Abstract
Dairy siphovirus φLmd1, which infects starter culture isolate Leuconostoc mesenteroides subsp. dextranicum A1, showed resistance to pasteurization and was able to grow on 3 of the 4 commercial starter cultures tested. Its 26,201-bp genome was similar to that of Leuconostoc phage of vegetable origin but not to those of dairy phages infecting Lactococcus.
TEXT
Bacteria of the genus Leuconostoc are incorporated into dairy starter cultures due to their ability to produce important metabolites such as diacetyl and CO2 from citric acid (6, 9). Diacetyl is the primary source of aroma and flavor compounds in a variety of fermented milk products, including buttermilk, butter, quarg, and various cheese types (6). Leuconostocs are important flavor producers in l-type and dl-type mesophilic starter cultures, in the latter case, together with Lactococcus lactis subsp. lactis biovar. diacetylactis. The different leuconostocs associated with dairy starters include L. mesenteroides subsp. cremoris, L. mesenteroides subsp. dextranicum, L. lactis, and L. pseudomesenteroides (5, 10).
Bacteriophages negatively affect dairy fermentations by inhibiting the growth of key lactic acid bacteria (LAB). Bacteriophages infecting Lactococcus have been extensively studied for decades due to their dramatic effect on milk acidification rates (24). Lactococcal phages are ubiquitous in dairy environments (26, 28, 32), and it has been shown that phages resident in the dairy plant are responsible for killing lactococcal starter bacteria early in the fermentation (18). Before phages become dairy residents, they are likely to enter dairies through contaminated milk (18, 20), and since natural habitats of Leuconostoc include green vegetation and silage (30), a similar route of entry is likely for Leuconostoc phages. Atamer and coworkers studied the thermal resistance of 77 Leuconostoc phages and found that commonly applied pasteurization conditions were insufficient to ensure complete inactivation of dairy Leuconostoc phages (5). Accordingly, Leuconostoc phages have been shown to be widely distributed in dairy products (5, 29). Phages infecting dairy leuconostocs have previously been characterized (5, 11), and the genome sequences of a virulent L. mesenteroides phage (φ1-A4) and a temperate L. pseudomesenteroides phage (φMH1), both isolated from vegetable fermentation, have been previously characterized (17, 19).
Knowledge on bacteriophages infecting dairy starter cultures is important for the continued improvement of phage countermeasures. In this study, we analyzed the complete genomic sequence of a Leuconostoc phage isolated from a Norwegian dairy producing Dutch-type cheese and characterized the phage with respect to its ability to affect dairy fermentation. Genomes of Leuconostoc phages from vegetable fermentations have been previously described but none from dairy fermentations.
Host strain L. mesenteroides subsp. dextranicum A1.
The host bacterium, L. mesenteroides subsp. dextranicum A1, was isolated from a commercial mixed mesophilic dl starter culture commonly employed in the industrial production of cultured butter and various cheese types. The bacterium was grown at 30°C in MRS (Oxoid, Baskingstoke, United Kingdom). The partial 16S rRNA gene sequence of isolate A1 (corresponding to positions 55 to 1387 in the Escherichia coli 16S rRNA gene) was 100.0% identical to that of leuconostocs belonging to ribospecies CHCC 2114 (27). Strains of this ribospecies have repeatedly been isolated from fermented dairy products and have been assigned to both L. mesenteroides and L. pseudomesenteroides species (27). The API50 CHL (bioMérieux, Lyon, France) sugar fermentation pattern of the host bacterium (acid production from d-ribose, d-galactose, d-glucose, d-fructose, d-mannose, methyl-α-d-glucopyranoside, N-acetylglucosamine, salicin, d-cellobiose, d-maltose, d-lactose, d-melibiose, sucrose, d-trehalose, d-raffinose, starch, gentibiose, and d-turanose) as well as its ability to grow in 6.5% NaCl were in best accordance with L. mesenteroides subsp. dextranicum (6, 13, 14). In the following, the host strain name is shortened to L. mesenteroides A1.
Bacteriophage isolation and characterization.
Bacteriophage Lmd1 was isolated from brine used in the production of Dutch-type cheese in a Norwegian dairy. Phage isolation and quantification were done by standard plaque assays performed in MRS soft agar supplemented with 5 mM CaCl2 (MRS-C). Before phage assays, the brine sample was dialyzed against TM buffer (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10 mM MgCl2, 10 mM CaCl2).
Bacteriophage Lmd1 belongs to the Siphoviridae family of tailed phages and is of the B1 morphotype (1, 2) (Fig. 1). It has a capsid diameter of 41 nm and a tail measuring 115 by 10 nm. The tail consists of 30 or 31 segments, and a distinct baseplate can be observed at the tail tip. The B1 morphotype is the most frequently encountered morphotype among the described Leuconostoc phages and also among dairy phages infecting Lactococcus lactis (2). φLmd1 produced large clear plaques on L. mesenteroides A1 lawns and had an average burst size (16) of about 50. Lysis was completed 30 min after adsorption. Thermal inactivation studies on φLmd1 revealed that the phage is unaffected by pasteurization, but its titer was reduced by more than 7 log upon exposure to a thermal inactivation scheme resembling commonly employed bulk starter vat sterilization schemes (96°C, 30 min). This was in accordance with thermal resistance of other Leuconostoc phages (5). Since pasteurization does not affect φLmd1, there is no barrier for the bacteriophage to enter cheese fermentation vats through contaminated milk. Entry into bulk starter vats would, however, require contamination during or after bulk starter milk cooling.
