proteins.
We then tested the localization of LAX2 in ProAUX1:LAX2 lines. As reported previously (Swarup et al., 2001; Swarup et al., 2005), AUX1 is expressed in columella, LRC, epidermis, and protophloem cells (Figure 6D). In ProAUX1:LAX2 lines, as expected, a strong LAX2 signal was seen in LAX2-expressing cells (endogenous LAX2; Figures 6E and 6F); however, in cells that normally also express AUX1, the transgene-derived LAX2 signal was either weak (LRC and columella; Figures 6F and 6G) or absent (epidermis; Figure 6H). Surprisingly, the LAX2 signal in columella and LRC cells accumulated inside the cell and was only occasionally found at the PM (Figures 6G and 6H, compare with inset in Figure 6D). Altogether, our data demonstrate that misexpressing LAX2 in AUX1-expressing cells results in targeting defects for the protein in these tissues. This was further supported by an analysis of Pro35S:LAX2-YFP lines. In these lines, YFP signal was clearly seen in AUX1-expressing cells, including the epidermis, but most of the signal is localized inside the cell. By contrast, LAX2-YFP seems to be correctly localized to the PM in the LAX2-expressing cells (Figures 6I and 6J). These results suggest that the subcellular distribution of LAX2 is distinct in different plant cells and tissues. To investigate whether other members of the family are also subject to such regulation, we expressed LAX3 under the control of the AUX1 promoter. The AUX1 promoter-driven LAX3 lines also failed to rescue aux1 mutant phenotypes. In situ immunolocalization revealed reduced LAX3 protein abundance and targeting defects that were similar in nature to those observed for ProAUX1:LAX2 lines (Figures 6K and 6L). We conclude that although AUX1/LAX family members may share auxin transport characteristics, these transport activities seem to be dependent on their unique cell- or tissue-type expression patterns.
The AUX1 N Terminus Is Required for Correct Localization in the AUX1 Expression Domain
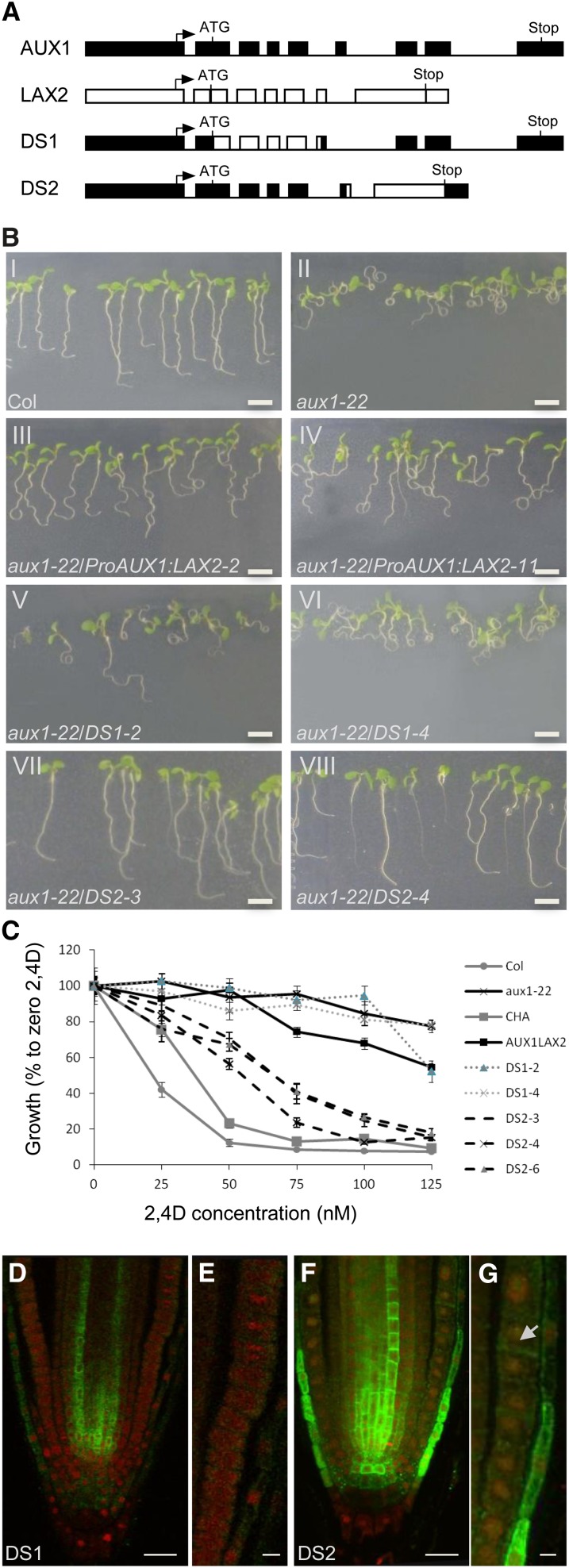
To further investigate the inability of LAX2 to correctly localize in the AUX1 expression domain, domain swap experiments were designed where either the N-terminal half of LAX2 was fused to the C-terminal half of AUX1 (DS1) or the N-terminal half of AUX1 was fused to the C-terminal half of LAX2 (DS2) to create chimeric genes driven by the AUX1 promoter (Figure 7A). These constructs were then introduced into an aux1 mutant allele, aux1-22. Homozygous T3 seedlings were then tested for the rescue of the aux1 mutant phenotype (root gravitropic response and sensitivity to 2,4-D). The results revealed that, like ProAUX1:LAX2 (Figures 7B, panels III and IV, and 7C), DS1 lines also failed to rescue root agravitropic phenotypes of aux1 seedlings (Figure 7B, panels V and VI) as well as 2,4-D–resistant root growth (Figure 7C). By contrast, DS2 lines rescued both the 2,4-D–resistant root growth (Figure 7C) and agravitropic phenotypes of aux1 seedlings (Figure 7B, panels VII and VIII).
