Abstract
Among the growing number of membrane protein structures in the Protein Data Bank, there are many transmembrane domains that appear to be native-like; at the same time there are others that appear to have less than complete native-like character. Hence there is an increasing need for validation tools that distinguish native-like from nonnative-like structures. Membrane mimetics used in protein structural characterizations differ in numerous physicochemical properties from native membranes and provide many opportunities for introducing nonnative-like features into membrane protein structures. One possible approach for validating membrane protein structures is based on the use of glycine residues in transmembrane domains. Here, we have reviewed the membrane protein structure database and identified a set of benchmark proteins that appear to be native like. In these structures, conserved glycine residues rarely face the lipid interstices, and many of them participate in close helix-helix packing. Glycine-based validation allowed the identification of nonnative-like features in several membrane proteins and also shows the potential for verifying the native-like character for numerous other membrane protein structures.
Alpha-helical membrane protein structures can be influenced by the membrane mimetic environments in subtle and not so subtle ways (1-6). Anfinsen in 1973 (7) stated “that the native conformation (of a protein) is determined by the totality of interatomic interactions and hence by the amino acid sequence in a given environment.” Therefore, the interactions from the heterogeneous membrane environment contribute to the sum of interactions responsible for defining the three-dimensional structure of a membrane protein. Here, we focus on the influence of the physical properties of membrane environments instead of the influence of specific lipids. Recently, a detailed description of how promiscuous membrane proteins can be in their interactions with different lipids has been published (8). A challenge for membrane protein structural biologists is to mimic the membrane environment adequately to stabilize the native protein structure, while preparing a sample that is appropriate for the specific structural technique. Only bacteriorhodopsin has been characterized in its native membrane environment (9). A few others have been characterized in liquid crystalline lipid bilayers (4, 10-14) and more in the presence of lipids.(15-17) The vast majority have been characterized in the presence of detergents, most in crystal lattices and others in detergent micelles. As the number of α-helical membrane protein structures increase in the Protein Data Bank (PDB), there is an increasing need for tools to evaluate their native-like structural quality. Here, we explore the basis for developing a tool to test the compatibility of membrane protein structures with a native membrane environment.
Large membrane proteins with cofactors in the transmembrane (TM) domains may have significant interactions with these cofactors to stabilize tertiary structures (18-20). However, the tertiary stability of a TM domain with only a few helices is often limited by very weak inter-helical interactions, due to a largely hydrophobic amino acid composition and the tendency for TM helices to have uniform backbone torsion angles resulting from strong hydrogen bonding in the low dielectric membrane environment. (21-24). In other words, the packing of a set of relatively rigid and uniform helical rods generates marginal tertiary stability. This minimal stability permits membrane proteins to adopt multiple tertiary conformations with different helical packing for various functional states. Consequently, there is a tendency to justify structural variations among different characterizations of a protein as reflecting different functional states. However, each of these structures should be compatible with the native membrane environment and therefore, the native-like character of these structures should be critically assessed, especially since TM domains with marginal tertiary stability are subject to distortion by membrane mimetics (1-4, 10).
Although membrane protein structural biologists have used functional assays to validate their structural conclusions, often these assays have to be performed in an environment different from that used for structural characterization, such as the assays for ion channel conductance, where a bilayer is needed. These assays validate the protein constructs, but not the structures. These assays are important in that they validate the non-native protein constructs often used for structural characterization. Site-directed mutagenesis, binding of antibodies, deletion of loops and termini, and the insertion of water soluble proteins into loops have frequently been used for recent structural characterizations (25-28). Yet, in addition to functional viability of the construct, it is important to validate the structure obtained from a membrane mimetic environment as to whether it is consistent with the native membrane environment. Functional misunderstanding of proteins influenced by the membrane mimetic environment can be propagated through the literature for many years (3, 24, 29-31).
Several approaches have been used to obtain detailed structural data for membrane proteins within a lipid environment and even a cellular membrane environment. Electron crystallography (32 depositions in the PDB) has been used to characterize multiple membrane proteins in a lipid environment matrix (17, 32-35). Solid-state NMR spectroscopy (52 depositions in the PDB) has been used to obtain structural restraints for membrane proteins in liposomal or planar bilayer environments (4, 10-14). Other techniques such as ESR have also provided important structural restraints for membrane proteins in proteoliposomes. Such data can be of great use in validating membrane protein structure, since these lipid environments are more native-like than the detergent environments typically used by x-ray crystallography and solution NMR spectroscopy that have determined the majority of the membrane protein structures in the PDB.
It is well recognized that charged and perhaps polar residues when exposed to the hydrophobic interstices of lipid bilayers will result in a significant energetic penalty that has to be compensated in order to achieve a stable structure (36, 37). Indeed, viewing the charged residues of a membrane protein is a useful way for evaluating the native-like quality of the structure. While polar residues like serine and threonine can form intra-helical hydrogen bonds to backbone amides and thereby shield both the polar sidechains and the polar backbone from the hydrophobic environment (38), even these polar residues have a tendency to avoid the lipid interstices (39). Other, hydrophilic residues are rarely exposed to the fatty acyl chains in a membrane environment.
Here, complementary to approaches based on the charged and polar residues, we suggest another approach for validating the native-like character of membrane protein structures, based on the location of glycine residues.
The Influence of Membrane Mimetic Environments
There are many ways that membrane mimetic environments can influence membrane protein structures and, conversely, for membrane proteins to influence the membrane mimetic environments. Synthetic bilayers used for solid state NMR, ESR, and electron crystallography studies may have a hydrophobic mismatch with membrane proteins (40-42), a lack of bilayer asymmetry, chemical and electrical gradients, inadequacy in lipid heterogeneity or curvature frustration (43, 44), and a nonnative lateral pressure profile (45). Nevertheless, synthetic bilayers are significantly better than other membrane mimetics. Detergent micelles provide a single highly curved hydrophilic surface that generates a different environment for amphipathic helices that bind at the lipid hydrophobic/hydrophilic interface (46). This hydrophilic surface can stabilize a distorted helical structure, by solvating hydrophilic residues in the middle of a curved helix. Moreover, the hydrophobic dimension of a detergent micelle can be easily changed in response to optimal packing of the helices that is potentially different from that in the native membrane (2).
Detergents also generate a weaker hydrophobic environment compared with lipid bilayers, with extensive water penetration into the micelle (35, 47-49) as well as a weaker and distorted lateral pressure profile (45, 50). While lipid headgroups retain considerable dynamics, the lateral pressure profile in membranes suggests close packing of the lipid headgroups/glycerol backbone around membrane solubilized proteins. In detergent micelles the headgroup region is even more dynamic due to the high curvature of the hydrophilic surface and consequently the lateral pressure profile is less dramatic and the headgroup region is less tightly packed (3). In crystal environments contacts between proteins within and between the unit cells can distort the protein structure. Hydrophilic organics and water molecules are often embedded in what would be the very low dielectric environment of the membrane interstices, thereby weakening the hydrophobic environment. Also, in crystal lattices detergents often appear to form a thin hydrophobic layer around membrane proteins. Consequently, if a membrane protein is dependent on its environment for defining the hydrophobic thickness, the structure may be distorted in such a crystalline environment.
