Abstract
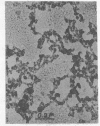
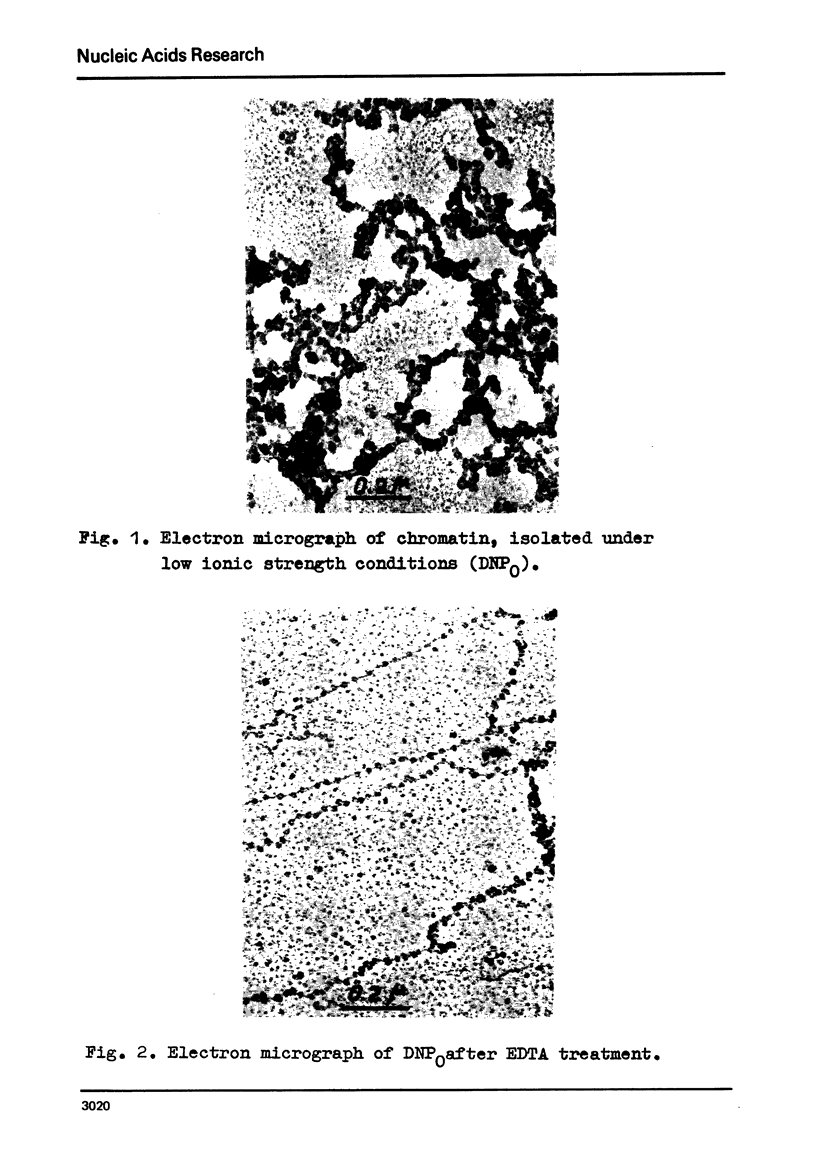
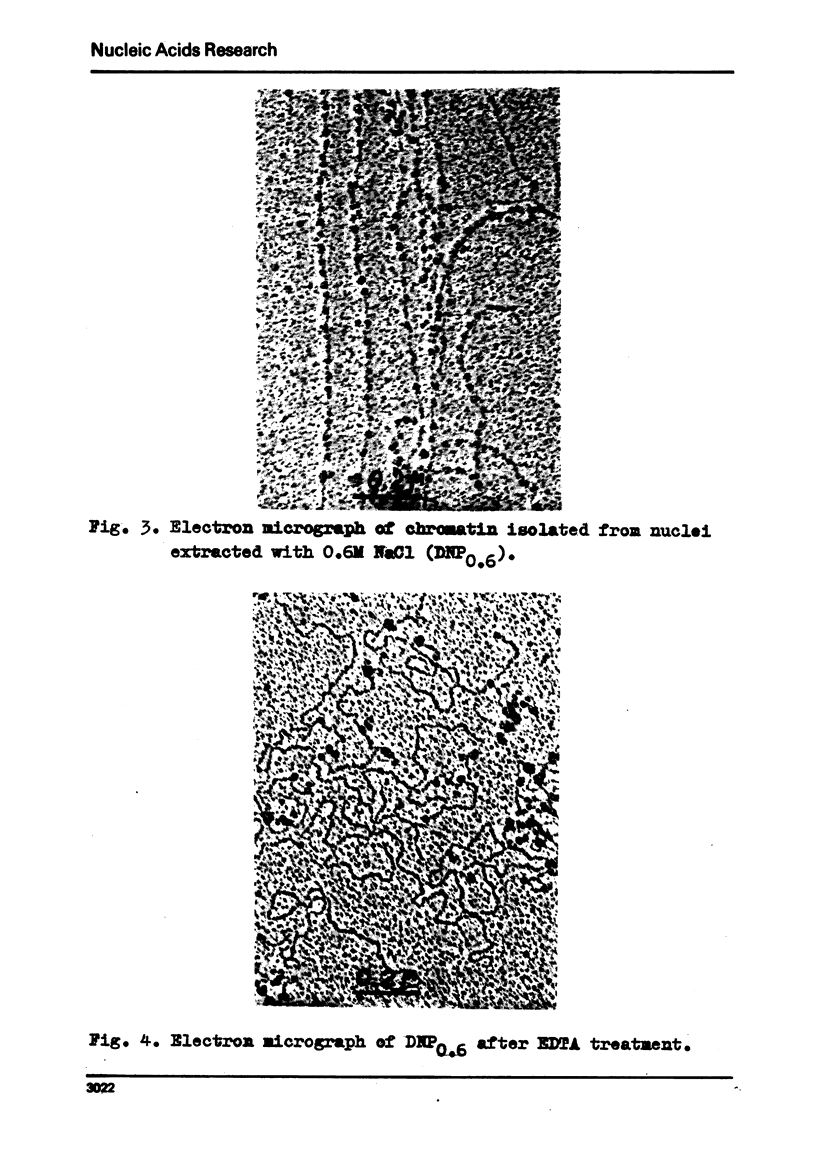
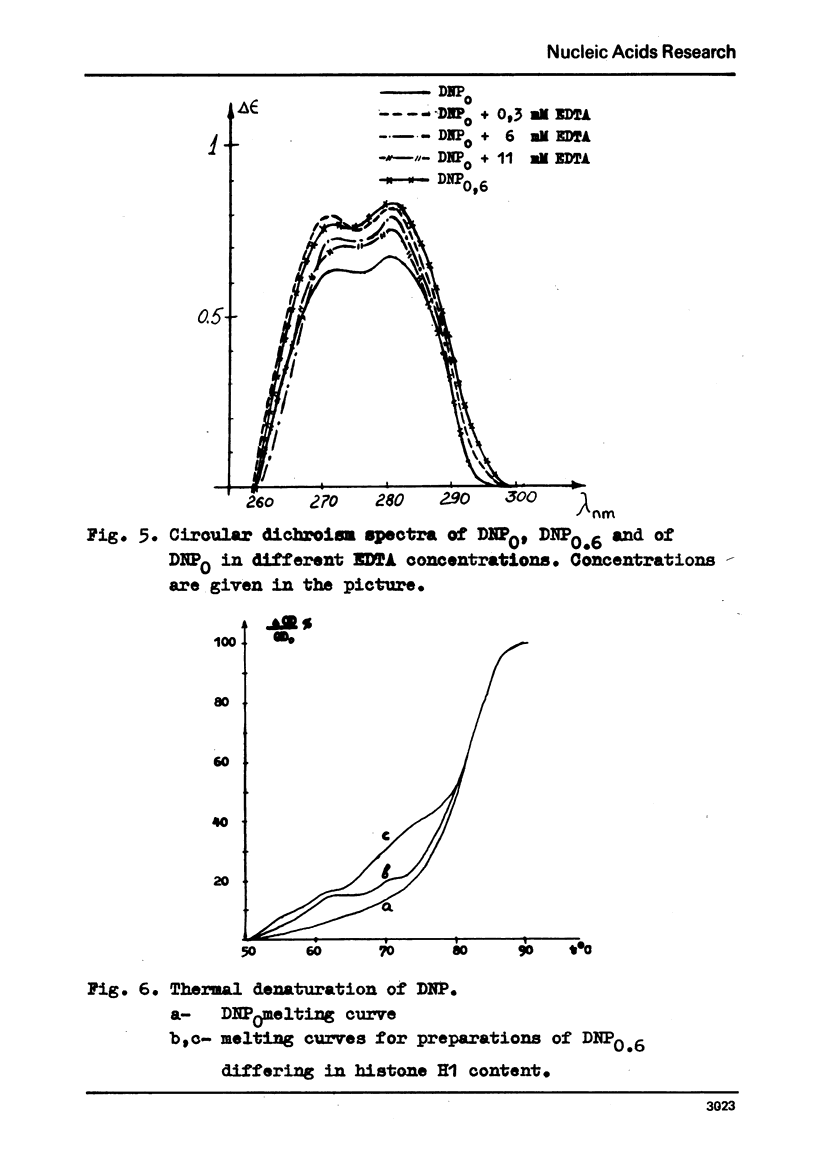
Electron microscopy shows that EDTA treatment or partial removal of histone HI converts 200-250 A chromatin fibres characteristic for native chromatin, isolated in low ionic strength conditions into fibres consisting of nucleosomes connected by segments of DNA. This structural transition is accompanied by an increase in the amplitude of positive band of CD spectra at 280 nm. Comparison of electron microscopic, thermal denaturation and electrophoretic data suggests that multiphasic character of melting curves, observed for chromatin, lacking histone HI is due to the removal of histone HI and destabilisation of the DNA segments, connecting nucleosomes. It is also shown that bivalent cations play an important part both in the stabilisation of 200 A globules and of nucleosomes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev G. P., Varshavskii A. Ia. Struktura Khromatina. (obzor nekotorykh novykh dannykh) Biokhimiia. 1976 Oct;41(10):1731–1741. [PubMed] [Google Scholar]
- Johnson R. S., Chan A., Hanlon S. Mixed conformations of deoxyribonucleic acid in intact chromatin isolated by various preparative methods. Biochemistry. 1972 Nov 7;11(23):4347–4358. doi: 10.1021/bi00773a023. [DOI] [PubMed] [Google Scholar]
- Kiryanov G. I., Manamshjan T. A., Polyakov V. Y., Fais D., Chentsov J. S. Levels of granular organization of chromatin fibres. FEBS Lett. 1976 Sep 1;67(3):323–327. doi: 10.1016/0014-5793(76)80557-1. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Lawrence J. J., Chan D. C., Piette L. H. Conformational state of DNA in chromatin subunits. Circular dichroism, melting, and ethidium bromide binding analysis. Nucleic Acids Res. 1976 Nov;3(11):2879–2893. doi: 10.1093/nar/3.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]