Abstract
During endomitosis, megakaryocytes undergo several rounds of DNA synthesis without division leading to polyploidization. In primary megakaryocytes and in the megakaryocytic cell line CHRF, loss or knock-down of p53 enhances cell cycling and inhibits apoptosis, leading to increased polyploidization. To support the hypothesis that p53 suppresses megakaryocytic polyploidization, we show that stable expression of wild-type p53 in K562 cells (a p53-null cell line) attenuates the cells' ability to undergo polyploidization during megakaryocytic differentiation due to diminished DNA synthesis and greater apoptosis. This suggested that p53's effects during megakaryopoiesis are mediated through cell cycle- and apoptosis-related target genes, possibly by arresting DNA synthesis and promoting apoptosis. To identify candidate genes through which p53 mediates these effects, gene expression was compared between p53 knock-down (p53-KD) and control CHRF cells induced to undergo terminal megakaryocytic differentiation using microarray analysis. Among substantially downregulated p53 targets in p53-KD megakaryocytes were cell cycle regulators CDKN1A (p21) and PLK2, proapoptotic FAS, TNFRSF10B, CASP8, NOTCH1, TP53INP1, TP53I3, DRAM1, ZMAT3 and PHLDA3, DNA-damage-related RRM2B and SESN1, and actin component ACTA2, while antiapoptotic CKS1B, BCL2, GTSE1, and p53 family member TP63 were upregulated in p53-KD cells. Additionally, a number of cell cycle-related, proapoptotic, and cytoskeleton-related genes with known functions in megakaryocytes but not known to carry p53-responsive elements were differentially expressed between p53-KD and control CHRF cells. Our data support a model whereby p53 expression during megakaryopoiesis serves to control polyploidization and the transition from endomitosis to apoptosis by impeding cell cycling and promoting apoptosis. Furthermore, we identify a putative p53 regulon that is proposed to orchestrate these effects.
Keywords: megakaryocytic, p53, polyploidization, microarray
megakaryocytes are giant, polyploid cells that reside in the bone marrow and are derived from hematopoietic stem cells under the influence of thrombopoietin (TPO) (36). Terminally differentiated megakaryocytes extend cytoplasmic protrusions, termed proplatelets, into the bone marrow microvasculature, which eventually fragment into platelets. Despite recent progress made in understanding the molecular events leading to megakaryocytic (Mk) polyploidization and proplatelet extension followed by platelet release into the bloodstream, many fundamental questions still remain, such as how to uncouple Mk cell apoptosis from polyploidization (35, 36).
Tumor suppressor p53 has roles in preventing genomic instability and aneuploidy or polyploidy (63). Work by others has documented the high expression of p53 in Mk cells undergoing polyploidization, but the functional role of p53 is still not clear (5, 19, 47, 54, 60, 72). We have also observed that total p53 and K382-acetylated p53, a transcriptionally active p53 form (71), are expressed at high levels in hematopoietic stem cells undergoing Mk differentiation in culture but are both expressed at lower levels in terminally differentiated polyploid Mk cells (27). Our recent studies (12, 23–25) have postulated the involvement of p53 and its downstream transcriptional targets in regulating DNA synthesis and inducing apoptosis during Mk differentiation. p53-knock-down (KD) in CHRF megakaryoblastic cells [a validated cell line model of human Mk differentiation (25)] induced with phorbol 12-myristate 13-acetate (PMA) to undergo terminal Mk differentiation results in higher Mk ploidy by means of enhancement of DNA synthesis and deferment of apoptosis (23). Furthermore, injection of TPO leads to increased ploidy of p53−/− bone marrow Mk cells in vivo compared with p53+/+ littermates (3). Moreover, the ploidy of Mk cells derived from murine p53−/− bone marrow hematopoietic progenitor cells in vitro was substantially increased (3, 23). Based on these findings we hypothesize that p53 is activated during megakaryopoiesis to control polyploidization and the transition from endomitosis to apoptosis by impeding cell cycling and promoting apoptosis.
To gain insight into potential p53 target genes that could be mediating the observed high ploidy in the p53-KD CHRF cells, the expression of a select set of p53 target genes was examined via quantitative (Q)-RT-PCR. p53-KD led to strong downregulation of the cell cycle inhibitor CDKN1A (p21); the proapoptotic genes TP53I3, TP53INP1, and BBC3; and the p53 regulator MDM2, while antiapoptotic BCL2 was upregulated in p53-KD CHRF cells (23). The proapoptotic gene BAX was also slightly downregulated, while there was hardly any impact on DNA damage-related GADD45A (23). However, a comprehensive gene expression analysis of p53 direct or indirect target genes in p53-KD vs. control Mk cells would provide much needed information for detailed studies to pursue mechanistic details of p53's role in megakaryopoiesis.
In this study, the first aim was to examine the previously formulated hypothesis (23) that p53 expression by Mk cells delimits their ability to become polyploid by inhibiting cell cycle progression and promoting apoptosis. For this aim, human wild-type p53 was expressed on K562 cells, a human p53− erythroleukemic cell line, to directly assess the effect of p53 expression on polyploidization and apoptosis during Mk differentiation induced by PMA treatment. The second aim of this study was to identify which of the known or predicted transcriptional targets of p53 are engaged in the Mk development, as well as identify indirect targets of p53 that in the context of Mk differentiation may mediate its ability to regulate cell cycle and apoptotic events. For this aim, gene expression microarray analysis was utilized to identify genes that are differentially expressed between p53-KD and control CHRF cells undergoing terminal Mk differentiation. The CHRF cell line was chosen for the microarray analysis because, first, it is a validated (25) model of human megakaryopoiesis, and second the p53-KD CHRF cells display enhanced polyploidization compared with control CHRF cells, similar to the hyperploid phenotype of the murine p53−/− Mk cells (23). While detailed validation of the role of the genes identified here is beyond the scope of this study for practical reasons, we provide strong arguments that the genes identified are likely to play a significant role in mediating the role of p53 in megakaryopoiesis.
MATERIALS AND METHODS
Culture of CHRF and K562 cells.
CHRF-288-11 (CHRF), provided by Dr. R. Smith [National Institutes of Health (NIH), Bethesda, MD] is a human megakaryoblastic cell line validated as a model for megakaryopoiesis (25). As described (23), p53-KD and control CHRF cell lines were previously generated. CHRF and K562 cells (American Type Culture Collection, Manassas, VA) were cultured in IMDM/10% FBS and treated with 10 ng/ml PMA (Sigma-Aldrich, St. Louis, MO) to induce Mk differentiation. At the designated time points, cells were washed with PBS and harvested using 1× trypsin-EDTA. Adherent and nonadherent fractions were combined for all analyses.
Measurement of ploidy, DNA synthesis, and apoptosis/viability using flow cytometry.
Ploidy analysis in CHRF and K562 cells was carried out as described (23). Measurement of DNA synthesis was carried out by assessing BrdU incorporation after incubation of the cells with 10 μM BrdU (BD Pharmingen, San Diego, CA) for 12 h at 37°C and 5% CO2 according to the manufacturer's guidelines and as reported previously (23). Measurement of apoptosis was carried out by staining the cells with annexin-PE (BD Pharmingen) for 15 min at room temperature according to the manufacturer's protocol. Measurement of viability was done by staining the cells with 1 μM TOPRO3 (Molecular Probes, Eugene, OR) for 15 min at room temperature.
Microarray analysis.
Cell samples taken from CHRF cell cultures [p53-KD and control cells (23)] on days 1, 3, 5, and 7 following stimulation with PMA and unstimulated cells were flash-frozen in liquid nitrogen to be used later for microarray and Q-RT-PCR analysis. RNA was isolated from cell pellets using the Total RNA Isolation Mini Kit (Agilent, Wilmington, DE). RNA purity and yield were estimated using a Nanodrop spectrophotometer (model ND-1000; Nanodrop, Wilmington, DE). The integrity of the isolated RNA was assessed by running the RNA 6000 Nano Assay (Agilent) on the Bioanalyzer (model 2100, Agilent). With the Quick Amp Labeling Kit (Agilent), fluorescently labeled complimentary RNA (cRNA) was generated and purified using the RNeasy Mini Spin Columns (Qiagen, Germantown, MD). Dye-swap replicates were performed for all comparisons to account for dye incorporation bias (14). Following fragmentation, the transcript mixture was hybridized on 4 × 44K whole human genome microarrays (cat. #G4112F, Agilent).The slides were scanned (Agilent Microarray Scanner) and Agilent's Feature Extraction Software (version 9.5.1, protocol GE2-v5_95_Feb07) was used to identify spots and feature outliers. To normalize signal intensity ratios for both Cy3 and Cy5, the SNNLERM algorithm developed in the Papoutsakis lab, which averages the log-transformed expression ratios for technical and biological replicates, was utilized (69). All raw and normalized microarray data are MIAME compliant and were deposited in the Gene Expression Omnibus (GEO accession number GSE30984). Subsequent analysis of the microarray data is described in detail in the results section.
Q-RT-PCR.
Q-RT-PCR was performed using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) to reverse transcribe RNA isolated from the same cell samples used in microarray hybridizations as described (23). The following Applied Biosystems Taqman Gene Expression Assays were used: Hs00153349_m1 for TP53, Hs00968432 for RRM2B, Hs00978338_m1 for TP63, Hs00368864 for CDT1, Hs00413187_m1 for NOTCH1, Hs00355782_m1 for CDKN1A (p21), Hs00180269_m1 for BAX, Hs00426835_g1 for ACTA2, Hs99999908_m1 for GUSB, and Hs99999902 for RPLP0.