Detergents also have a monomeric concentration that is as much as or more than six orders of magnitude greater than that for monomeric lipids (51). As a result water soluble and/or dynamic domains as well as pores through the membrane protein structure may be altered when detergents are used as a membrane mimetic (2, 52), because of the relatively high concentration of monomeric detergents in the bulk aqueous environment. Thus, membrane mimetics provide many opportunities for introducing nonnative-like structural perturbations.
Nonnative-like Structural Perturbations
The need for validation tools is apparent on many different levels. Observed structural influences by membrane mimetics can be minor, such as charged sidechains that are oriented toward what would be the bilayer interstices instead of toward the aqueous environment, or nearly complete disruption of the tertiary structure as in the voltage sensing domain of the well-known initial structure of KvAP (PDB entry 1ORQ) (26). In between there are multiple examples of TM domains that appear to have less than complete native-like character. This is not to say that these structures are not significant stepping stones towards structural and functional understanding of these proteins, but if the nonnative-like structural perturbations are unrecognized these structures can also be misleading.
In a few cases there are multiple structures of the same or similar membrane protein obtained under differing conditions that provide possible insights into the structural abnormalities. Because of the potential for multiple functional conformations care must be taken in such interpretations. Therefore, the goal here is not so much to validate these TM domains as representing functional states, but rather to validate these structures as compatible with native-membrane environments. While being heterogeneous these environments still have well-defined properties, such as a very low dielectric constant in the hydrophobic interior and a significant hydrophobic dimension.
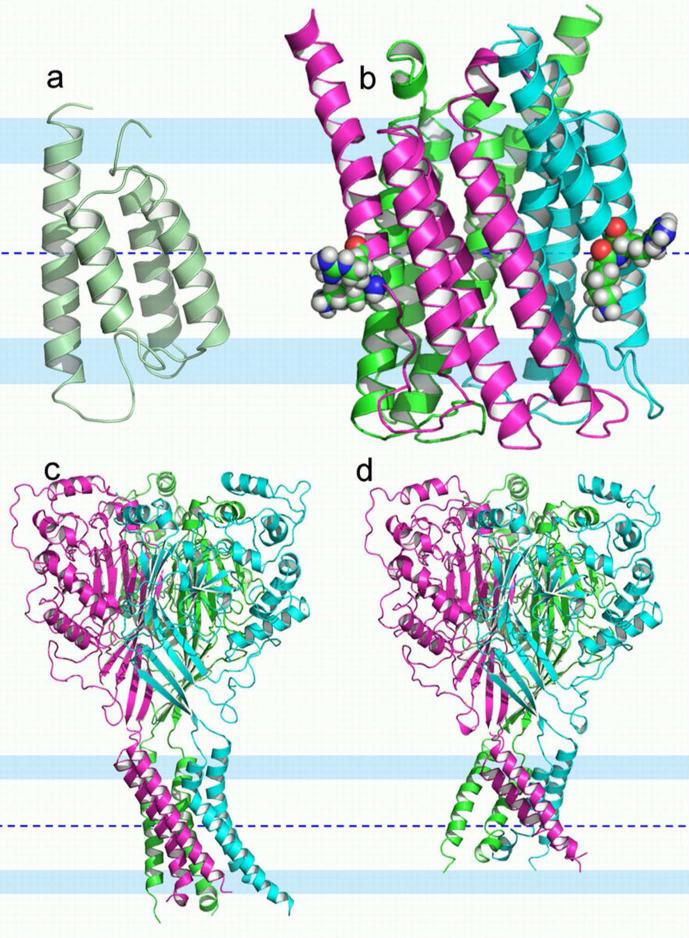
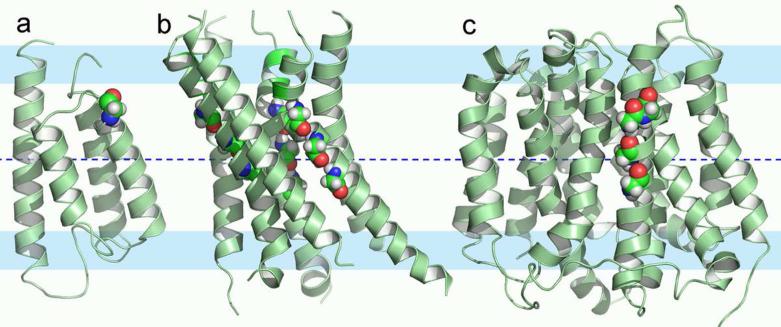
Potential nonnative-like features are illustrated by three structures in Fig. 1. KdpD is a histidine kinase receptor for regulating the operon that codes for the K+ transporter, Kdp. The solution NMR structure (2KSF) of the TM domain of KdpD is a four-helix bundle in detergent micelles (53) (Fig. 1a). A striking feature of these helices is that two of them, helices 2 and 3, are not nearly long enough to span the approximate 30 Å hydrophobic dimension of the native membrane, suggesting that the hydrophobic span of the membrane mimetic is insufficient. Throughout much of helices 1, 3 and 4 there is hydrogen/deuterium exchange in the amide backbone suggesting exposure to water and consequently a weak hydrophobic environment. In addition, numerous hydrophilic sidechains (Ser409, Thr413, Ser448, Thr452, and Asn493) are exposed to what would be the lipid interstices as opposed to being oriented toward the interior of the helical bundle. Indeed, it appears as if there is a minimum number of hydrophilic sidechains oriented toward the interior of the bundle.
Figure 1.
Putative nonnative-like structural perturbations of three membrane proteins. The membrane central plane was located as described in Supplemental Material. This membrane central plane is shown as a blue dashed line; interfacial regions are represented by 8 Å wide pale blue colored bands; and the conservative hydrophobic thickness in between is 25 Å. (a) The histidine kinase receptor KdpD TM domain (solution NMR structure, PDB entry 2KSF). This four-helix bundle has two very short helices and multiple hydrophilic residues exposed to the hydrophobic region of the would-be membrane. The short helices dictate that hydrophilic backbone amides of the inter-helical loops are also exposed to the membrane interstices. (b) 5-lipoxygenase-activating protein (4.0-Å resolution structure, PDB entry 2Q7M). The three chains are displayed in different colors. Helix 4 appears shifted along the helical axis, exposing two charged residues (Lys116 and Arg117 in space filling mode with carbon atoms green, nitrogen atoms blue, oxygen atoms red, and hydrogen atoms white) to the very center of the membrane and the inter-helical loop between helices 3 and 4 is drawn into the lipid interstices exposing more hydrophilic sites to the hydrophobic region of the membrane. (c-d) Two structures of trimeric acid-sensing ion channel (1.9-Å resolution structure, PDB entry 2QTS in (c); and 3-Å resolution structure, PDB entry 3HGC in (d)). They have similar symmetric extramembranous domains but different TM domains. The TM domain in (c) has a sufficient hydrophobic dimension but is asymmetric, probably the result of substantial crystal contacts, while the TM domain in (d) is more symmetric, but does not span the hydrophobic dimension of native membranes.