Figure 7.
N-Terminal Half of AUX1 Is Required for Correct Localization in the AUX1 Expression Domain.
(A) Gene constructs used for domain swap experiments (boxes represent promoter, 5′, and 3′ untranslated regions and exons; lines represent introns).
(B) Root gravitropic responses of the wild type (Col-0), aux1-22, and aux1-22 complemented by ProAUX1:LAX2, DS1, or DS2 transgenes (n = 40).
(C) Growth responses of the wild type (Col-0), aux1-22, CHA-AUX1 (CHA), and aux1-22 complemented by ProAUX1:LAX2, DS1, or DS2 transgenes grown at various concentrations of 2,4-D (n = 40). Error bars represent se.
(D) In situ immunodetection of chimeric DS1 protein (green) by anti-HA antibody in primary roots counter stained with propidium iodide (red).
(E) Close-up of LRC and epidermal cells in DS1 roots.
(F) In situ immunodetection of chimeric DS2 protein (green) by anti-LAX2 antibody in primary roots counter stained with propidium iodide (red).
(G) Close up of LRC and epidermal cells in DS2 roots showing localization of DS2 protein (green) in epidermal (arrow) and LRC cells.
(E) to (H) Expression profile of ProLAX1:LAX1-VENUS [(E) to (I)] and ProLAX2:LAX2-VENUS [(J) to (N)] during lateral root primordium development.
Bar in (B) = 5 cm; bars in (D) and (F) = 20 μm; bar in (G) = 5 μm.
To probe the molecular basis of rescue, in situ immunolocalization experiments were done using either anti-HA antibody (for DS1) or anti-LAX2 antibody (for DS2). As shown in Figures 7F and 7G, DS2 lines show strong signal in AUX1 expression domains in the LRC and epidermal cells besides endogenous LAX2 signal in the vascular tissues. By contrast, DS1 lines show almost no signal in the LRC and the epidermal cell (Figures 7D and 7E), but a surprisingly strong signal is seen in the vascular tissues (Figure 7D). On the basis of these results, we conclude that the N-terminal half of AUX1 is required for correct localization in the AUX1 expression domain.
DISCUSSION
During evolution, gene family members acquire mutations that alter one or more subfunctions of the single gene progenitor. The fate of duplicated genes can encompass pseudogenization (loss of function), subfunctionalization, and neofunctionalization (Moore and Purugganan, 2005). This study shows that AUX/LAX family members in Arabidopsis have not undergone pseudogenization or neofunctionalization but have experienced subfunctionalization.