Western blot analysis for p53 targets CDKN1A (p21) and BCL2.
CHRF cells were either untreated or induced to undergo terminal Mk differentiation with 10 ng/ml PMA before harvesting with a cell scraper in cell dissociation buffer (PBS + 2 mM EDTA). For total cell lysates, cell pellets from unstimulated (day 0) and PMA stimulated p53-KD and control CHRF cells (days 3, 5, and 9 after PMA), were lysed by boiling in 2× SDS sample buffer. For nuclear lysates, cell pellets were lysed using the Nuclear Extraction kit (Panomics, Fremont, CA) following the manufacturer's protocol. Whole cell lysate proteins (30 μg for BCL2) and nuclear lysate proteins [25 μg for CDKN1A(p21)] were separated by SDS-PAGE and transferred to nitrocellulose by standard techniques. Western blots were probed with an antibody to BCL2 (mouse MAb, clone: C-2, 1:5,000 dilution) followed by a chicken anti-mouse secondary IgG-horseradish peroxidase (HRP) (1:10,000 dilution) or an antibody to CKDN1A (p21) (rabbit PAb, 1:2,500 dilution) followed by a chicken-anti-rabbit secondary IgG-HRP (1:10,000 dilution) (antibodies from Santa Cruz Biotechnology, Santa Cruz, CA). An antibody against GAPDH (mouse MAb, clone: 9484, 1:2,500 dilution; Abcam, Cambridge, MA) with a chicken-anti-mouse secondary IgG-HRP (1:10,000 dilution, Santa Cruz Biotechnology) was used as loading control. Densitometry analysis was performed using ImageJ Software version 1.38 (NIH, Bethesda, MD).
Expression of p53 in K562 and CHRF cells.
The wild-type human p53 transcript (NM_000546), developed by the Vogelstein lab (6) and cloned into a pCMV-Neo-Bam vector, was purchased from Addgene (plasmid 16434; Cambridge, MA). Sequencing primers for p53 are summarized in Table 1. The p53 transcript (1,212 base pairs) was PCR amplified with the AmpliTaq Gold 360 DNA polymerase (Applied Biosystems) using primers 5′-ATTGGCAGCCAGA CTGCCTTC-3′ (forward primer) and 5′-GTCTGAGTCAGGCCCTTCTGTCTT-3′ (reverse primer) with the following cycling parameters: 95°C for 7 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 59°C for 30 s, and extension at 72°C for 1 min, followed by 1 cycle of final extension at 72°C for 7 min. The amplicon was then cloned into the PCR8/GW/TOPO vector (Invitrogen, Carlsbad, CA). Next, the LR recombination reaction was carried out using the LR Clonase II enzyme mix (Invitrogen) to recombine the p53 sequence into the pLenti7.3/V5-DEST destination vector (Invitrogen). The construct was then transformed into One Shot Stbl3 E-coli, and two of the p53 clones (p53-1 and p53-2) were grown in liquid LB with ampicillin and plasmid DNA was isolated using the Purelink Hipure Plasmid Filter Midiprep Kit (Invitrogen). DNA was also isolated from empty destination vector (MT) propagated in ccdB-resistant competent E. coli (Invitrogen). Lentivirus production and cell transduction were carried out as described (23). One- to two-week posttransduction K562 or CHRF cells were sorted for EmGFP expression in a FACSAria (Becton-Dickinson) achieving postsort purities of roughly 95% to generate two K562 or CHRF cell lines expressing or overexpressing p53 (p53-1 and p53-2) and one CHRF or K562 cell line expressing the expression cassette included in the empty pLenti vector (MT).
Table 1.
Sequencing primers for human wild-type p53
| Primer Sequence | Site of Hybridization |
|---|---|
| 5′-CTGGTCATCATCCTGCCTTT-3′ | on the pCMV-Neo-Bam vector prior to exon 1 of p53 |
| 5′-TTCACTGAAGACCCAGGTCCAGAT-3′ | on exon 4 of p53 |
| 5′-CACAGAGGAAGAGAATCTCCGCAAG-3′ | on exon 8 of p53 |
| 5′-ATGTTCCGAGAGCTGAATGAGGC-3′ | on exon 11 of p53 |
RESULTS
Stable expression of p53 in K562 cells suppresses polyploidization upon Mk differentiation.
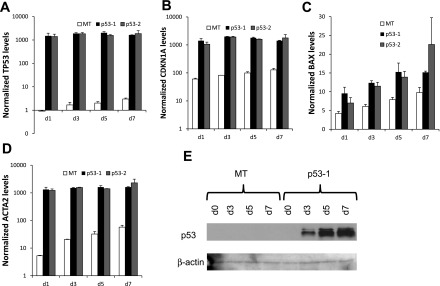
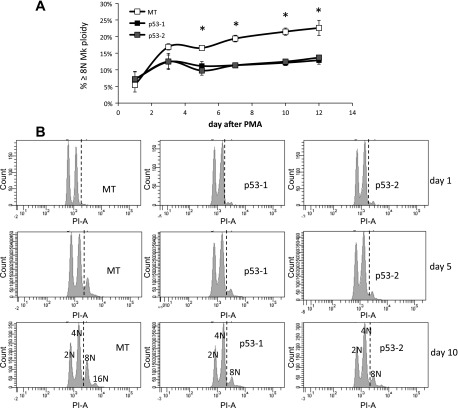
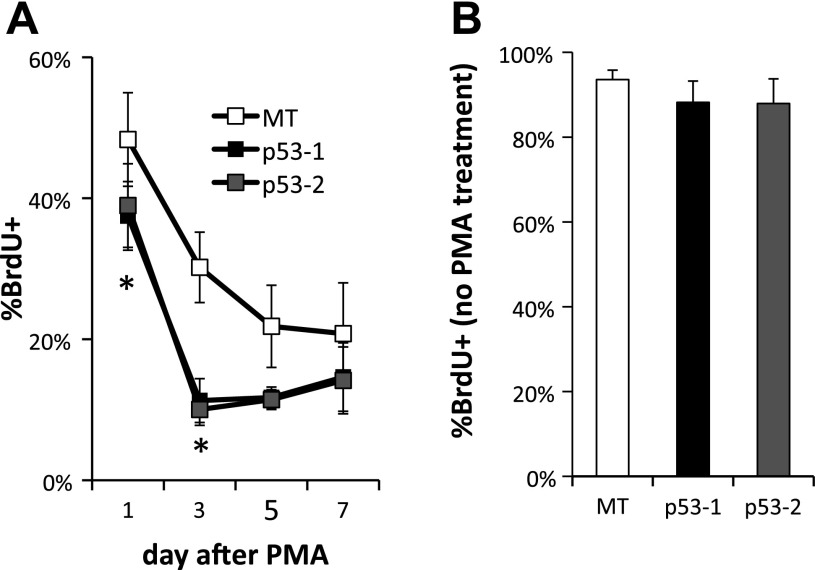
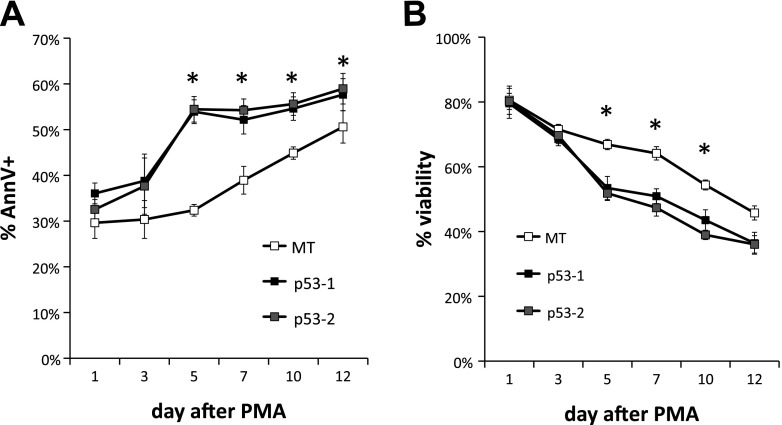
To test the hypothesis that p53 suppresses polyploidization in megakaryocytes, we examined the impact of wild-type p53 expression in K562 cells, which are p53−, as they fail to produce p53 protein due to a frame-shift insertion (41). K562 is an erythroleukemic cell line able to undergo terminal Mk differentiation upon PMA treatment (45), exhibiting substantial polyploidization (37) and apoptosis (15). Two stable p53-expressing K562 cell lines (p53-1 and p53-2) were generated by transduction of K562 cells with lentiviral constructs originating from two different clones of competent cells expressing the p53 construct followed by sorting of the transduced K562 cell population. Additionally, one stable empty vector-expressing (MT) K562 cell line was generated after transduction with an empty construct and sorting the transduced K562 cells. Expression of p53 and higher expression of p53 targets CDKN1A (p21), BAX, and ACTA2 in p53-1 and p53-2 K562 cells was confirmed at the transcript level in the p53-1 and p53-2 K562 cells (Fig. 1, A–D). Expression of the p53 protein was confirmed in p53-1 and p53-2 K562 cells by Western analysis (Fig. 1E and data not shown). Upon PMA-induced Mk differentiation, polyploidization was suppressed in p53-1 and p53-2 relative to control MT K562 cells: the percentage of K562 cells undergoing polyploization (≥8N ploidy) was on average 36–75% higher in MT cells relative to p53-1 and p53-2 cells between days 3 and 12 after PMA (Fig. 2). Additionally, p53 expression induced both G1/S and G2/M cell cycle arrest in K562 cells undergoing Mk differentiation on day 1 of PMA treatment (Fig. 2B); however, in the absence of Mk differentiation, the cell cycle profiles of p53-expressing and control K562 cells were similar (data not shown). Reduced polyploidization in p53-expressing Mk cells was in part due to substantially diminished DNA synthesis after PMA treatment: DNA synthesis, assessed by BrdU incorporation, was higher in MT cells by an average of 184 and 89% on days 3 and 5 after PMA, respectively, relative to p53-1 and p53-2 cells (Fig. 3A). Importantly, in the absence of Mk differentiation (no PMA), DNA synthesis was statistically similar in the three cell lines (Fig. 3B). Reduced polyploidization in p53-expressing Mk cells was also due to increased apoptosis and cell death. Apoptosis, assessed by annexin V binding, was on average 23–67% greater in p53-1 and p53-2 cells relative to MT cells 3–10 days after PMA-induced Mk differentiation (Fig. 4A). Furthermore, viability, measured by exclusion of TOPRO3, a DNA-binding dye taken up by dead and late apoptotic cells, was on average reduced by 20–28% in p53-1 and p53-2 cells, relative to MT cells 5–12 days after PMA (Fig. 4B).