Trimeric 5-lipoxygenase activating protein (FLAP) has four TM helices per monomer. FLAP (Fig. 1b) has been characterized by x-ray crystallography (2Q7M), showing both Lys116 and Arg117 in what would be the middle of the hydrophobic region (54). Both the guanidinium group of Arg117 and the backbone amide of Phe138 form electrostatic contacts with a neighboring trimer in the crystal. As a result, helix 4 (including Lys116 and Arg117 residues) appears to be shifted along the direction of the helical axis to the extent that the inter-helical loop (residues 108-115) extends to the center of what would be the membrane, exposing a large number of hydrophilic amides to the hydrophobic environment of the would-be membrane interior. Moreover, the C-terminal residues, 138-149, of the neighboring trimer have their amides completely exposed to the would-be membrane interior surrounding the first trimer. Another member of this protein family, microsomal prostaglandin E synthase 1, has been characterized in a lipid environment by electron crystallography (3DWW) and shows 4 complete TM helices with no exposure of charged residues or amides from non-helical segments exposed to the fatty acyl chains of the lipid environment (55).
There are two crystal structures of the acid sensing ion channel (ASIC) using somewhat different constructs (56, 57). These structures are also trimers with each monomer contributing a pair of TM helices. The very large symmetric extracellular domains from the two structures superimpose well, but while the TM domain of the 2009 structure has approximate three-fold symmetry (Fig. 1d), the 2007 structure lacks this symmetry (Fig. 1c), possibly due to substantial crystal packing interactions. This latter structure has long helices that would readily span the membrane, while the 2009 structure has a very short hydrophobic dimension resulting from helices that are both kinked and tilted at too large an angle to the symmetry axis. The result is that the 2009 structure is not consistent with the hydrophobic dimension of native membranes and the 2007 structure deviates from expected three-fold symmetry.
Glycine Residues in Transmembrane Helices
Glycine and proline residues are surprisingly common in TM α-helices (24, 58), even though these residues are known to be helix-destabilizing in water soluble proteins (59-62). There have been several reasons identified for their presence in TM helices. In the TM environment helical hydrogen bonds are stronger due to the low dielectric of this environment (21-23, 63). The resulting helical regularity would limit helix packing opportunities and hence glycine (having access to a much greater φ,ψ torsional space) and proline (lacking an amide proton for hydrogen bonding within the helix) are present to induce kinks. These two residues, although destabilizing the helical structure, permit enhanced tertiary structural interactions from an increased surface area between helices and hence enhanced tertiary structural stability that would otherwise be very limited (20).
During functional processes membrane proteins frequently undergo significant structural rearrangement involving kinking and repacking of helices (64, 65). The presence of glycine and proline residues can facilitate such rearrangements. Indeed, TM helices can be remarkably regular even in the presence of these residues, leading to the moniker that these residues can be pro-kink sites, i.e. they induce kinks in some functional states and not in others (23). Kinks also lead to exposure of amide hydrogen bonding sites for structural or functional purposes. Such exposure is needed, since there are few hydrophilic sidechains that can provide chemically active sites and hence the backbone is a very important source for functional activity, for instance in the solvation of ions in the gramicidin A channel and in the KcsA channel (11, 66). The exposure of backbone amides through helix kinks may also lead to the binding of water in the bilayer interstices where its presence is very limited. Water is known to play important roles in TM domains, including the facilitation of structural interconversion through hydrogen bond rearrangements (65, 67, 68) and proton wires (69, 70); the exposure of backbone amides through kinks could greatly enhance these functional activities.
Moreover, glycine residues are known to be important for helix packing by allowing close approach of the helical backbones (71). The pioneering studies on glycophorin identified GxxxG motifs that permitted close packing of a dimer and increased helix-helix stability associated with increased van der Waals contacts (20, 72), increased long-range electrostatic interactions between helices, and the potential for Cα-H hydrogen bonding (63, 73). Since then GxxxG motifs and permutations involving alanine, serine and threonine have been widely identified (74-77). In addition, glycine zippers (GxxxGxxxG) have been described for helices that pack with a modest crossing angle (78).
This extensive use of glycine in TM helices could facilitate β-strand formation, but the frequent presence of proline residues would counter this tendency toward the formation of β-strands (77, 79, 80), resulting in the assurance that TM helices are formed despite the extensive use of glycine. Consequently, it would seem that glycine is used judiciously for facilitating tertiary and quaternary structural stability.
A Benchmark of Native-Like Structures
Using two criteria, an adequate hydrophobic dimension to span the hydrophobic dimension of the bilayer and a lack of hydrophilic site exposure to what would be the interstices of native membranes, an initial set of benchmark proteins from the PDB were identified. From these proteins, we extracted distributions and rules that are expected to be followed by native-like structures. Structures that deviate from these norms can then be suggested to contain nonnative-like features.
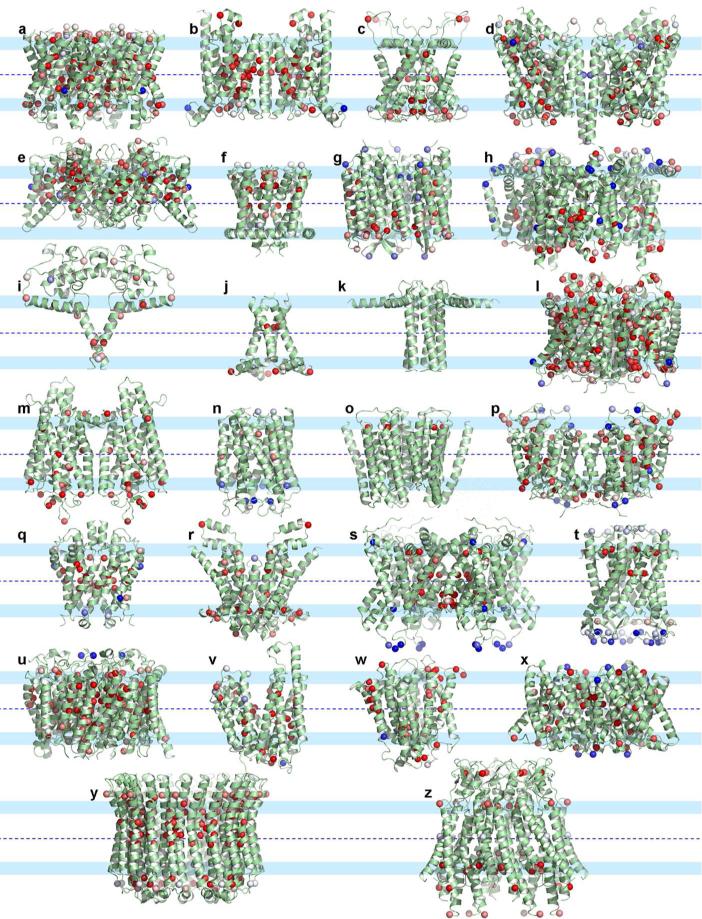
The 26 proteins comprising our benchmark are listed in Table 1 and displayed in Fig. 2. These structures span many of the structure/function families identified for TM helical proteins on the “Membrane Proteins of Known 3D Structure” website (http://blanco.biomol.uci.edu/mpstruc/listAll/list). In identifying structures for our benchmark set we did not include ones with multiple cofactors stabilizing the TM domains as these cofactors contribute significantly to structural stability. For the remaining structure/function families, nearly half are represented in our benchmark set. We have considerable confidence that these and many other proteins from the same structure/function families have native-like structures. (Many structures from the other structure/function families may also be native-like, but we have chosen conservatively in order not to distort the statistics of native-like structures.) None of the benchmark structures have charged residues in the hydrophobic interstices and they all have helices that appear to span a hydrophobic dimension consistent with native membrane environments. These proteins range in size from the solid state NMR structure of the tetrameric conductance domain (2L0J) of the Influenza A M2 protein with a single TM helix per monomer (10), and the x-ray crystallographic structure of dimeric cytochrome C quinol dehydrogenase (2J7A) also with a single TM helix per monomer (81), to the crystallographic structure of the dimeric HCL exchange transporter (3ND0) (82).