Fig 1.
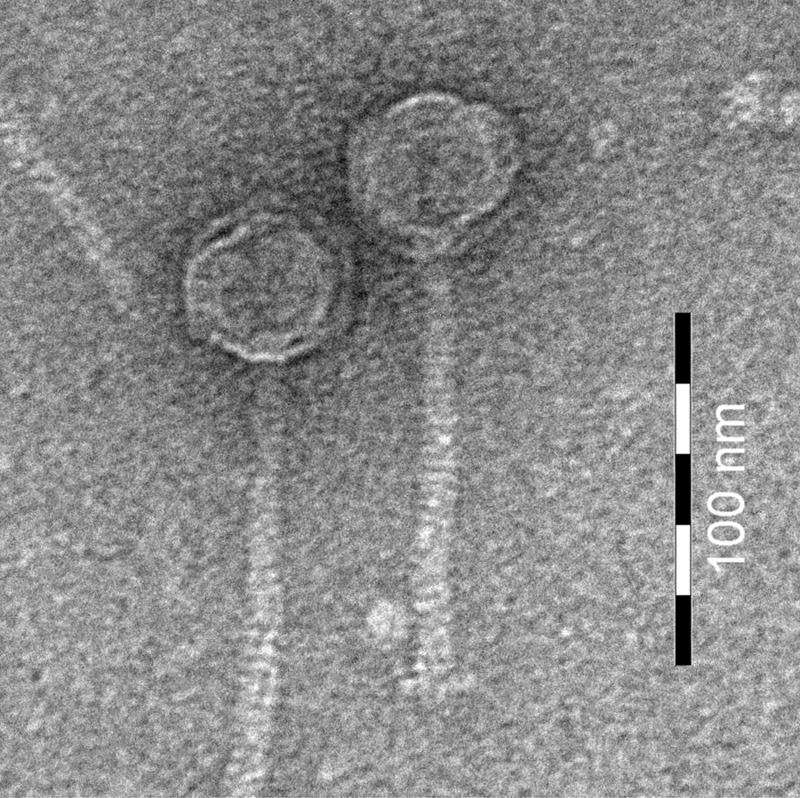
Transmission electron micrograph of φLmd1. Phage particles were purified on CsCl gradients according to Boulanger (7), negatively stained with 2% (wt/vol) uranyl acetate on a carbon-Formvar membrane grid, and examined on a FEI Morgagni 268 (FEI Company B.V., Eindhoven, The Netherlands) microscope at an accelerating voltage of 100 kV. Bar, 100 nm.
Many dairies practice rotation of different phage-unrelated starter cultures in order to reduce the impact of bacteriophages (26). We tested the ability of φLmd1 to multiply on 4 commercial starter cultures commonly used in the production of Dutch-type cheese and found that 3 of the 4 starters contained hosts for φLmd1 proliferation (see Fig. S1 in the supplemental material). This finding emphasizes the importance of assaying for Leuconostoc phages during selection of starter cultures for rotation schemes.
The genome of φLmd1.
The sequence of the 26,201-bp linear genome of Leuconostoc phage Lmd1 was found by a combination of shotgun sequencing and primer walking. Briefly, genomic DNA was isolated from purified phage particles (7) by standard phenol-chloroform extraction, and a shotgun library was prepared after partial digestion with AluI. Sequencing was performed using BigDye 3.1 chemistry (Applied Biosystems, Foster City, CA), and sequence assembly and analysis were done using CLC Main Workbench version 6.5 (CLC bio, Aarhus, Denmark). Homology searches were done using BLASTP and PSI-BLAST build 2.2.26+ (3, 4), and conserved domains were found by searching the Conserved Domains Database (21–23) (June 2012).
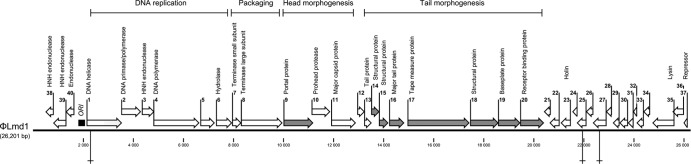
Cohesive genome ends (23 bp; 5′-TCGTGCAATAGTAGGCGTTTTAA-3′) were identified by restriction analysis and sequencing as described by others (8, 19). The G+C content of the φLmd1 genome is 36.4%. A putative origin of replication (ori) was found between positions 1639 and 1873. This region comprises an A-T-rich region and multiple repeats and hairpin structures typical of phage replication origins (33). Forty open reading frames (ORFs) were predicted using Prodigal (15). These constitute 91.7% of the genomic sequence. Starting with the ORF immediately downstream of ori, ORFs were given consecutive numbers (Fig. 2). By homology searches, putative functions were assigned to 24 ORFs. Eight proteins, ORF9 and ORF14 through 20, were identified as structural proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry, performed essentially as described elsewhere (25) (Fig. 2; see also Fig. S2 in the supplemental material). Similar to Leuconostoc phage 1-A4 (19), none of the major protein bands seen by SDS-PAGE were identified as the in silico-predicted major capsid protein. The function of this protein remains to be elucidated. Predicted ribosomal binding sites, start codons, and putative gene functions are shown in Table S1 in the supplemental material. As with most characterized bacteriophage genomes, the genome of φLmd1 is organized in functional modules. Four modules are clearly identifiable: the DNA replication module, the DNA packaging module, and the head and tail morphogenesis modules (Fig. 2).