Genetic evidence presented in this and other articles demonstrates that each AUX/LAX family member regulates an auxin-dependent development process. For example, several studies support a role for AUX1 in auxin-mediated developmental programs, including root gravitropism (Swarup et al., 2001; Swarup et al., 2005; Dharmasiri et al., 2006), root hair development (Grebe et al., 2002; Jones et al., 2009), and leaf phyllotaxy (Reinhardt et al., 2003; Bainbridge et al., 2008), whereas both AUX1 and LAX3 are required for lateral root development (Swarup et al., 2008) and apical hook formation (Vandenbussche et al., 2010). Although a role for LAX1 and LAX2 in auxin-regulated root development is limited, evidence is growing that they are both required for Arabidopsis aerial development. In our study, we have provided evidence that LAX2 regulates vascular development, whereas LAX1 and LAX2 are required for leaf phyllotactic patterning (Bainbridge et al., 2008).
There is also no evidence to support neofunctionalization of AUX/LAX genes. Instead, all four AUX/LAX proteins retain an auxin influx carrier function. Using either heterologous oocyte or yeast expression systems or complementation of aux1 mutant root phenotypes, we demonstrated that AUX1 (Yang et al., 2006), LAX3 (Swarup et al., 2008), and LAX1 and LAX2 (this study) encode a family of auxin uptake transporters.
Our study provides clear evidence for subfunctionalization of AUX/LAX sequences. As a result of divergence to their regulatory sequences, we observed that AUX/LAX spatial expressions differ considerably within and between plant tissues (Figure 1; see Supplemental Figure 2 online) (Bainbridge et al., 2008; Swarup et al., 2008; Jones et al., 2009). We also report subfunctionalization of AUX/LAX coding sequences that regulate intracellular trafficking. When ectopically expressed, in situ immunolocalization revealed that LAX2 and LAX3 proteins were unable to be correctly targeted to the PMs of AUX1-expressing root cells. In wild-type roots, AUX1 is localized in cells that are involved in gravity perception (columella), signal transmission (LRC), and gravity response (epidermis) (Swarup et al., 2001; Swarup et al., 2005). The PM targeting defect of LAX2 and LAX3 is particularly severe in epidermal cells, where almost no LAX2 or LAX3 could be detected (Figures 6E to 6L). The simplest explanation for the observed tissue-specific intracellular targeting defect is the requirement of LAX2 and LAX3 for additional trafficking factors that are coexpressed in stele cells but absent in outer root tissues.
Domain swap experiments designed to test this support this notion and suggest that intramolecular trafficking signals are located in the N-terminal half of AUX1. Besides, the ability of DS2 to rescue the aux1 mutant phenotype clearly suggests that the C-terminal half of LAX2 in the chimeric DS2 protein must play a key role in its overall function as auxin influx carrier, because several missense loss-of-function aux1 alleles are located in the C-terminal half of AUX1 (Swarup et al., 2004).