Fig. 1.
Expression of p53 in K562 cells, a p53− cell line. P53-1 and p53-2, 2 stable wild-type p53+ K562 cell lines, and MT, an empty vector-transduced K562 cell line, were generated. K562 cells were stimulated with 10 ng/ml PMA to induce terminal megakaryocytic differentiation. Transcript level expression of p53 (A) and p53 targets CDKN1A (p21) (B), BAX (C), and ACTA2 (D) was measured by Q-RT-PCR on days 1, 3, 5, and 7 following PMA stimulation. Error bars: SE, n = 2. Western blot confirming p53 protein expression in p53-1 (E) K562 cells relative to MT cells on days 1, 3, 5, and 7 following PMA stimulation. Shown is 1 of 2 biological replicates.
Fig. 2.
p53 expression in K562 cells suppresses polyploidization upon PMA-induced terminal megakaryocytic (Mk) differentiation. A: % polyploidy (≥8N ploidy) among p53-expressing (p53-1 and p53-2) and control (MT) K562 cells undergoing Mk differentiation. Error bars: SE, n = 4. *P ≤ 0.02 (2-tailed paired t-test). B: representative Mk ploidy histograms of MT, p53-1, and p53-2 K562 cells undergoing Mk differentiation on days 1, 5, and 10 after PMA treatment.
Fig. 3.
The block in Mk polyploidization in PMA-treated K562 cells upon p53 expression is in part due to diminished DNA synthesis. A: cultures of p53-expressing (p53-1 and p53-2) and control (MT) K562 cells were incubated with 10 μM BrdU for 12 h on days 1, 3, 5, and 7 after PMA to assess DNA synthesis. Error bars: SE, n = 3. *P ≤ 0.03 (2-tailed paired t-test). B: in the absence of PMA-induced Mk differentiation, DNA synthesis, measured after a 12-h incubation with 10 μM BrdU, is not affected by p53 expression. Error bars: SE, n = 3.
Fig. 4.
Expression of p53 in K562 cells promotes apoptosis and cell death upon PMA-induced Mk differentiation. A: binding of annexin V was measured in cultures of p53-expressing (p53-1 and p53-2) and control (MT) K562 cells on days 1, 3, 5, 7, 10, and 12 after PMA treatment. Error bars: SE, n = 4. B: viability assessed by exclusion of TOPRO3 was measured at the same time points. Error bars: SE, n = 4. *P < 0.04 (2-tailed paired t-test).
Identifying the p53 Mk regulon: experimental design of gene expression microarray analysis and data validation.
The first objective of this microarray analysis was to identify which p53-controlled genes may impact Mk differentiation: this is the direct Mk p53 regulon. For this purpose, a list of p53 target genes was curated from the literature, as described below and focused on the strongly differentially expressed p53 target genes between p53-KD and control CHRF cells. Furthermore, because the complete, general p53 regulon may not yet be fully identified and because p53 direct targets could regulate other genes related to megakaryopoiesis, a second objective was to identity additional strongly differentially expressed genes affected by p53 in this megakaryopoietic phenotype. Therefore, a large set of genes was explored including several p53 targets coding for proteins, whose function has been studied in the context of megakaryopoiesis and platelet production.
To examine which p53-related genes impact Mk differentiation, we used gene expression microarray analysis based on the CHRF cell line, a human megakaryoblastic cell line carrying the wild-type version of the p53 gene. Upon PMA stimulation, CHRF cells undergo polyploidization and extend proplatelet-like cytoplasmic protrusions combined with apoptosis (32). We have previously generated a stable p53-KD CHRF cell line by lentiviral delivery of a microRNA-adapted-short hairpin sequence targeting p53, achieving 50–90% p53-KD on the transcript level, as well as a suitable scrambled-control cell line (previously referred to as p53-B and Neg-A, respectively) (23). This analysis compared the p53-KD CHRF cells against the control CHRF cells in the absence of PMA treatment (day 0) and on days 1, 3, 5, and 7 after PMA treatment (5 time-points). p53-KD and control samples were hybridized against each other (direct comparison) (61). Two biological replicate experiments were carried out and thus 10 dye-swap (total of 20) measurements of gene expression for each microarray probe. Probes for which ≥50% of measurements were missing were discarded and not included in the analysis of differential expression.
Differentially expressed genes were identified using the significance analysis of microarrays (65) with a false discovery rate of <5%, as implemented in the MultiExperiment Viewer (56). For the data presented in Figs. 5 and 6, the extra constraint of ≥1.5-fold higher or lower gene expression in at least two time-points between p53-KD and control CHRF cells was chosen to narrow down the set of genes that had the greatest extent of differential expression, similar to what was done previously (52). The Agilent Probe IDs together with the help of the Array-Check bioinformatics tool, as implemented in Splice Center (55), were used to differentiate between redundant microarray probes targeting the same transcript vs. probes for splice variants.
Fig. 5.
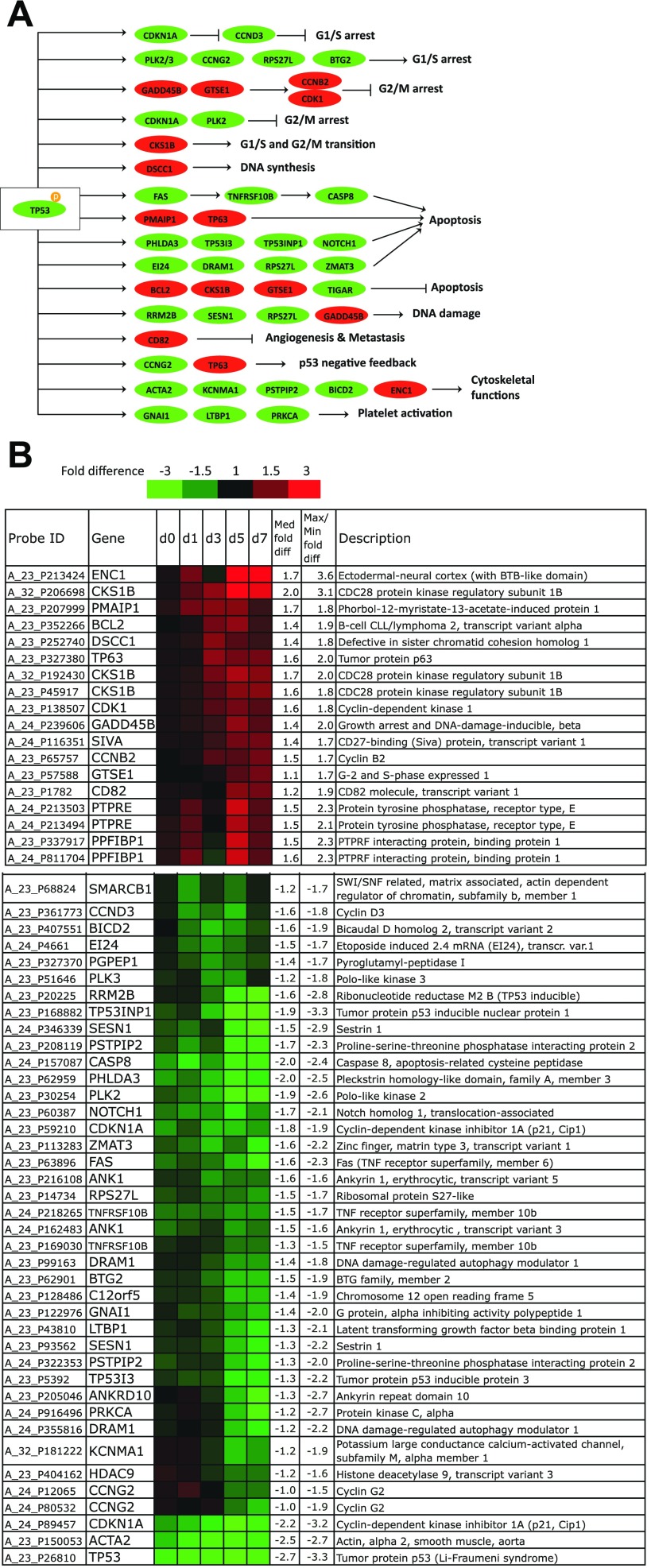
Differentially expressed p53 target genes. A: mapping of differentially expressed p53 target genes to known functions. Red, upregulated; green, downregulated. The figure was modified from the KEGG p53 signaling pathway by including recently discovered p53 targets. B: highly upregulated p53 targets in p53-knock-down (KD) CHRF cells and highly downregulated p53 targets in p53-KD CHRF cells relative to control CHRF cells. Rows show the probe ID on the Agilent 4 × 44K whole human genome microarray, expression ratios between p53-KD, and control CHRF cells on day 0 (unstimulated) and days 1, 3, 5, and 7 after PMA stimulation, the gene name, median, and maximal/minimal fold difference, and gene description. Saturated red indicates 3-fold upregulation in p53-KD cells relative to control cells, while saturated green indicates 3-fold downregulation in p53-KD cells relative to control cells. p53-KD and control CHRF cells were either unstimulated or treated with 10 ng/ml PMA. Experiments 1 and 2 are averaged.