Table 1.
The benchmark set of 26 membrane proteins
| ID | PDB | Resolution/method | Name | Oligomeric state |
|---|---|---|---|---|
| a | 1FX8 | 2.2 Å | Escherichia coli Glycerol facilitator | Tetramer |
| b | 1L7V | 3.2 Å | Escherichia coli Vitamin B12 ABC transporter | Dimer |
| c | 1P7B | 3.65 Å | Potassium channel KirBac1.1 | Tetramer |
| d | 2A65 | 1.65 Å | Bacterial homologue of Na+/Cl--dependent neurotransmitter transporter | Dimer |
| e | 3ND0 | 3.5 Å | Cyanobacterial HCl exchange antiporter | Dimer |
| f | 2AHY | 2.4 Å | Bacillus cereus NaK channel | Tetramer |
| g | 2EI4 | 2.1 Å | Archaerhodopsin-2 | Tetramer |
| h | 2GIF | 2.9 Å | Escherichia coli Multidrug efflux transporter AcrB | Trimer |
| i | 2J7A | 2.3 Å | Cytochrome c quinol dehydrogenase | Dimer |
| j | 2L0J | solid state NMR | Influenza A M2 proton channel | Tetramer |
| k | 2KYV | solid-state/solution NMR | Phospholamban | Pentamer |
| l | 2NS1 | 1.96 Å | Escherichia coli GlnK bound Ammonia channel | Trimer |
| m | 3DQB | 3.2 Å | G-protein peptide bound G-protein-coupled receptor opsin | Dimer |
| n | 3DWW | electron crystallography | Glutathione bound Microsomal prostaglandin E synthase 1 | Trimer |
| o | 3EAM | 2.9Å | Gloeobacter violaceus ligand gated ion channel | Pentamer |
| p | 3NCY | 3.2 Å | Arginine agmatine antiporter | Dimer |
| q | 2YVX | 3.5 Å | Thermus thermophilus Magnesium transporter | Dimer |
| r | 2ONK | 3.1Å | ModA bound Archaeoglobus fulgidus Molybdate ABC transporter | Dimer |
| s | 2R9R | 2.4 Å | Chimera Kv2.1 and Kv1.2 potassium channel | Tetramer |
| t | 2OAR | 3.5Å | Mycobacterium tuberculosis Large mechanosensitive channel | Pentamer |
| u | 3HD6 | 2.1 Å | Homo sapiens Ammonium transporter Rh type C | Trimer |
| v | 3O7Q | 3.1 Å | Escherichia coli Fucose transporter | Monomer |
| w | 3MKT | 3.65 Å | Vibrio cholera Multidrug and toxic compound extrusion transporter | Monomer |
| x | 3M71 | 1.2 Å | Haemophilus influenzae SLAC1 channel | Trimer |
| y | 2BL2 | 2.1 Å | Enterococcus hirae V-Type Na+-ATPase rotor | Decamer |
| z | 2ZW3 | 3.5 Å | Homo sapiens Connexin-26 gap junction channel | Hexamer |
Figure 2.
Transmembrane domains of benchmark membrane protein structures. Cα atoms of glycines are shown as spheres and color-coded according to their conservation scores (red: highly conserved; blue; not conserved; pale colors: intermediate; see Supplemental Material for details). Default parameters for residue conservation were used for all the proteins except for the ligand-gated ion channel (PDB entry 3EAM), where the minimal sequence identity for sequence alignment was lowered from the default 35% to 25%. (a-z) Structures corresponding to entries a-z in Table 1. Note that the outward facing surface of the helices in these proteins is rarely interrupted by a glycine sphere.
Conserved Glycines and Their Structural Roles
The 26 benchmark proteins have a total of 673 glycines, compared to 837 alanines and 244 aspartates. Below we contrast these residues in terms of sequence conservation, solvent accessibility, and distance (z) from the would-be bilayer center (presented as the absolute value, |z|). The bilayer center was defined (see Supplemental Material) based on the distribution of the Cα carbons of charged residues in each protein, and could have a significant error. As a result the hydrophobic region of the bilayer was conservatively limited to ±10 Å from the bilayer center for evaluating the characteristics of glycine residues. Sequence conservation scores were obtained from the ConSurf web server (83); we refer to residues with the top 20% conservation scores as conserved.
Of the 673 glycines, 264 (39%) reside in the hydrophobic region, illustrating just how common glycine residues are in TM helices. In comparison, 40% of the alanines, but only 8% of the aspartates have |z| < 10 Å. Glycine thus has a similar tendency to locate in the hydrophobic region as alanine, while aspartate avoids the hydrophobic region of the bilayer. Therefore, based on this sampling, glycine, despite the nearly lowest helix propensity (proline is the only residues with a lower propensity), has the same likelihood of being in a TM helix as alanine, the residue with the highest helix propensity (61).
Of the 264 glycines in the hydrophobic region of the bilayer, 147 (56%) are conserved; in contrast only 31% of the glycines outside the hydrophobic region are conserved. For the alanines, 29% are conserved in the hydrophobic region, and 19% are conserved outside the hydrophobic region. There are too few aspartates with |z| < 10 Å to contrast the two regions in terms of conservation, but overall 75% of these residues are conserved. The fact that a very high fraction of the glycines are conserved further confirms that the glycines have important roles in these helices, as described above. Indeed, without a good reason to be present, and hence conserved, one might expect an evolutionary pressure to remove these glycine residues, since they destabilize helical structures.