All DS2 (ProAUX1:NAUX1-CLAX2 chimeric protein fusion) lines can rescue the root agravitropic defect and 2,4-D–resistant root growth of aux1 seedlings plus show correct localization of chimeric DS2 protein in LRC and epidermal cells when probed using anti-LAX2 antibodies (Figure 7). By contrast, none of the DS1 (ProAUX1:NLAX2-CHA-AUX1 chimeric protein fusion) lines rescued aux1 mutant phenotypes or showed much signal in LRC and epidermal cells (Figure 7). It has been previously shown that these expression domains of AUX1 are crucial for its function (Swarup et al., 2005), and the inability of DS1 but not DS2 to correctly localize in these expression domains provides strong evidence that the N-terminal half of AUX1 is required for correct localization in these expression domains. Surprisingly, when probed using anti-HA antibodies, although almost no signal was detected in the LRC and epidermal cells, strong DS1 signal was detected in the vascular cells. As mentioned above, the DS1 protein is a translational fusion between the N-terminal half of LAX2 and C-terminal half of AUX1 (Figure 7A), and vascular tissues are the natural/endogenous expression domain of LAX2. Although we do not currently understand the molecular basis for this differential localization of DS1 protein, it is tempting to speculate that, because the N-terminal half of AUX1 is required for correct localization in the AUX1 expression domain, the N-terminal half of LAX2 contains molecular signals that are recognized by trafficking factors in those tissues. However, compared with endogenous LAX2, DS1 chimeric protein does not seem to be correctly targeted to the PM, suggesting that, in contrast with AUX1, in the case of LAX2, the N-terminal part is still not sufficient for proper membrane targeting. AUX1 intracellular targeting is known to be regulated by AXR4, which encodes a putative endoplasmic reticulum (ER) chaperone that has been proposed to facilitate the correct folding of AUX1 and its export from ER to golgi (Dharmasiri et al., 2006). Hence, the failure of LAX2 and LAX3 to be properly targeted in AUX1-expressing cells may simply reflect a need for their own specific ER chaperones. Future identification of such trafficking factors and of intramolecular trafficking signals within AUX/LAX coding sequences will help reveal how and why they have undergone subfunctionalization during evolution.
METHODS
Plant Materials and Growth Conditions
The aux1-22 allele was used throughout this study (Swarup et al., 2004). lax1, lax2, and lax3 insertion lines have been described previously (Bainbridge et al., 2008; Swarup et al., 2008). Plants were grown on vertical Murashige and Skoog plates at 23°C under continuous light at 150 μmol m−2 s−1. Gravitropic assays were performed as previously described (Swarup et al., 2005). Lateral root numbers were determined on 6-d-old plants using a stereomicroscope. Primary root length was measured using the NeuronJ plugin of ImageJ software.
Isolation of the LAX dSpm Insertion Lines and RT-PCR Analysis
Insertion lines for the Arabidopsis thaliana LAX1, LAX2, and LAX3 were identified in the Sainsbury Laboratory Arabidopsis thaliana population as described previously (Swarup et al. 2008). LAX2 RT-PCR analysis on RNA isolated from the wild type (ecotype Columbia [Col-0]) and dSpm lax2-1 allele was performed using primers Lax2F2 (5′-GAGAACGGTGAGAAAGCAGC-3′) and Lax2R4 (5′-CGCAGAAGGCAGCGTTAGCG-3′).
Isolation of LAX2 GABI-Kat Allele (lax2-2) and RT-PCR Analysis
The lax2-2 allele (line ID GK_345D11; Nottingham Arabidopsis Stock Centre ID N433071) of LAX2 was identified from the GABI-Kat T-DNA insertional population (Kleinboelting et al., 2012). T-DNA insertion was confirmed by PCR using primer pairs 0849 (left border primer 5′-ATATTGACCATCATACTCATTGC-3′) and Lx2-25 (gene-specific primer 5′-CACAAAGTAGAGTGGCGTG-3′). The homozygous line was confirmed by the absence of a LAX2-specific band using primers Lx2-19 (5′-GGCACAAGTGCTGTTGAC-3′) and Lx2-28 (5′-CAGACGCAGAAGGCAGCG-3′). LAX2 RT-PCR analysis of RNA isolated from the wild type (Col-0) and GABI-kat lax2-2 allele was performed using primers Lx2-19 (5′-GGCACAAGTGCTGTTGAC-3′) and Lx2-28.