Fig. 6.
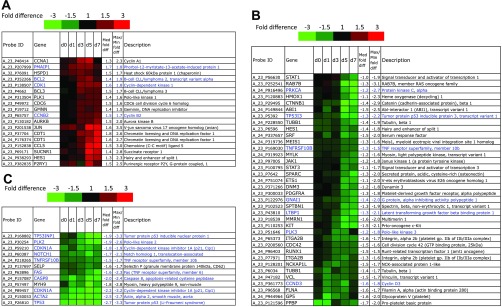
Megakaryopoiesis-associated differentially expressed genes. Rows and color-coding of gene-expression data are similar to what is described for Fig. 5. p53-KD and control CHRF cells were either unstimulated or treated with 10 ng/ml PMA. Experiments 1 and 2 are averaged. A: cluster 1: Mk-related genes upregulated in p53-KD CHRF cells. B: Mk-related genes downregulated in late p53-KD CHRF cultures. C: Mk-related genes constantly downregulated in p53-KD CHRF cells relative to control CHRF cells. P53 target genes found in this list of differentially expressed Mk-related genes are in blue.
In the context of a systems analysis like the present one, it is not practical to functionally validate even a small fraction of the data. However, assurance that the set of genes identified in the microarray analysis is a robust, valid set of p53 target genes, is essential for the bioinformatics analyses described below. To this effect, the quality of the microarray data was assessed using three independent but partially overlapping methods. First, we examined if the new data identified the six strongly differentially expressed genes that are direct p53 targets that were previously identified to belong to the Mk p53 regulon (12, 23). Second, we determined if this analysis was able to identify a high percentage of genes that are p53 targets, and whose function has been studied in the context of megakaryopoiesis. Third, a select set of microarray findings from the current study were validated using targeted Q-RT-PCR or Western blotting.
Direct Mk p53 regulon.
The first aim was to identify which p53-controlled genes may impact Mk differentiation. a list of experimentally validated p53 target genes (30, 42, 53, 64, 68) and predicted p53 targets based on the presence of consensus p53 binding motifs (68) was compiled. Thus, we constructed a list of 227 genes carrying p53 response elements (Supplemental Table S1), which are represented by 373 probes on the Agilent microarrays used in this study.1 Using the differential expression analysis delineated above, this list was narrowed down to 47 p53 target genes (58 Agilent probes; Fig. 5; each probe is shown separately) as strongly differentially expressed between the p53-KD and control CHRF cells.
First, as part of our broad validation strategy, using the aforementioned stringent metrics for determining differential expression, we found that this current microarray analysis identified four (CDKN1A, BCL2, TP53I3, TP53INP1) out of six (the four above plus MDM2 and BBC3) of the previously identified p53 target genes (23) as strongly differentially expressed (meeting the 1.5-fold differential expression in at least two time-points) between p53-KD and control CHRF cells (the other two genes, BAX and GADD45A in Ref. 23, do not meet the current criteria). This 67% success rate suggests that the current genome-scale study has the potential to identify many Mk-related p53 target genes.
A functional analysis of biological processes that involve the strongly differentially expressed p53 targets shown in Fig. 5 using Gene Ontology associations implemented in the DAVID functional association database found a significant enrichment in functions related to apoptosis and the cell cycle (Table 2). An overview of the differentially expressed p53 targets and the functions they are known to mediate is shown in Fig. 5A.
Table 2.
GO-based analysis of biological processes found enriched in differentially expressed p53 target genes between p53-KD and control CHRF cells
| Term | Count | FDR |
|---|---|---|
| GO:0043065∼positive regulation of apoptosis | BCL2, FAS, NOTCH1, SIVA1, CASP8, CDKN1A, EI24, PMAIP1, PHLDA3, KCNMA1, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT3 | 1.78E-13 |
| GO:0043068∼positive regulation of programmed cell death | BCL2, FAS, NOTCH1, SIVA1, CASP8, CDKN1A, EI24, PMAIP1, PHLDA3, KCNMA1, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT3 | 1.78E-13 |
| GO:0010942∼positive regulation of cell death | BCL2, FAS, NOTCH1, SIVA1, CASP8, CDKN1A, EI24, PMAIP1, PHLDA3, KCNMA1, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT4 | 1.78E-13 |
| GO:0006917∼induction of apoptosis | FAS, SIVA1, CASP8, CDKN1A, EI24, PMAIP1, PHLDA3, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT3 | 1.70E-11 |
| GO:0012502∼induction of programmed cell death | FAS, SIVA1, CASP8, CDKN1A, EI24, PMAIP1, PHLDA3, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT4 | 1.77E-11 |
| GO:0042981∼regulation of apoptosis | BCL2, BTG2, FAS, NOTCH1, SIVA1, CASP8, CDK1, CDKN1A, EI24, PMAIP1, PHLDA3, KCNMA1, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT3 | 3.13E-11 |
| GO:0043067∼regulation of programmed cell death | BCL2, BTG2, FAS, NOTCH1, SIVA1, CASP8, CDK1, CDKN1A, EI24, PMAIP1, PHLDA3, KCNMA1, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT4 | 3.76E-11 |
| GO:0010941∼regulation of cell death | BCL2, BTG2, FAS, NOTCH1, SIVA1, CASP8, CDK1, CDKN1A, EI24, PMAIP1, PHLDA3, KCNMA1, PRKCA, RRM2B, RPS27L, TNFRSF10b, TP53, TP53INP1, TP53I3, TP63, ZMAT5 | 4.02E-11 |
| GO:0008629∼induction of apoptosis by intracellular signals | CDKN1A, PHLDA3, PRKCA, RPS27L, TP53, TP53I3, TP63 | 1.94E-05 |
| GO:0007049∼cell cycle | BCL2, CKS1B, GTSE1, SMARCB1, CDK1, CCNB2, CCND3, CCNG2, CDKN1A, DSCC1, PLK2, PLK3, SESN1, TP53, TP53INP1 | 4.86E-05 |
| GO:0006974∼response to DNA damage stimulus | BTG2, GTSE1, CDK1, CDKN1A, PHLDA3, RRM2B, RPS27L, SESN1, TP53, TP63, ZMAT3 | 1.86E-04 |
The false discovery rate (FDR) was used to detect Gene Ontology (GO) terms. KD, knock-down.
Among the upregulated p53 target genes (Fig. 5B) in p53-KD CHRF cells was the antiapoptotic BCL2 and the antiapoptotic CKS1B, a component of cyclin-dependent kinases, with a role in promoting G1/S and G2/M cell cycle transition (67). GTSE1, which has reported antiapoptotic functions, is a negative regulator of p53 that promotes G2 phase checkpoint recovery by shuttling p53 out of the nucleus (46). Also upregulated was TP63, which has dual roles in cell-cycle inhibition and in promoting apoptosis. TP63, a member of the p53 family of tumor suppressors, may compensate for the reduced activity of p53 (26). CCNB2 and CDK1 are part of the cyclin B/CDK1 complex, which promotes the G2/M cell cycle transition and is activated during Mk endomitosis (7). GADD45b, although traditionally thought to mediate cell cycle arrest and apoptotic signals, may induce survival especially in hematopoietic cells following UV irradiation (44). DSCC1 is essential for DNA synthesis (51), PPFIBP1 codes for a protein promoting cell adhesion (58), while ENC1 (PIG10) codes for a component of the actin cytoskeleton (70).
The majority of the downregulated p53 targets (Fig. 5B) have well-established roles in apoptosis, cell cycle arrest, and DNA repair or are related to the cytoskeleton. FAS (42), CASP8 (42), EI24 (42), RPS27L (43), PHLDA3 (38), TP53INP1 (33), TP53I3 (42), DRAM1 (FLJ11259) (18), NOTCH1 (31), and ZMAT3 (42) have roles in promoting apoptosis, while C12orf5 (TIGAR) (9) may regulate apoptosis. Among the cell cycle-related p53 targets, CDKN1A (42), PLK2 (4), PLK3 (4), RPS27L (43), CCNG2 (22), and BTG2 (20) are known to regulate the G1/S cell cycle transition, while CCND3 promotes the G1/S transition (2). CDKN1A (1) and PLK2 (62) may also regulate the G2/M cell cycle transition. Moreover, SESN1 (42), RPS27L (43), and RRM2B (42) are induced in a p53-dependent fashion in response to DNA damage. Notably, cytoskeleton-associated genes were also found downregulated in p53-KD CHRF cells; ACTA2 is a component of smooth muscle actin (28), KCNMA1 (11) and PSTPIP2 (13) both code for actin-binding proteins, while BICD2 has a role in promoting transport along microtubules (29). Moreover, GNAI1 (66), LTBP1 (48), and PRKCA (39) have roles in platelet activation.