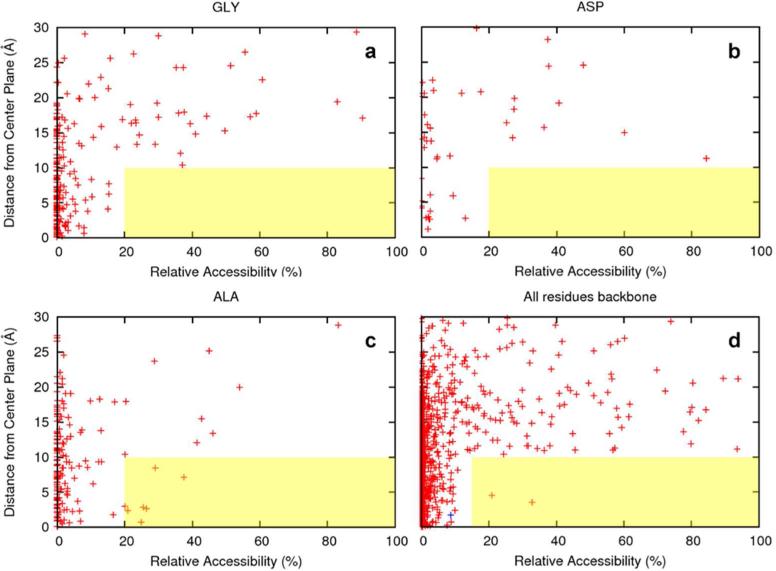
In Fig. 3a-c, we present scatter plots for the conserved glycines, alanines, and aspartates showing their whole-residue (i.e. including the sidechains) relative accessibility as a function of |z| values. Surprisingly, in the hydrophobic region (i.e., |z| < 10 Å), the glycines are similar to the aspartates in avoiding exposure to the lipid fatty acyl chains. No residues of either type have > 20% accessibility to the hydrophobic interstices of the bilayer. In comparison, 4% of the alanines in the hydrophobic region have > 20% accessibility to the hydrophobic interstices. Here 20% was used as the threshold in whole-residue relative accessibility for considering a residue as exposed. As a reference, residues in ideal poly-Gly, poly-Asp, and poly-Ala helices would have 64%, 66%, and 59% whole-residue exposure, respectively.
Figure 3.
Scatter plots displaying relative accessibility and distance (expressed as |z|, i.e. the absolute value of z) from the membrane central plane for the conserved residues in the benchmark proteins. Relative accessibility (i.e., percentage of the nominal maximum area (irrespective of secondary structure); see Supplemental Material) was calculated for either a whole residue or for the backbone polar atoms (C, O, and N) only. Oligomeric protein structures were used to calculate the solvent accessibility of each residue. However, for each oligomeric protein, only a single chain was used to count the number of glycines and other residues as well as for the backbone statistics. (a-c) Whole-residue relative accessibility for glycine, aspartate, and alanine residues. The hydrophobic region was conservatively defined as ±10 Å from the bilayer center. Surface exposure above 20% was considered significant. (d) Backbone relative accessibility for all 20 types of residues. Lipid-facing surface exposure of the backbone above 15% was considered significant. The two backbone sites in the entire benchmark set that have significant exposure are both proline residues (Pro315 of the B12 ABC transporter, PDB entry 1LV7; and Pro300 of a ligand gated channel, PDB entry 3EAM). The glycine residue with the greatest exposure (Gly87 of the NaK channel, PDB entry 2AHY) is highlighted as a blue ‘+’.
Glycine thus exhibits a strong tendency to avoid exposure to the hydrophobic interstices of the bilayer. While the exact numbers for accessibility may be subject to variation due to calculation details (e.g., the nominal maximum areas used for calculating relative accessibility and the value, 20%, used here as the accessibility threshold for considering a residue as exposed), the avoidance of exposure for glycine is unmistakable. This is likely driven by two factors: the inability of glycine residues to perform their function (summarized above as promoting tertiary structure) while exposed to the hydrophobic environment, and the need to avoid exposure of hydrophilic backbone atoms (N, C, and O) to the hydrophobic environment. Whereas glycines in an ideal poly-Gly helix have as much as 31% backbone exposure, Fig. 3d shows that, in the hydrophobic region (i.e., |z| < 10Å) the greatest exposure of the backbone by any glycine residue in our benchmark set is only 9%. Furthermore, all hydrophilic backbone atoms, regardless of sidechain type, are well shielded from the hydrophobic interstices of the bilayer -- all but two residues (two prolines) have ≤ 15% accessibility. While other types of residues can rely on sidechains to partially shield their hydrophilic backbone atoms (backbone exposure is down from 31% in a poly-Gly helix to 15% in a poly-Ala helix and to only 2% in a poly-Asp helix), the only recourse for glycine is tertiary contacts, which means avoiding lipid-facing positions.
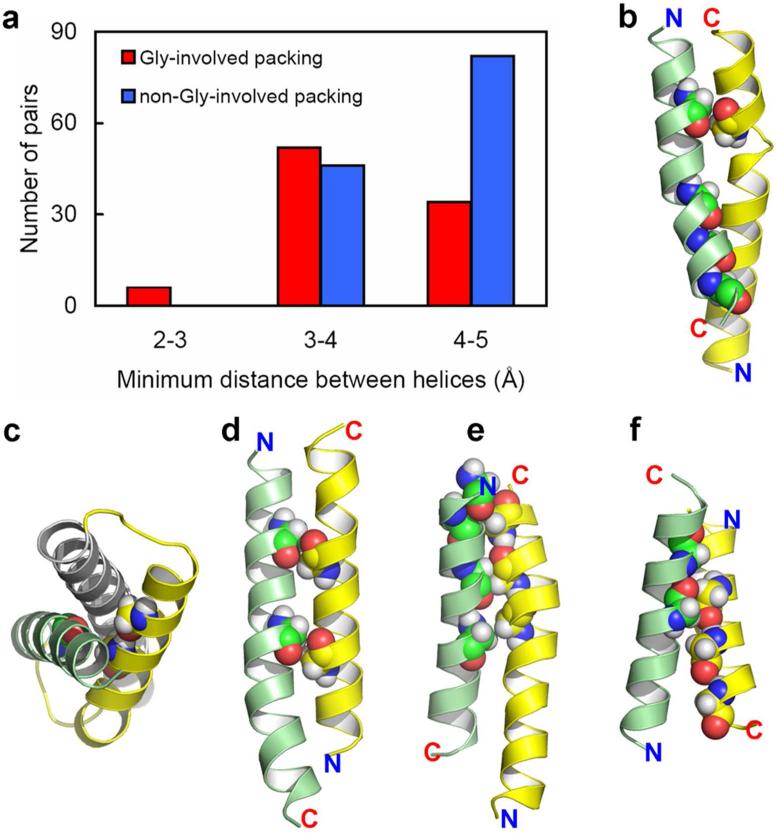
Of the 147 conserved glycines with |z| < 10Å in our benchmark, 96 (65%) are found in close helix-helix contacts, meaning that these glycines are separated from the partner helix by no more than 110% of the minimum distance of a helix pair. Presumably some of the other glycines could be in helix-helix contacts in alternative functional states of these proteins. In Fig. 4a, we further show that, among the 26 benchmark proteins, 92 of the 220 helix-helix pairs that have heavy atom contacts with distances < 5 Å and located in the hydrophobic region involve at least one glycine residue in these contacts. Furthermore, the results suggest that when glycine residues are involved the minimum distance between helices is somewhat decreased. Such a decrease in distance promotes not only additional van der Waals interactions, but potentially additional electrostatic interactions.