Generation of Transgenic Lines
The promoter GUS lines ProAUX1:GUS (Marchant et al., 2002), ProLAX1:GUS, ProLAX2:GUS (Bainbridge et al., 2008), and ProLAX3:GUS (Swarup et al., 2008) have been described before. Similarly, N- or C-terminal HA-AUX1 (NHA-AUX1 or CHA-AUX1) have been described before (Swarup et al., 2001). For genetic complementation of aux1, AUX1 and LAX2 genomic sequences were PCR amplified and fused with the Arabidopsis AUX1 promoter (1.7 kb) and terminator (0.3 kb) in a pMOG402 binary vector (MOGEN International) as previously described (Péret et al., 2007). For creation of domain swap constructs (DS1 and DS2), CHA-AUX1 (Swarup et al., 2001) and LAX2 genomic sequences were cloned into Gateway entry vector pENTR11 (Invitrogen). Both these vectors were then cut with SphI (internal unique site at identical position in both CHA-AUX1 and LAX2) and XhoI (in the vector), and the resulting inserts were swapped to create DS1 and DS2. The resulting chimeric constructs DS1 and DS2 were cut out with BamHI and XhoI and fused with the Arabidopsis AUX1 promoter (1.7 kb) and terminator (0.3 kb) in a pMOG402 binary vector (MOGEN International) as previously described (Péret et al., 2007). The LAX3-FLAG line was created by fusing the 2× FLAG epitope tag (MDYKDHDIDYKDDDDK) to the C-terminal of LAX3. The upstream activating sequence was then fused upstream of LAX3-FLAG. This UAS:LAX3-FLAG line was then crossed to the ProAUX1:GAL4 line (Swarup et al., 2005) to transactivate LAX3-FLAG in AUX1-expressing cells. The VENUS fluorescent protein (Tursun et al., 2009) fusions of LAX1 and LAX2 were generated by a recombineering approach (Zhou et al., 2011). VENUS was fused in frame after the codon 122 for ProLAX1:LAX1-VENUS and codon 110 for ProLAX2:LAX2-VENUS. Transformation of Agrobacterium (C58) and Arabidopsis was done as described before (Péret et al., 2007). Transgene-specific cDNA sequences of these lines were PCR-amplified and sequenced to ensure against rearrangements of the transgenes. All complementation experiments were performed on two independent homozygous T3 lines.
Histochemical GUS Staining
GUS staining was done as described previously (Péret et al., 2007). Plants were cleared for 24 h in 1 M chloral hydrate and 33% glycerol. Seedlings were mounted in 50% glycerol and observed with a Leica DMRB microscope.
Quantitative RT-PCR
Total RNA was extracted from roots using the Qiagen RNeasy Plant Mini Kit with on-column DNase treatment (RNase free DNase set, Qiagen). Poly(dT) cDNA was prepared from 3 μg total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Quantitative PCR was performed using SYBR Green Sensimix (Quantace) on a Stratagene Mx3005P apparatus. PCR was performed in 96-well optical reaction plates heated for 5 min to 95°C, followed by 40 cycles of denaturation for 10 s at 95°C and annealing-extension for 30 s at 60°C. Target quantifications were performed with the following specific primer pairs: AtAUX1F 5′-tgctctgatcaaagtcttctcct-3′ and AtAUX1R 5′-gaagagaagaacccagaaatgtg-3′. Expression levels were normalized to UBA using the following primers: UBAforward 5′-agtggagaggctgcagaaga-3′ and UBAreverse 5′-ctcgggtagcacgagcttta-3′. All quantitative RT-PCR experiments were performed in triplicate, and the values presented represent means ± se.
Production of LAX2 Antibody
For generation of LAX2 antibody, a peptide containing the C-terminal 15 amino acids of LAX2 (PPPISHPHFNHTHGL) plus an added Cys (for attachment to carrier protein KLH) was conjugated to KLH and was injected to rabbits in complete Freunds adjuvant. Boosters were given on days 14, 28, 42, 56, and 70. Immune serum was collected on days 49, 63, and 77. Affinity purification of the antiserum was done against the LAX2 peptide that was coupled to Pierce SulfoLink resin as per manufacturer’s instruction. The column was washed twice with 10 mM Tris-Cl buffer (pH 7.5) containing 0.5 M NaCl, once with 100 mM Gly (pH 2.5), and finally with two more washes with 10 mM Tris-Cl buffer (pH 7.5). Crude LAX2 antiserum (10 mL) buffered in 100 μL 1 M Tris-Cl (pH 7.5) was then applied on the column and rotated at 4°C overnight. Flow-through was passed twice on the column at room temperature followed by a wash each with 10 mM Tris-Cl buffer (pH 7.5) and 10 mM Tris-Cl buffer (pH 7.5) containing 0.5 M NaCl. Purified antibodies were eluted in 250-μL fractions with 100 mM Gly (pH 2.5) and neutralized with 50 μL of Tris-Cl buffer (pH 8.0).