Which known Mk genes are differentially expressed as a result of the p53-KD?
The next aim was to identify Mk genes whose expression is altered substantially in p53-KD CHRF cells undergoing Mk differentiation. This analysis aimed to identify genes directly or indirectly controlled by p53 that mediate the enhanced polyploidization observed in p53-KD Mk cells. To this effect, on the basis of an exhaustive literature search, we curated a list of 279 human genes (429 Agilent probes) (Supplemental Table S2) associated with Mk differentiation and platelet production. After applying the differential expression analysis as described above, we narrowed this list down to 60 genes (66 Agilent probes, Fig. 6) that are thus apparently directly or indirectly regulated by p53. Each probe is shown separately as differentially expressed between the p53-KD and control CHRF cells.
Hierarchical clustering of the 66 differentially expressed Mk probes revealed three clusters of gene expression: cluster 1 (Fig. 6A, genes upregulated in p53-KD CHRF cell cultures), cluster 2 (Fig. 6B, genes downregulated in late p53-KD CHRF cell cultures), and cluster 3 (Fig. 6C, genes constantly downregulated in p53-KD CHRF cell cultures). Of the differentially expressed Mk genes, 19/60 (32%) were also p53 targets (Fig. 6, in blue). As part of our validation strategy (see materials and methods), we conclude that the strong enrichment of known p53 targets among known Mk-related genes identified by this study provides further support for the validity of our approach and quality of data. While in the first two clusters there is low enrichment in known p53 target genes [cluster 1: 4/18 (22%) and cluster 2: 7/33 (21%)], 8/10 (80%) of Mk genes in cluster 3 are also p53 transcriptional targets (TP53INP1, PLK2, CDKN1A, NOTCH1, TNFRSF10B, FAS, CASP8, ACTA2). This suggests that these eight strongly downregulated p53 targets are part of p53's regulon in Mk development. While a set of the rest of these 60 genes may represent still unrecognized bona fide p53 transcriptional targets, others may interact with p53 targets or their expression may be dictated by the enhanced polyploidization observed in p53-KD Mk cultures.
To shed light into functions that could be affected by the differential modulation of these 60 Mk-related genes, a Gene Ontology analysis of biological processes was performed using the DAVID functional association database. This analysis reveals (Table 3) that the functions affected were mostly related to apoptosis, cell growth, immune system biology, and hematopoiesis. Because of the preponderance of functions related primarily to apoptosis and secondarily to the cell cycle among the Mk genes, focus was put on ontologies related to these functions. By examining the largest apoptosis-related category, the “regulation of programmed cell death,” we found that while 10/12 genes associated with promotion of apoptosis roles in Mk development were downregulated (STAT1, NOTCH1, CASP8, TNFRSF10B, TP53, TP53INP1, TP53I3, CDKN1A, FAS, PRKCA), 3/5 antiapoptosis genes were upregulated (JUN, BCL2, BCL3). Moreover, by examining the largest cell growth-related category, the “cell cycle,” we found that 7/9 genes whose low levels are associated with increased Mk ploidy were downregulated (PLK1, PLK2, PLK3, MYH9, TP53, TP53INP1, CDKN1A) and that 6/7 genes whose high levels are associated with increased Mk ploidy (BCL2, CCNA1, CDC6, CDK1, CCNB2, CDT1) were upregulated. In conclusion, the ontological analysis of Mk development-associated genes indicated that KD of p53 promotes transcriptional programs leading to polyploidization and prevention of apoptosis in the context of Mk differentiation, thus confirming independently what we had previously reported (23).
Table 3.
GO-based analysis of biological processes found enriched in differentially expressed Mk-related genes between p53-KD and control CHRF cells
| Term | Genes | FDR |
|---|---|---|
| GO:0043065∼positive regulation of apoptosis | PRKCA, TP53, PMAIP1, STAT1, NOTCH1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, JUN, BCL2, CASP8, BCL3, HSPD1, FAS, TP53INP1 | 2.38E-08 |
| GO:0043068∼positive regulation of programmed cell death | PRKCA, TP53, PMAIP1, STAT1, NOTCH1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, JUN, BCL2, CASP8, BCL3, HSPD1, FAS, TP53INP1 | 2.64E-08 |
| GO:0010942∼positive regulation of cell death | PRKCA, TP53, PMAIP1, STAT1, NOTCH1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, JUN, BCL2, CASP8, BCL3, HSPD1, FAS, TP53INP1 | 2.83E-08 |
| GO:0034097∼response to cytokine stimulus | PRKCA, JUN, BCL2, CASP8, PDGFRA, CCL5, STAT1, SRF, STAT3, CTNNB1 | 5.84E-08 |
| GO:0043067∼regulation of programmed cell death | PRKCA, CDK1, TP53, KIT, PMAIP1, STAT1, NOTCH1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, JUN, BCL2, CASP8, BCL3, FAS, HSPD1, TP53INP1 | 4.70E-06 |
| GO:0010941∼regulation of cell death | PRKCA, CDK1, TP53, KIT, PMAIP1, STAT1, NOTCH1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, JUN, BCL2, CASP8, BCL3, FAS, HSPD1, TP53INP1 | 4.99E-06 |
| GO:0006917∼induction of apoptosis | PRKCA, TP53, PMAIP1, STAT1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, CASP8, BCL3, FAS, TP53INP1 | 1.12E-05 |
| GO:0012502∼induction of programmed cell death | PRKCA, TP53, PMAIP1, STAT1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, CASP8, BCL3, FAS, TP53INP1 | 1.16E-05 |
| GO:0007049∼cell cycle | CDK1, CDC6, GMNN, TP53, AURKB, MYH9, CTNNB1, CDT1, CDKN1A, CCNB2, PLK3, CCND3, PLK2, PLK1, BCL2, TUBB1, CCNA1, TP53INP1 | 1.74E-05 |
| GO:0002520∼immune system development | CDC42, CCNB2, BCL2, CASP8, TP53, BCL3, KIT, FAS, HSPD1, MYH9, RUNX1, CTNNB1 | 2.76E-05 |
| GO:0042981∼regulation of apoptosis | PRKCA, CDK1, TP53, PMAIP1, STAT1, NOTCH1, TP53I3, CDKN1A, TNFRSF10B, ETS1, HMOX1, JUN, BCL2, CASP8, BCL3, FAS, HSPD1, TP53INP1 | 2.95E-05 |
| GO:0010604∼positive regulation of macromolecule metabolic process | PRKCA, CDK1, TP53, KIT, SRF, STAT3, CTNNB1, HES1, NOTCH1, CCND3, ETS1, PLK1, HMOX1, JUN, BCL2, PDGFRA, BCL3, RUNX1 | 7.55E-05 |
| GO:0002521∼leukocyte differentiation | CDC42, BCL2, CASP8, TP53, BCL3, KIT, FAS, MYH9, CTNNB1 | 1.14E-04 |
| GO:0048534∼hemopoietic or lymphoid organ development | CDC42, CCNB2, BCL2, CASP8, TP53, BCL3, KIT, FAS, MYH9, RUNX1, CTNNB1 | 1.86E-04 |
| GO:0010033∼response to organic substance | PRKCA, SELP, PMAIP1, STAT1, CCL5, SRF, STAT3, CTNNB1, CDKN1A, HMOX1, BCL2, JUN, CASP8, PDGFRA, HSPD1, FAS | 3.07E-04 |
| GO:0051301∼cell division | CDK1, CDC42, CDC6, NOTCH1, CCNB2, CCND3, PLK1, AURKB, KIT, MYH9, CCNA1 | 6.00E-04 |
| GO:0030097∼hemopoiesis | CDC42, BCL2, CASP8, TP53, BCL3, KIT, FAS, MYH9, RUNX1, CTNNB1 | 9.19E-04 |
| GO:0042127∼regulation of cell proliferation | PRKCA, CDC6, TP53, ABI1, SPARC, KIT, STAT1, CTNNB1, HES1, NOTCH1, CDKN1A, ETS1, HMOX1, BCL2, JUN, PDGFRA | 9.46E-04 |
| GO:0014070∼response to organic cyclic substance | PRKCA, CDKN1A, HMOX1, JUN, BCL2, STAT1, STAT3, CTNNB1 | 0.001157 |
| GO:0042110∼T cell activation | CCND3, BCL2, TP53, BCL3, FAS, HSPD1, MYH9, CTNNB1 | 0.001522 |
The FDR was used to detect GO terms. Mk, megakaryocytic.
Further validation of differentially expressed genes by Q-RT-PCR and Western analysis.