Figure 4.
Involvement of glycines in helix packing. A total of 220 helix-helix pairs in the 26 benchmark proteins were identified, (a) Number of helix-helix pairs binned according to distance of the closest contact and grouped according to whether glycine is involved. (b-f) Examples of helix pairs showing helix packing facilitated by glycine residues, highlighted here in space-filling mode. (b) Helix 1 (yellow) residue 27 and helix 6 (green) residues 204, 211, 214, and 218 from PDB entry 2NS1. Gly27 and Gly211 both appear to induce helix kinks that facilitate helix-helix interactions along the entire length of the TM helices despite substantial crossing angles at both crossing points. (c) Helix 7 (gray), helix 8 (yellow) residues 264 and 268, and helix 9 (green) residue 288 from PDB entry 2NS1. (d) Helix 3 (yellow) residues 97 and 104 and helix 4 (green) residues 123 and 130 from PDB entry 3O7Q. The i to i + 7 glycine residues on both helices, along with a small crossing angle, result in a large van der Waals interaction surface. (e) Helix 11 (yellow) residues 402, 406, and 410 and helix 12 (green) residues 421, 424, and 428 from PDB entry 3MKT. Here a GxxxGxxxG motif interacts with a GxxGxxxG motif. (f) Helix 4 (yellow) residues 151, 155, and 159 and helix 5 (green) residues 176 and 180 from PDB entry 3ND0. Here a GxxxGxxxG motif interacts with a GxxxG motif.
Glycine residues facilitate helix packing in a wide variety of ways (Fig. 4b-f). Helices pack at different crossing angles. When the crossing angle is relatively small various glycine motifs facilitate interactions over a considerable helical length (e.g. Fig. 4d, e). When the crossing angle is larger glycine residues also facilitate packing, sometimes with them on both helices (Fig. 4f), but frequently on a single helix (Fig. 4b, c). Glycines also facilitate helix kinking such as in Fig. 4b, thereby increasing the helical contact. Without such a kink the interactions between this pair of helices would be significantly reduced.
Surface Exposure of Glycines
The forgoing results show that in native-like structures conserved glycine residues are primarily involved in enhancing helix-helix interactions and are not likely to be exposed to the lipid environment. The results led us to posit that the lack of exposure of conserved glycine residues to the fatty acyl chains of native membranes may be used as a criterion for assessing the native-like quality of membrane protein structures.
As a simple test of our glycine-based validation approach, we searched for exposed, conserved glycines in the three structures in Fig. 1, which we already recognized as nonnative-like based on insufficient helix lengths, exposure of hydrophilic sidechains, and lack of oligomeric symmetry. Indeed, a number of conserved glycines are exposed in all these structures, including G444 located in helix 2 of KdpD (Fig. 5a), and G435, G439, and G443 in TM helix 2 of ASIC (Fig. 5b). A conserved glycine, G100, in helix 3 of FLAP was also exposed as a result of the helix 3-helix 4 loop being drawn into the membrane interstices.
Figure 5.
Application of the glycine-based validation tool. Conserved glycine residues that are exposed to the lipid interstices are highlighted in a space-filling mode. (a) Gly444 from PDB entry 2KSF. (b) Glycine residues 435, 439, and 443 from each monomer of PDB entry 2QTS. (c) Glycine residues 333, 336, 340, and 341 of the multi-drug transporter, EmrD (3.5-Å resolution structure, PDB entry 2GFP).
We also found exposed conserved glycines in the structure of EmrD (PDB entry 2GFP) (84), four conserved glycine residues, G333, G336, G340, and G341, in helix 11 are all exposed to the hydrophobic environment (Fig. 5c), suggesting that there is a local packing problem. The loop between helices 11 and 12 contain only hydrophilic, not charged residues, so the boundaries between the loop and these helices as well as the rotational orientation of these helices maybe sensitive to a presumably weak hydrophobic environment in the crystal lattice. Indeed, several hydrophilic residues in these two helices are exposed (residues Gln343, Thr360, and Ser364), although both Gln343 and Thr360 are relatively close to the bilayer interfacial region. Consequently, these hydrophilic residues alone might not generate vey much concern for this structure, while the exposed glycine residues generates a more significant concern. Other than the potential problem with helices 11 and 12, much of the rest of this large structure appears to be native-like. These results displayed in Fig. 5 collectively support a need for validating whether a given membrane protein structure is compatible with the native membrane environment.
Concluding Remarks
A structural validation approach based on the exposure of conserved glycine residues has been suggested and supported through the development of a set of benchmark proteins representing approximately half of the helical membrane protein structure/function families and the identification of several distorted proteins having conserved glycine residues exposed to what would be the membrane interstices. Glycine exposure to the lipid interstices is avoided in native-like structures, both to prevent exposing the helix backbone to the lipid hydrophobic environment and to allow for strengthened helix-helix packing. This validation approach was effective both in confirming putative nonnative-like structures and in identifying a previously unrecognized nonnative-like structure. Wide applicability is thus expected.
It should be recognized that, if a protein formed an oligomeric state or some other protein-protein complex, there might be a reason to have conserved glycine residues at the monomer-monomer interface. Many single TM helical proteins have glycine residues and it can be expected that these proteins will either form oligomers or interact with other membrane proteins such that in complex the glycine residues will not be exposed. We would further anticipate that exposing conserved glycine residues to the membrane interstices should drive the binding reaction towards complex formation (20). Interestingly, in the set of benchmark membrane proteins analyzed here the monomeric units of oligomeric proteins appear to utilize conserved glycine residues only infrequently at the monomer-monomer interfaces, while alanine residues appear to be more common at these interfaces.
Given their destabilizing influence on water soluble helices, how can the large number of glycine residues be tolerated in TM helices? These helices are primarily composed of aliphatic residues embedded in a low dielectric environment that is largely devoid of water, leading to strengthened intra-helical hydrogen bonds (21, 23). Therefore, secondary structural stability is substantially increased in the membrane environment and consequently glycine residues can be tolerated at some cost to helical stability. However, the glycine residues facilitate helix-helix packing, allowing for increased electrostatic and van der Waals interactions between the helices. So the result of the increased presence of glycine residues in TM helices is that excess secondary structural stability is sacrificed for increased tertiary structural stability, demonstrating an ingenious adaptation of membrane proteins to their environment.