Immunolocalization
Four-d-old seedlings were fixed, and immunolocalization experiments were performed as described previously (Swarup et al., 2005) using various primary and secondary antibodies. Localization was visualized using confocal microscopy. Primary antibodies anti-HA (Roche) and anti-FLAG (Sigma-Aldrich) were used at a dilution of 1:200, whereas anti-LAX2 was used at a dilution of 1:100. Oregon Green or Alexa Fluor–coupled secondary anti-rat, anti-mouse, or anti-rabbit antibodies (Invitrogen) were used at a dilution of 1:200. Background staining was performed with propidium iodide (Sigma-Aldrich).
Auxin Transport Assays
Auxin transport assays in oocytes were performed as previously described (Swarup et al., 2008). For uptake experiments in Schizosaccharomyces pombe, the LAX2 cDNA was amplified from pOO2-LAX2 using primers ccacLAX5 (CACCATGGAGAACGGTGAGAA) and LAX2nonstop3 (AAGGCCGTGAGTGTGATTGA), cloned into Gateway cloning vector pENTR/TOPO (Invitrogen), and confirmed by sequencing. LAX2 cDNA was subsequently cloned into the S. pombe expression vector pREP41GWHA (Yang and Murphy, 2009). LAX2 expression in S. pombe vat3 cells (Yang and Murphy, 2009) was confirmed by immunoblot using anti-HA primary antibody (1:500 dilution; Santa Cruz Biotechnology).
Accession Numbers
Atg nomenclature gene accession numbers of The Arabidopsis Information Resource database (http://www.Arabidopsis.org) are: At2g38120 (AUX1), At5g01240 (LAX1), At2g21050 (LAX2), At1g77690 (LAX3), and UBA (At1g04850).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The AUX/LAX Genes Represent a Highly Conserved Family of Auxin Influx Transporters.
Supplemental Figure 2. AUX/LAX Genes Exhibit Complementary Expression Patterns.
Supplemental Figure 3. Promoter Analysis of AUX/LAX Genes.
Supplemental Figure 4. LAX1 and LAX3 Genes Are Induced by Auxin.
Supplemental Figure 5. lax1, lax2, and lax3 Mutants Exhibit Normal Gravitropic Response.
Supplemental Figure 6. lax1 and lax2 Mutants Exhibit a Normal Response to 2,4-D.
Supplemental Figure 7. lax1 and lax2 Mutants Exhibit Normal Root Growth and Lateral Root Growth.
Supplemental Figure 8. lax1 and lax2 Mutants Do Not Enhance aux1 Mutant Phenotypes.
Supplemental Figure 9. ProAUX1:AUX1 and ProAUX1:LAX2 Seedlings Exhibit Comparable Transgene Expression Levels.
Supplemental Data Set 1. Alignment Used to Generate the Phylogeny Presented in Supplemental Figure 1A Online.
Supplementary Material
Acknowledgments
This study was supported by a Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme PIEF-GA-2008-220506 (B.P.), Biotechnology and Biological Science Research Council (M.J.B. and A.F.), U.S. Department of Energy grant DE-FG02-07ER15887 (E.N.), U.S. Department of Energy grant DE-FG02-06ER15804 (A.M.), a grant from the Bijzonder Onderzoeksfonds Methusalem project (BOF08/01M00408) of Ghent University, by Interuniversity Attraction Poles grants 6/33 and 7/29 from the Belgian Science Policy Office (M.J.B., G.T.S.B., and S.D.), National Science Foundation Grant DBI0820755 (J.M.A.), and Ayudas de Movilidad de la Junta de Extremadura, Spain GRU09054 (I.C.).
AUTHOR CONTRIBUTIONS
R.S. and B.P. designed the research, performed research, analyzed data, wrote, and edited the article. K.S., A.F., M.S., Y.Y., S.D., N.J., I.C., P.P., A.S., H.Y., and J.R., performed research and analyzed data. E.V., C.H., M.A.P.-A., J.Y., and J.A. contributed new genetic tools (recombineering technology). L.L., A.M., G.T.S.B., and E.N. analyzed data. M.J.B. designed the research and edited the article.