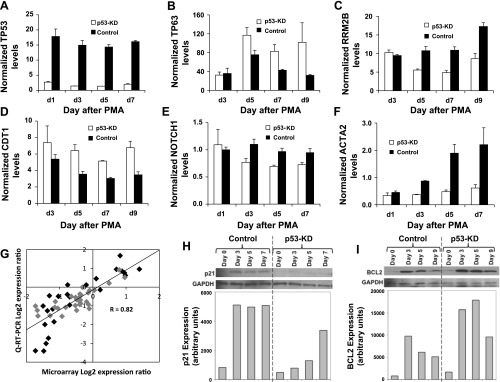
In addition to the two data-validation approaches already discussed, as the third component of our validation strategy (see materials and methods), we validated the transcript levels of p53 and five key genes [TP63, RRM2B, NOTCH1, ACTA2 (Fig. 5), and CDT1 (Fig. 6)] identified by the microarray study as strongly differentially expressed between p53-KD and control cells undergoing Mk differentiation. These five genes were chosen because they represent distinct p53 target gene functions and because they were found to be very strongly differentially expressed between p53-KD and control CHRF cells. p53 transcript levels were up to 10.6-fold higher in control CHRF cells compared with p53-KD cells on day 3 of PMA stimulation (Fig. 7A), which is consistent with our previous findings (23) and microarray data shown here. TP63 transcript levels were up to 3.2-fold higher in p53-KD compared with control CHRF cells by day 7 of PMA stimulation (Fig. 7B). RRM2B transcript levels were up to 2.2-fold lower in p53-KD compared with control CHRF cells on day 5 of PMA treatment (Fig. 7C). CDT1 mRNA levels were up to 1.9-fold higher in p53-KD relative to control CHRF cells by day 7 of PMA-induced Mk differentiation (Fig. 7D). Furthermore, NOTCH1 mRNA levels were up to 1.4-fold higher in control relative to p53-KD CHRF cells (Fig. 7E). Lastly, ACTA2 mRNA levels were up to fourfold higher in control relative to p53-KD CHRF cells (Fig. 7F). In our previous study (23) the expression of a set of eight p53 target genes (BBC3, CDKN1A, BCL2, GADD45A, BAX, TP53I3, TP53INP1, MDM2) was examined via Q-RT-PCR. We sought to further validate the current microarray data for these genes by examining their correlation to the previously published Q-RT-PCR data (gray points) in addition to the Q-RT-PCR data presented in this study (black points). Overall, there was good correlation of the microarray data and the Q-RT-PCR results (R2 = 0.82, Fig. 7G).
Fig. 7.
Q-RT-PCR and Western blot validation of key microarray findings. p53-KD and control CHRF cells were either unstimulated or treated with 10 ng/ml PMA. Q-RT-PCR validation for TP53 (A), TP63 (B), RRM2B (C), CDT1 (D), NOTCH1 (E), and ACTA2 (F) transcript expression in PMA-stimulated p53-KD and control CHRF cells. Starting quantity (SQ) amounts were normalized to the average SQ of 2 housekeeping genes (GUSB and RPLP0). Error bars: SE, n = 2. G: correlation of gene expression between microarray data presented here and Q-RT-PCR data for genes BBC3, CDKN1A, BCL2, GADD45A, BAX, TP53I3, TP53INP1, MDM2 from our previous study (23) (gray points) and genes validated with Q-RT-PCR in the present study (black points). Overall, there is good correlation between the microarray and Q-RT-PCR data (R2 = 0.82). Western blots and densitometry analysis confirming differential expression of p53 target proteins p21 (CDKN1A) using nuclear extracts (H) and Bcl2 using total cell extracts (I) in p53-KD and control CHRF cells. Representative blots and corresponding densitometry analysis are shown; n = 2 for both.
Finally, we validated the impact of p53-KD on the expression of key p53 target proteins cell cycle arrest-related CDKN1A (p21) and antiapoptotic BCL2. As anticipated based on the microarray data (Fig. 5B), p21 expression quickly increased in control CHRF cells upon PMA stimulation, and p53 KD significantly delayed this upregulation (Fig. 7H). In addition, as expected, antiapoptotic BCL2 was highly expressed in early Mk-differentiated control CHRF cells, but expression dropped as Mk maturation continued (Fig. 7I). The protein expression patterns for p21 and BCL2 match perfectly the previously published Q-RT-PCR data for these two genes (23) and the microarray data (Fig. 5B).
DISCUSSION
Tumor suppressor p53 is a master transcription factor that becomes activated as a result of a variety of stress signals and responds by orchestrating its downstream genes to effect outcomes such as arrest of DNA synthesis, senescence, or apoptosis (8). Our previous studies have shown that KD and loss of p53 leads to enhanced Mk polyploidization in cultures of Mk cell lines and mouse primary Mk cells and that increased Mk polyploidization is also observed in bone marrow-resident Mk cells in vivo upon treatment of p53−/− mice with thrombopoietin (3, 23). Another group recently demonstrated that in p53−/− mice, platelet recovery 25 days following myelosuppressive treatment with 5-fluorouracil led to twofold higher platelet counts by comparison to p53+/+ mice that had undergone the same treatment (49).
Here, a genome-wide gene expression comparison of p53-KD and control CHRF cells undergoing terminal Mk differentiation was conducted to comprehensively identify genes that may be transcriptionally regulated by p53. The initial focus of the microarray analysis was on annotated p53 target genes to identify specific genes and transcriptional responses affected by the reduction of p53 expression in megakaryopoiesis. Overall, out of a total of 42 p53 target genes that are known to have roles in the biology of Mk development, this analysis identified 19 genes (45%) as differentially expressed (Supplemental Table S3). This suggests that these differentially expressed p53 target genes could be part of p53's regulon in Mk differentiation.
These data correctly identified several core p53 target genes that have known functions in megakaryopoiesis, thus supporting the accuracy of our approach. For instance, CDKN1A (p21) was identified as downregulated; overexpression of CDKN1A in vitro serves to obstruct Mk polyploidization (5). Also downregulated was the TNF receptor FAS, whose ligation induces Mk proplatelet protrusions and platelet production (16). The downregulation of NOTCH1 in p53-KD CHRF cells is in agreement with an older study in PMA-stimulated K562 cells, where NOTCH1 was shown to inhibit Mk differentiation and polyploidization and induce apoptosis (31). The downregulation of PLK3 in p53-KD CHRF cells is consistent with a recent study that reported low levels of PLK3 in a Mk cell line undergoing polyploidization (40). The downregulation of TNFRSF10B in p53-KD cells is in agreement with a proposed role of TNFRSF10B in inhibiting Mk cell growth (50).
However, and more importantly, a large number of other genes directly or indirectly regulated by p53 that have not been previously associated with megakaryopoiesis were also identified. These include a number of proapoptotic p53 targets: DRAM1, ZMAT3, RPS27L, PHLDA3, and EI24, whose function in Mk development remains to be investigated, were also strongly downregulated in p53-KD cells and may represent novel effectors of p53-mediated Mk apoptosis. Because it is likely that several p53 target genes have not yet been identified and that p53 target genes exert their effects on Mk development through other genes unrelated to p53, we examined whether genes previously associated with megakaryopoiesis were affected by the KD of p53. For instance, CDC6 and CDT1, two components of the origin of replication complex, essential for initiation of DNA replication, were strongly upregulated in p53-KD cells (Fig. 6). Stabilization of CDC6 and CDT1 was shown to be essential for Mk endomitosis (10). Of note, CDT1 overexpression combined with loss of p53 can lead to spontaneous polyploidization in a lung cancer cell line (57). Additionally, among the downregulated Mk genes (Fig. 6) were MYH9, CTNNB1, and FLNA, three genes associated with the cytoskeleton. MYH9, which codes for nonmuscle myosin II A, participates in a complex with actin that restricts microtubule polymerization (21), and its inhibition can lead to enhanced polyploidization in cultured Mk cells (59). β-Catenin (CTTNB1) links the actin cytoskeleton to intercellular adherens junctions (17). FLNA, which codes for the alpha isoform of filamin, binds actin and organizes actin filaments into a three-dimensional structure conferring structural stability to the cells; loss of FLNA leads to lower Mk ploidy in vitro coupled with abnormal platelet generation leading to macrothrombocytopenia (34). The pronounced downregulation of genes coding for actin-related components ACTA2 (Fig. 5B), MYH9, FLNA, and CTNNB1 in p53-KD cells (Fig. 6B), suggests that the enhanced polyploidization in p53-deficient Mk cells could be explained by diminished actomyosin-mediated force. Notably, other actin cytoskeletal components, such as p53 targets KCNMA1 and PSTPIP2, and microtubule components TUBB1 (β1-tubulin) and BICD2 were also downregulated in p53-KD cells. Future work will also examine the influence that the elasticity of the matrix of adhesion has on the polyploidization of normal and p53-deficient Mk cells (59).
CHRF cells already express the wild-type version of the p53 gene, and the p53 protein is strongly induced upon PMA treatment (23). Therefore, predictably, CHRF cell lines generated to stably overexpress p53 did not exhibit suppressed polyploidization compared with control CHRF cells undergoing PMA-induced terminal Mk differentiation (data not shown). However, we sought to validate our hypothesis that p53 expression suppresses polyploidization in the context of terminal megakaryopoiesis by employing K562 cells, a p53-negative cell line, which undergoes terminal Mk differentiation upon PMA treatment.
A previous study, which examined K562 cells transfected with a temperature-sensitive form of p53, found that p53 expression led to enhanced erythroid differentiation upon hemin treatment, whereas it did not affect Mk-specific surface marker expression upon PMA treatment (15). However, the authors did not examine polyploidization during PMA-induced Mk differentiation of K562 cells. In agreement with that study, it is shown that DNA synthesis was not affected in unstimulated K562 cells (Fig. 3B), which maintain the more primitive erythroleukemic phenotype. By contrast, upon PMA treatment, polyploidization was reduced in p53-expressing K562 cells (Fig. 2). This reduction in Mk polyploidization could be attributed to diminished DNA synthesis (Fig. 3A) and increased apoptosis (Fig. 4). Thus, these results suggest that while p53 expression does not influence commitment to Mk differentiation, it specifically affects the process of polyploidization by inhibiting DNA synthesis and inducing apoptosis in megakaryocytes.