Membrane protein structures are fundamentally important for many scientific communities including those interested in understanding cellular physiology and the development of pharmaceuticals. Our inability to characterize protein structures in the native membrane environment leads to the use of membrane mimetics that may or may not be good models of the native environment for a given membrane protein. The validation approach described here can facilitate the verification of native-like structures and the recognition of nonnative-like features that could otherwise mislead researchers who depend on the high fidelity of these important membrane protein structures.
Supplementary Material
Acknowledgments
Financial Support
This research has been supported in part by the National Institutes of Health through Grants AI-074805 and GM-088187 and the National Science Foundation through Cooperative Agreement 0654118 between the Division of Materials Research and the State of Florida.
Footnotes
SUPPORTING INFORMATION AVAILABLE:
The supplemental information provides details of how the structures from the Protein Data Bank were processed and analyzed as well as reference helical structures. Details for the calculation of solvent accessible surface areas and conservation scores are presented. The calculations for locating the center plane of the bilayer are described as well as the calculations for helix-helix contacts. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Tate CG. Comparison of three structures of the multidrug transporter EmrE. Curr Opin Struct Biol. 2006;16:457–464. doi: 10.1016/j.sbi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Cross TA, Sharma M, Yi M, Zhou HX. Influence of solubilizing environments on membrane protein structures. Trends Biochem Sci. 2011;36:117–125. doi: 10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinothkumar KR, Henderson R. Structures of membrane proteins. Q Rev Biophys. 2010;43:65–158. doi: 10.1017/S0033583510000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci U S A. 2011;108:9101–9106. doi: 10.1073/pnas.1016535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botelho AV, Huber T, Sakmar TP, Brown MF. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botelho AV, Gibson NJ, Thurmond RL, Wang Y, Brown MF. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 7.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 8.Sanders CR, Mittendorf KF. Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry. 2011;50:7858–7867. doi: 10.1021/bi2011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y. The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution. J Mol Biol. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- 10.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ketchem RR, Roux B, Cross TA. High-Resolution Polypeptide Structure in a Lamellar Phase Lipid Environment from Solid-State NMR Derived Orientational Constraints. Structure. 1997;5:1655–1669. doi: 10.1016/s0969-2126(97)00312-2. [DOI] [PubMed] [Google Scholar]
- 12.Das BB, Ajithkumar TG, Sinha N, Opella SJ, Ramanathan KV. Cross- and axial-peak intensities in 2D-SLF experiments based on cross-polarization--the role of the initial density matrix. J Magn Reson. 2007;185:308–317. doi: 10.1016/j.jmr.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Park SH, Marassi FM, Black D, Opella SJ. Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly. Biophys J. 2010;99:1465–1474. doi: 10.1016/j.bpj.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SH, De Angelis AA, Nevzorov AA, Wu CH, Opella SJ. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys J. 2006;91:3032–3042. doi: 10.1529/biophysj.106.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 16.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hite RK, Gonen T, Harrison SC, Walz T. Interactions of lipids with aquaporin-0 and other membrane proteins. Pflugers Arch. 2008;456:651–661. doi: 10.1007/s00424-007-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popot JLE, D.M. Helical Membrane Protein Folding, Stability and Evolution. Annu. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 19.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKenzie KR, Fleming KG. Association energetics of membrane spanning alpha-helices. Curr Opin Struct Biol. 2008;18:412–419. doi: 10.1016/j.sbi.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Cross TA. Uniformity, ideality, and hydrogen bonds in transmembrane alpha-helices. Biophys J. 2002;83:2084–2095. doi: 10.1016/S0006-3495(02)73969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page RC, Li C, Hu J, Gao FP, Cross TA. Lipid bilayers: an essential environment for the understanding of membrane proteins. Magn Reson Chem. 2007;45:S2–S11. doi: 10.1002/mrc.2077. [DOI] [PubMed] [Google Scholar]
- 23.Page RC, Kim S, Cross TA. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eilers M, Patel AB, Liu W, Smith SO. Comparison of helix interactions in membrane and soluble alpha-bundle proteins. Biophys J. 2002;82:2720–2736. doi: 10.1016/S0006-3495(02)75613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutzler R, Campbell EB, MacKinnon R. Gating the selectivity filter in ClC chloride channels. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 27.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano-Vega MJ, Magnani F, Shibata Y, Tate CG. Conformational thermostabilization of the beta1-adrenergic receptor in a detergent-resistant form. Proc Natl Acad Sci U S A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butterwick JA, MacKinnon R. Solution structure and phospholipid interactions of the isolated voltage-sensor domain from KvAP. J Mol Biol. 2010;403:591–606. doi: 10.1016/j.jmb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci U S A. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargas E, Bezanilla F, Roux B. In search of a consensus model of the resting state of a voltage-sensing domain. Neuron. 2011;72:713–720. doi: 10.1016/j.neuron.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg MF, O'Ryan LP, Hughes G, Zhao Z, Aleksandrov LA, Riordan JR, Ford RC. The cystic fibrosis transmembrane conductance regulator (CFTR): three-dimensional structure and localization of a channel gate. J Biol Chem. 2011;286:42647–42654. doi: 10.1074/jbc.M111.292268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abe K, Tani K, Fujiyoshi Y. Conformational rearrangement of gastric H(+),K(+)-ATPase induced by an acid suppressant. Nature communications. 2011;2:155. doi: 10.1038/ncomms1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oshima A, Tani K, Toloue MM, Hiroaki Y, Smock A, Inukai S, Cone A, Nicholson BJ, Sosinsky GE, Fujiyoshi Y. Asymmetric configurations and N-terminal rearrangements in connexin26 gap junction channels. J Mol Biol. 2011;405:724–735. doi: 10.1016/j.jmb.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J Mol Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 36.Hessa T, White SH, von Heijne G. Membrane insertion of a potassium-channel voltage sensor. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 37.Bond PJ, Wee CL, Sansom MS. Coarse-grained molecular dynamics simulations of the energetics of helix insertion into a lipid bilayer. Biochemistry. 2008;47:11321–11331. doi: 10.1021/bi800642m. [DOI] [PubMed] [Google Scholar]
- 38.Gray TM, Matthews BW. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J Mol Biol. 1984;175:75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- 39.Adamian L, Nanda V, DeGrado WF, Liang J. Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins. 2005;59:496–509. doi: 10.1002/prot.20456. [DOI] [PubMed] [Google Scholar]