Glossary
- IAA
indole-3-acetic acid
- 2,4-D
2,4 dichlorophenoxyacetic acid
- GUS
β-glucuronidase
- PM
plasma membrane
- YFP
yellow fluorescent protein
- LRC
lateral root cap
- [3H]IAA
tritium-labeled IAA
- ER
endoplasmic reticulum
- Col-0
ecotype Columbia
- cRNA
complementary RNA
References
- Bainbridge K., Guyomarc’h S., Bayer E., Swarup R., Bennett M., Mandel T., Kuhlemeier C. (2008). Auxin influx carriers stabilize phyllotactic patterning. Genes Dev. 22: 810–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.J., Marchant A., Green H.G., May S.T., Ward S.P., Millner P.A., Walker A.R., Schulz B., Feldmann K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Carrier D.J., Bakar N.T., Swarup R., Callaghan R., Napier R.M., Bennett M.J., Kerr I.D. (2008). The binding of auxin to the Arabidopsis auxin influx transporter AUX1. Plant Physiol. 148: 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M., Lee S.H., Cho H.T. (2007). P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri S., Swarup R., Mockaitis K., Dharmasiri N., Singh S.K., Kowalchyk M., Marchant A., Mills S., Sandberg G., Bennett M.J., Estelle M. (2006). AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312: 1218–1220 [DOI] [PubMed] [Google Scholar]
- Geisler M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Grebe M., Friml J., Swarup R., Ljung K., Sandberg G., Terlou M., Palme K., Bennett M.J., Scheres B. (2002). Cell polarity signaling in Arabidopsis involves a BFA-sensitive auxin influx pathway. Curr. Biol. 12: 329–334 [DOI] [PubMed] [Google Scholar]
- Jones A.R., Kramer E.M., Knox K., Swarup R., Bennett M.J., Lazarus C.M., Leyser H.M., Grierson C.S. (2009). Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11: 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N., Huep G., Kloetgen A., Viehoever P., Weisshaar B. (2012). GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Swarup R., Bennett M., Friml J. (2006). Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell 18: 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A., Bhalerao R., Casimiro I., Eklöf J., Casero P.J., Bennett M., Sandberg G. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.C., Purugganan M.D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Péret B., Swarup R., Jansen L., Devos G., Auguy F., Collin M., Santi C., Hocher V., Franche C., Bogusz D., Bennett M., Laplaze L. (2007). Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol. 144: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Petrásek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Reinhardt D. (2003). Vascular patterning: More than just auxin? Curr. Biol. 13: R485–R487 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15: 2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Kramer E.M., Perry P., Knox K., Leyser H.M., Haseloff J., Beemster G.T., Bhalerao R., Bennett M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Swarup R., et al. (2004). Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10: 946–954 [DOI] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I., Hobert O. (2009). A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One 4: e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Petrásek J., Zádníková P., Hoyerová K., Pesek B., Raz V., Swarup R., Bennett M., Zazímalová E., Benková E., Van Der Straeten D. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606 [DOI] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Vieten A., Sauer M., Brewer P.B., Friml J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12: 160–168 [DOI] [PubMed] [Google Scholar]
- Vieten A., Vanneste S., Wisniewska J., Benková E., Benjamins R., Beeckman T., Luschnig C., Friml J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertová D., Brewer P.B., Ruzicka K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Yang H., Murphy A.S. (2009). Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 59: 179–191 [DOI] [PubMed] [Google Scholar]
- Yang Y., Hammes U.Z., Taylor C.G., Schachtman D.P., Nielsen E. (2006). High-affinity auxin transport by the AUX1 influx carrier protein. Curr. Biol. 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Young G.B., Jack D.L., Smith D.W., Saier M.H., Jr (1999). The amino acid/auxin:proton symport permease family. Biochim. Biophys. Acta 1415: 306–322 [DOI] [PubMed] [Google Scholar]
- Zhou R., Benavente L.M., Stepanova A.N., Alonso J.M. (2011). A recombineering-based gene tagging system for Arabidopsis. Plant J. 66: 712–723 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.