These data and analysis are consistent with and provide further support for the proposed model whereby p53 is called upon to play the role of Mk ploidy regulator throughout the course of Mk differentiation. It is tempting to speculate that the activation of p53 is due to cellular stress that endomitosis imparts on a cell as it aborts cytokinesis. For example, increased acetylation of p53, denoting p53 activation, was found in immature cultured Mk cells treated with nicotinamide to substantially enhance polyploidization (27). The implication would be that p53's role is in reaction to the onset of polyploidization. However, p53 levels and its DNA binding activity diminished in mature polyploid Mk cells taken from a late time time-point of culture (27). This is reminiscent of the increased Mk ploidy observed in the absence of p53 (3, 23). Taken together, our data suggest that downregulation of p53 in terminally differentiated Mk cells is needed to give way to endomitosis leading to polyploidization.
Conclusions
The current study has extended our knowledge of the biology of p53 during megakaryopoiesis 1) by testing and validating the hypothesis that p53 expression in a p53− cell line suppresses Mk polyploidization. We also concluded that p53 expression specifically affects the process of polyploidization possibly by inhibiting DNA synthesis and inducing apoptosis in megakaryocytes; 2) by identifying putative direct targets of p53 that are strongly differentially expressed in cells undergoing terminal Mk differentiation upon p53 downregulation, which may be involved in regulating polyploidization in Mk cells; and 3) by identifying a set of cell cycle-, apoptosis-, and cytoskeleton-related genes, not known to be transcriptional targets of p53, that could also participate in regulating p53-dependent responses during megakaryopoiesis.
GRANTS
This work was supported in part by funding to E. T. Papoutsakis through the Delaware Biotechnology Institute. P. A. Apostolidis acknowledges personal funding through the Delaware Biotechnology Institute and the A. S. Onassis foundation. S. Lindsey was supported by National Heart, Lung, and Blood Institute (NHLBI) Grant F32HL-091620. Additional support was received from NHLBI Grant R21HL-106397. The content is solely the responsibility of the authors and does not necessarily represent the official views of NHLBI or NIH.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: P.A.A., W.M.M., and E.T.P. conception and design of research; P.A.A. and S.L. performed experiments; P.A.A., S.L., W.M.M., and E.T.P. analyzed data; P.A.A., S.L., W.M.M., and E.T.P. interpreted results of experiments; P.A.A. and S.L. prepared figures; P.A.A., W.M.M., and E.T.P. drafted manuscript; P.A.A., W.M.M., and E.T.P. edited and revised manuscript; P.A.A., W.M.M., and E.T.P. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Dr. Ryan S. Senger (Biological Systems Engineering, Virginia Tech) for help in the biocomputational processing of raw microarray data. Dr. Yili Chen is acknowledged for help with gene annotation using Gene Ontology. Dr. Min Wang is acknowledged for providing hands-on expertise in performing the microarray experiments. We thank Dr. John Crispino for helpful comments on the manuscript.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA 92: 8493–8497, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ando K, Ajchenbaum-Cymbalista F, Griffin JD. Regulation of G1/S transition by cyclins D2 and D3 in hematopoietic cells. Proc Natl Acad Sci USA 90: 9571–9575, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Apostolidis PA, Woulfe DS, Chavez M, Miller WM, Papoutsakis ET. Role of tumor suppressor p53 in megakaryopoiesis and platelet function. Exp Hematol 40: 131–42, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 10: 265–275, 2009. [DOI] [PubMed] [Google Scholar]
- 5. Baccini V, Roy L, Vitrat N, Chagraoui H, Sabri S, Le Couedic JP, Debili N, Wendling F, Vainchenker W. Role of p21(Cip1/Waf1) in cell-cycle exit of endomitotic megakaryocytes. Blood 98: 3274–3282, 2001. [DOI] [PubMed] [Google Scholar]
- 6. Baker SJ, Markowitz S, Fearon ER, Willson JK, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249: 912–915, 1990. [DOI] [PubMed] [Google Scholar]
- 7. Bassini A, Pierpaoli S, Falcieri E, Vitale M, Guidotti L, Capitani S, Zauli G. Selective modulation of the cyclin B/CDK1 and cyclin D/CDK4 complexes during in vitro human megakaryocyte development. Br J Haematol 104: 820–828, 1999. [DOI] [PubMed] [Google Scholar]
- 8. Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2: a000935, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126: 107–120, 2006. [DOI] [PubMed] [Google Scholar]
- 10. Bermejo R, Vilaboa N, Cales C. Regulation of CDC6, geminin, and CDT1 in human cells that undergo polyploidization. Mol Biol Cell 13: 3989–4000, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brainard AM, Miller AJ, Martens JR, England SK. Maxi-K channels localize to caveolae in human myometrium: a role for an actin-channel-caveolin complex in the regulation of myometrial smooth muscle K+ current. Am J Physiol Cell Physiol 289: C49–C57, 2005. [DOI] [PubMed] [Google Scholar]
- 12. Chen C, Fuhrken PG, Huang LT, Apostolidis P, Wang M, Paredes CJ, Miller WM, Papoutsakis ET. A systems-biology analysis of isogenic megakaryocytic and granulocytic cultures identifies new molecular components of megakaryocytic apoptosis. BMC Genomics 8: 384, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chitu V, Pixley FJ, Macaluso F, Larson DR, Condeelis J, Yeung YG, Stanley ER. The PCH family member MAYP/PSTPIP2 directly regulates F-actin bundling and enhances filopodia formation and motility in macrophages. Mol Biol Cell 16: 2947–2959, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet 32, Suppl: 490–495, 2002. [DOI] [PubMed] [Google Scholar]
- 15. Chylicki K, Ehinger M, Svedberg H, Bergh G, Olsson I, Gullberg U. p53-mediated differentiation of the erythroleukemia cell line K562. Cell Growth Differ 11: 315–324, 2000. [PubMed] [Google Scholar]
- 16. Clarke MC, Savill J, Jones DB, Noble BS, Brown SB. Compartmentalized megakaryocyte death generates functional platelets committed to caspase-independent death. J Cell Biol 160: 577–587, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conacci-Sorrell M, Zhurinsky J, Ben-Ze'ev A. The cadherin-catenin adhesion system in signaling and cancer. J Clin Invest 109: 987–991, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126: 121–134, 2006. [DOI] [PubMed] [Google Scholar]
- 19. Datta NS, Long MW. Modulation of MDM2/p53 and cyclin-activating kinase during the megakaryocyte differentiation of human erythroleukemia cells. Exp Hematol 30: 158–165, 2002. [DOI] [PubMed] [Google Scholar]
- 20. Duriez C, Falette N, Audoynaud C, Moyret-Lalle C, Bensaad K, Courtois S, Wang Q, Soussi T, Puisieux A. The human BTG2/TIS21/PC3 gene: genomic structure, transcriptional regulation and evaluation as a candidate tumor suppressor gene. Gene 282: 207–214, 2002. [DOI] [PubMed] [Google Scholar]
- 21. Even-Ram S, Yamada KM. Of mice and men: relevance of cellular and molecular characterizations of myosin IIA to MYH9-related human disease. Cell Adh Migr 1: 152–155, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Faradji F, Bloyer S, Dardalhon-Cumenal D, Randsholt NB, Peronnet F. Drosophila melanogaster cyclin G coordinates cell growth and cell proliferation. Cell Cycle 10: 805–818, 2011. [DOI] [PubMed] [Google Scholar]
- 23. Fuhrken PG, Apostolidis PA, Lindsey S, Miller WM, Papoutsakis ET. Tumor suppressor protein p53 regulates megakaryocytic polyploidization and apoptosis. J Biol Chem 283: 15589–15600, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fuhrken PG, Chen C, Apostolidis PA, Wang M, Miller WM, Papoutsakis ET. Gene Ontology-driven transcriptional analysis of CD34+ cell-initiated megakaryocytic cultures identifies new transcriptional regulators of megakaryopoiesis. Physiol Genomics 33: 159–169, 2008. [DOI] [PubMed] [Google Scholar]
- 25. Fuhrken PG, Chen C, Miller WM, Papoutsakis ET. Comparative, genome-scale transcriptional analysis of CHRF-288-11 and primary human megakaryocytic cell cultures provides novel insights into lineage-specific differentiation. Exp Hematol 35: 476–489, 2007. [DOI] [PubMed] [Google Scholar]
- 26. Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 21: 1874–1887, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giammona LM, Panuganti S, Kemper JM, Apostolidis PA, Lindsey S, Papoutsakis ET, Miller WM. Mechanistic studies on the effects of nicotinamide on megakaryocytic polyploidization and the roles of NAD+ levels and SIRT inhibition. Exp Hematol 37: 1340–1352, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heminger K, Markey M, Mpagi M, Berberich SJ. Alterations in gene expression and sensitivity to genotoxic stress following HdmX or Hdm2 knockdown in human tumor cells harboring wild-type p53. Aging (Albany NY) 1: 89–108, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, Galjart N. Mammalian Golgi-associated bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J 20: 4041–4054, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet 3: e127, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishiko E, Matsumura I, Ezoe S, Gale K, Ishiko J, Satoh Y, Tanaka H, Shibayama H, Mizuki M, Era T, Enver T, Kanakura Y. Notch signals inhibit the development of erythroid/megakaryocytic cells by suppressing GATA-1 activity through the induction of HES1. J Biol Chem 280: 4929–4939, 2005. [DOI] [PubMed] [Google Scholar]