- 40.Parton DL, Klingelhoefer JW, Sansom MS. Aggregation of model membrane proteins, modulated by hydrophobic mismatch, membrane curvature, and protein class. Biophys J. 2011;101:691–699. doi: 10.1016/j.bpj.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer LV, de Jong DH, Holt A, Rzepiela AJ, de Vries AH, Poolman B, Killian JA, Marrink SJ. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc Natl Acad Sci U S A. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Planque MR, Greathouse DV, Koeppe RE, 2nd, Schafer H, Marsh D, Killian JA. Influence of lipid/peptide hydrophobic mismatch on the thickness of diacylphosphatidylcholine bilayers. A 2H NMR and ESR study using designed transmembrane alpha-helical peptides and gramicidin A. Biochemistry. 1998;37:9333–9345. doi: 10.1021/bi980233r. [DOI] [PubMed] [Google Scholar]
- 43.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 44.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantor RS. Lipid composition and the lateral pressure profile in bilayers. Biophys J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-Induced Curvature in a Water-Insoluble HIV-1 Env Peptide Revealed by NMR Dipolar Coupling Measurement in Stretched Polyacrylamide Gel. J. Am. Chem. Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 47.Kalyanasundaram K, Thomas JK. Environmental Effects on Vibronic Band Intensities in Pyrene Monomer Flourescence and Their Application in Studies of Micellar Systems. J Am Chem Soc. 1977;99:2039–2044. [Google Scholar]
- 48.Fendler JH, Fendler EJ. Catalysis in Micellar and Macromolecular Systems. Academic Press; New York: 1975. [Google Scholar]
- 49.Podo F, Ray A, Nemethy G. Structure and Hydration of Nonionic Detergent Micelles. High Resolution Nucelar Magnetic Resonance Study. J Am Chem Soc. 1973;95:6164–6171. [Google Scholar]
- 50.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc Natl Acad Sci U S A. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cevc G, Marsh D. Phospholipid Bilayers: Physical Principles and Models. Vol. 5. John Wiley & Sons; New York: 1987. [Google Scholar]
- 52.Higman VA, Varga K, Aslimovska L, Judge PJ, Sperling LJ, Rienstra CM, Watts A. The Conformation of Bacteriorhodopsin Loops in Purple Membranes Resolved by Solid-State MAS NMR Spectroscopy. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201100730. [DOI] [PubMed] [Google Scholar]
- 53.Maslennikov I, Klammt C, Hwang E, Kefala G, Okamura M, Esquivies L, Mors K, Glaubitz C, Kwiatkowski W, Jeon YH, Choe S. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. Proc Natl Acad Sci U S A. 2010;107:10902–10907. doi: 10.1073/pnas.1001656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferguson AD, McKeever BM, Xu S, Wisniewski D, Miller DK, Yamin TT, Spencer RH, Chu L, Ujjainwalla F, Cunningham BR, Evans JF, Becker JW. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science. 2007;317:510–512. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 55.Jegerschold C, Pawelzik SC, Purhonen P, Bhakat P, Gheorghe KR, Gyobu N, Mitsuoka K, Morgenstern R, Jakobsson PJ, Hebert H. Structural basis for induced formation of the inflammatory mediator prostaglandin E2. Proc Natl Acad Sci U S A. 2008;105:11110–11115. doi: 10.1073/pnas.0802894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 57.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Heijne G. Membrane proteins: the amino acid composition of membrane-penetrating segments. Eur J Biochem. 1981;120:275–278. doi: 10.1111/j.1432-1033.1981.tb05700.x. [DOI] [PubMed] [Google Scholar]
- 59.O'Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 60.Lyu PC, Liff MI, Marky LA, Kallenbach NR. Side chain contributions to the stability of alpha-helical structure in peptides. Science. 1990;250:669–673. doi: 10.1126/science.2237416. [DOI] [PubMed] [Google Scholar]
- 61.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chou PY, Fasman GD. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 63.Senes A, Ubarretxena-Belandia I, Engelman DM. The Calpha --- H...O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci U S A. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grottesi A, Domene C, Hall B, Sansom MS. Conformational dynamics of M2 helices in KirBac channels: helix flexibility in relation to gating via molecular dynamics simulations. Biochemistry. 2005;44:14586–14594. doi: 10.1021/bi0510429. [DOI] [PubMed] [Google Scholar]
- 65.Yi M, Cross TA, Zhou HX. Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc Natl Acad Sci U S A. 2009;106:13311–13316. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 67.Xu F, Wang A, Vaughn JB, Jr., Cross TA. A Catalytic Role for Protic Solvents in Conformational Interconversion. J. Am. Chem. Soc. 1996;118:9176–9177. [Google Scholar]
- 68.Xu F, Cross TA. Water: foldase activity in catalyzing polypeptide conformational rearrangements. Proc Natl Acad Sci U S A. 1999;96:9057–9061. doi: 10.1073/pnas.96.16.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brewer ML, Schmitt UW, Voth GA. The formation and dynamics of proton wires in channel environments. Biophys J. 2001;80:1691–1702. doi: 10.1016/S0006-3495(01)76140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pomès RR, B. Structure and dynamics of a proton wire: A theoretical study of H+ translocation along the single-file water chain in the gramicidin A channel. Biophys. J. 1996;71:19–39. doi: 10.1016/S0006-3495(96)79211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Javadpour MM, Eilers M, Groesbeek M, Smith SO. Helix packing in polytopic membrane proteins: Role of glycine in transmembrane helix association. Biophys. J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacKenzie KR, Prestegard JH, Engelman DM. A Transmembrane Helix Dimer: Structure and Implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 73.Yohannan S, Faham S, Yang D, Grosfeld D, Chamberlain AK, Bowie JU. A C alpha-H...O hydrogen bond in a membrane protein is not stabilizing. J Am Chem Soc. 2004;126:2284–2285. doi: 10.1021/ja0317574. [DOI] [PubMed] [Google Scholar]
- 74.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 75.Melnyk RA, Kim S, Curran AR, Engelman DM, Bowie JU, Deber CM. The affinity of GXXXG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J Biol Chem. 2004;279:16591–16597. doi: 10.1074/jbc.M313936200. [DOI] [PubMed] [Google Scholar]
- 76.Kleiger G, Grothe R, Mallick P, Eisenberg D. GXXXG and AXXXA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry. 2002;41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- 77.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 78.Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc Natl Acad Sci U S A. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wigley WC, Corboy MJ, Cutler TD, Thibodeau PH, Oldan J, Lee MG, Rizo J, Hunt JF, Thomas PJ. A protein sequence that can encode native structure by disfavoring alternate conformations. Nat Struct Biol. 2002;9:381–388. doi: 10.1038/nsb784. [DOI] [PubMed] [Google Scholar]
- 80.Deber CM, Therien AG. Putting the beta-breaks on membrane protein misfolding. Nat Struct Biol. 2002;9:318–319. doi: 10.1038/nsb0502-318. [DOI] [PubMed] [Google Scholar]
- 81.Rodrigues ML, Oliveira TF, Pereira IA, Archer M. X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J. 2006;25:5951–5960. doi: 10.1038/sj.emboj.7601439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jayaram H, Robertson JL, Wu F, Williams C, Miller C. Structure of a slow CLC Cl/H+ antiporter from a cyanobacterium. Biochemistry. 2011;50:788–794. doi: 10.1021/bi1019258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.