- 32. Jiang F, Jia Y, Cohen I. Fibronectin- and protein kinase C-mediated activation of ERK/MAPK are essential for proplateletlike formation. Blood 99: 3579–3584, 2002. [DOI] [PubMed] [Google Scholar]
- 33. Jiang PH, Motoo Y, Iovanna JL, Pebusque MJ, Xie MJ, Okada G, Sawabu N. Tumor protein p53-induced nuclear protein 1 (TP53INP1) in spontaneous chronic pancreatitis in the WBN/Kob rat: drug effects on its expression in the pancreas. JOP 5: 205–216, 2004. [PubMed] [Google Scholar]
- 34. Jurak Begonja A, Hoffmeister KM, Hartwig JH, Falet H. FlnA-null megakaryocytes prematurely release large and fragile platelets that circulate poorly. Blood 118: 2285–2295, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaushansky K. Determinants of platelet number and regulation of thrombopoiesis. Hematology Am Soc Hematol Educ Program: 147–152, 2009. [DOI] [PubMed] [Google Scholar]
- 36. Kaushansky K. Historical review: megakaryopoiesis and thrombopoiesis. Blood 111: 981–986, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kawasaki A, Matsumura I, Miyagawa J, Ezoe S, Tanaka H, Terada Y, Tatsuka M, Machii T, Miyazaki H, Furukawa Y, Kanakura Y. Downregulation of an AIM-1 kinase couples with megakaryocytic polyploidization of human hematopoietic cells. J Cell Biol 152: 275–287, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kawase T, Ohki R, Shibata T, Tsutsumi S, Kamimura N, Inazawa J, Ohta T, Ichikawa H, Aburatani H, Tashiro F, Taya Y. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell 136: 535–550, 2009. [DOI] [PubMed] [Google Scholar]
- 39. Konopatskaya O, Gilio K, Harper MT, Zhao Y, Cosemans JM, Karim ZA, Whiteheart SW, Molkentin JD, Verkade P, Watson SP, Heemskerk JW, Poole AW. PKCalpha regulates platelet granule secretion and thrombus formation in mice. J Clin Invest 119: 399–407, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kostyak JC, Naik UP. Calcium- and integrin-binding protein 1 regulates endomitosis and its interaction with Polo-like kinase 3 is enhanced in endomitotic Dami cells. PLoS One 6: e14513, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Law JC, Ritke MK, Yalowich JC, Leder GH, Ferrell RE. Mutational inactivation of the p53 gene in the human erythroid leukemic K562 cell line. Leuk Res 17: 1045–1050, 1993. [DOI] [PubMed] [Google Scholar]
- 42. Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ 13: 1027–1036, 2006. [DOI] [PubMed] [Google Scholar]
- 43. Li J, Tan J, Zhuang L, Banerjee B, Yang X, Chau JF, Lee PL, Hande MP, Li B, Yu Q. Ribosomal protein S27-like, a p53-inducible modulator of cell fate in response to genotoxic stress. Cancer Res 67: 11317–11326, 2007. [DOI] [PubMed] [Google Scholar]
- 44. Liebermann DA, Hoffman B. Gadd45 in the response of hematopoietic cells to genotoxic stress. Blood Cells Mol Dis 39: 329–335, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Limb JK, Yoon S, Lee KE, Kim BH, Lee S, Bae YS, Jhon GJ, Kim J. Regulation of megakaryocytic differentiation of K562 cells by FosB, a member of the Fos family of AP-1 transcription factors. Cell Mol Life Sci 66: 1962–1973, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu XS, Li H, Song B, Liu X. Polo-like kinase 1 phosphorylation of G2 and S-phase-expressed 1 protein is essential for p53 inactivation during G2 checkpoint recovery. EMBO Rep 11: 626–632, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lordier L, Chang Y, Jalil A, Aurade F, Garcon L, Lecluse Y, Larbret F, Kawashima T, Kitamura T, Larghero J, Debili N, Vainchenker W. Aurora B is dispensable for megakaryocyte polyploidization, but contributes to the endomitotic process. Blood 116: 2345–2355, 2010. [DOI] [PubMed] [Google Scholar]
- 48. Maynard DM, Heijnen HF, Gahl WA, Gunay-Aygun M. The α-granule proteome: novel proteins in normal and ghost granules in gray platelet syndrome. J Thromb Haemost 8: 1786–1796, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGowan KA, Pang WW, Bhardwaj R, Perez MG, Pluvinage JV, Glader BE, Malek R, Mendrysa SM, Weissman IL, Park CY, Barsh GS. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood 118: 3622–3633, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Melloni E, Secchiero P, Celeghini C, Campioni D, Grill V, Guidotti L, Zauli G. Functional expression of TRAIL and TRAIL-R2 during human megakaryocytic development. J Cell Physiol 204: 975–982, 2005. [DOI] [PubMed] [Google Scholar]
- 51. Merkle CJ, Karnitz LM, Henry-Sanchez JT, Chen J. Cloning and characterization of hCTF18, hCTF8, and hDCC1. Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J Biol Chem 278: 30051–30056, 2003. [DOI] [PubMed] [Google Scholar]
- 52. Ramsborg CG, Papoutsakis ET. Global transcriptional analysis delineates the differential inflammatory response interleukin-15 elicits from cultured human T cells. Exp Hematol 35: 454–464, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412, 2008. [DOI] [PubMed] [Google Scholar]
- 54. Ritchie A, Braun SE, He J, Broxmeyer HE. Thrombopoietin-induced conformational change in p53 lies downstream of the p44/p42 mitogen activated protein kinase cascade in the human growth factor-dependent cell line M07e. Oncogene 18: 1465–1477, 1999. [DOI] [PubMed] [Google Scholar]
- 55. Ryan MC, Zeeberg BR, Caplen NJ, Cleland JA, Kahn AB, Liu H, Weinstein JN. SpliceCenter: a suite of web-based bioinformatic applications for evaluating the impact of alternative splicing on RT-PCR, RNAi, microarray, and peptide-based studies. BMC Bioinformatics 9: 313, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378, 2003. [DOI] [PubMed] [Google Scholar]
- 57. Saxena S, Dutta A. Geminin and p53: deterrents to rereplication in human cancer cells. Cell Cycle 2: 283–286, 2003. [PubMed] [Google Scholar]
- 58. Serra-Pages C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem 273: 15611–15620, 1998. [DOI] [PubMed] [Google Scholar]
- 59. Shin JW, Swift J, Spinler KR, Discher DE. Myosin-II inhibition and soft 2D matrix maximize multinucleation and cellular projections typical of platelet-producing megakaryocytes. Proc Natl Acad Sci USA 108: 11458–11463, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sigurjonsson OE, Gudmundsson KO, Haraldsdottir V, Rafnar T, Agnarsson BA, Gudmundsson S. Flt3/Flk-2 ligand in combination with thrombopoietin decreases apoptosis in megakaryocyte development. Stem Cells Dev 13: 183–191, 2004. [DOI] [PubMed] [Google Scholar]
- 61. Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol 224: 111–136, 2003. [DOI] [PubMed] [Google Scholar]
- 62. Syed N, Coley HM, Sehouli J, Koensgen D, Mustea A, Szlosarek P, McNeish I, Blagden SP, Schmid P, Lovell DP, Hatzimichael E, Crook T. Polo-like kinase Plk2 is an epigenetic determinant of chemosensitivity and clinical outcomes in ovarian cancer. Cancer Res 71: 3317–3327, 2011. [DOI] [PubMed] [Google Scholar]
- 63. Talos F, Nemajerova A, Flores ER, Petrenko O, Moll UM. p73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol Cell 27: 647–659, 2007. [DOI] [PubMed] [Google Scholar]
- 64. Tarcea VG, Weymouth T, Ade A, Bookvich A, Gao J, Mahavisno V, Wright Z, Chapman A, Jayapandian M, Ozgur A, Tian Y, Cavalcoli J, Mirel B, Patel J, Radev D, Athey B, States D, Jagadish HV. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res 37: D642–D646, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Geet C, Izzi B, Labarque V, Freson K. Human platelet pathology related to defects in the G-protein signaling cascade. J Thromb Haemost 7, Suppl 1: 282–286, 2009. [DOI] [PubMed] [Google Scholar]
- 67. van Zon W, Ogink J, ter Riet B, Medema RH, te Riele H, Wolthuis RM. The APC/C recruits cyclin B1-Cdk1-Cks in prometaphase before D box recognition to control mitotic exit. J Cell Biol 190: 587–602, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219, 2006. [DOI] [PubMed] [Google Scholar]
- 69. Yang H, Haddad H, Tomas C, Alsaker K, Papoutsakis ET. A segmental nearest neighbor normalization and gene identification method gives superior results for DNA-array analysis. Proc Natl Acad Sci USA 100: 1122–1127, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhao L, Gregoire F, Sul HS. Transient induction of ENC-1, a Kelch-related actin-binding protein, is required for adipocyte differentiation. J Biol Chem 275: 16845–16850, 2000. [DOI] [PubMed] [Google Scholar]
- 71. Zhao Y, Lu S, Wu L, Chai G, Wang H, Chen Y, Sun J, Yu Y, Zhou W, Zheng Q, Wu M, Otterson GA, Zhu WG. Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21(Waf1/Cip1). Mol Cell Biol 26: 2782–2790, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhao Y, Zhang Y, Wang S, Hua Z, Zhang J. The clock gene Per2 is required for normal platelet formation and function. Thromb Res 127: 122–130, 2011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.