Fig 2.
Genome map of φLmd1. Positions of the predicted open reading frames are indicated by arrows. Putative functions and functional modules are indicated above. Structural proteins identified by mass spectrometry in this study are indicated by gray arrows. The putative origin of replication is indicated by a black square, and the three EcoRI recognition sites used in the cos site analysis are marked by crosses. The scale bar marks genome positions at 2,000-bp intervals.
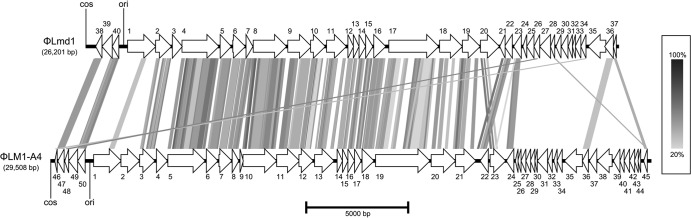
The genome of φLmd1 closely resembles that of L. mesenteroides phage 1-A4 (19), with respect to both sequence similarity and genome organization (Fig. 3). Through a functional distribution analysis, Lu and coworkers showed that Leuconostoc phage 1-A4 clusters most closely with several lactococcal phages, including Q54-like, c2-like, and 936-like phages (12), but they suggested that φ1-A4 should form a separate functional cluster based on the relatively large distance between it and its closest relatives (19). This is in agreement with the low number of significant BLAST hits we found to phage sequences other than φ1-A4.
Fig 3.
Genome comparison between Leuconostoc mesenteroides phages Lmd1 and 1-A4 (GenBank accession no. GQ451696). ORFs are indicated by numbered arrows. Gray connecting lines between ORFs indicate identities. Light gray indicates 20% identities and black lines 100%, according to the grayscale bar on the right. For details of BLASTP scores between ORFs, see Table S1 in the supplemental material. The locations of putative origins of replication (ori) and cos sites (cos) are indicated. The scale bar below indicates 5,000 bp. Genome comparison was carried out using Easyfig software version 1.2.1 (31) with the following cutoff settings: minimum alignment length = 20, and maximum tblastx e-value = 0.001. This corresponded to a minimum sequence identity of 19.23%.
Almost half of the predicted proteins in φLmd1 did not show any similarity to φ1-A4 ORFs (Fig. 3). The dissimilar ORFs were mostly found on the negative strand in both phages, in modules possibly involved in transcription regulation or host interaction. This putative functional assignment is supported by the presence of homologs of conserved Leuconostoc and Weissella prophage genes in this region.
There was generally low sequence similarity at the DNA level between φLmd1 and φ1-A4, even within orfs encoding homologous proteins (not shown). The genome sequence of φLmd1 might thus be useful in the development of DNA-based detection methods for dairy Leuconostoc phages.
Nucleotide sequence accession number.
The complete genome sequence of Leuconostoc phage Lmd1 has been deposited in GenBank under accession number JQ659259.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by TINE SA and The Research Council of Norway.
We are grateful to Morten Skaugen for excellent technical assistance with the mass spectrometry analysis.
Footnotes
Published ahead of print 13 July 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ackermann H-W. 1998. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ackermann HW. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843–857 [DOI] [PubMed] [Google Scholar]
- 3. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altschul SF, et al. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272:5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atamer Z, Ali Y, Neve H, Heller KJ, Hinrichs J. 2011. Thermal resistance of bacteriophages attacking flavour-producing dairy Leuconostoc starter cultures. Int. Dairy J. 21:327–334 [Google Scholar]
- 6. Björkroth J, Holzapfel W. 2006. Genera Leuconostoc, Oenococcus and Weissella, p 267–319 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 7. Boulanger P. 2009. Purification of bacteriophages and SDS-PAGE analysis of phage structural proteins from ghost particles. Methods Mol. Biol. 502:227–238 [DOI] [PubMed] [Google Scholar]
- 8. Casjens SR, Gilcrease EB. 2009. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol. 502:91–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cogan TM, Jordan KN. 1994. Metabolism of Leuconostoc bacteria. J. Dairy Sci. 77:2704–2717 [Google Scholar]
- 10. Daly C. 1983. The use of mesophilic cultures in the dairy industry. Antonie Van Leeuwenhoek 49:297–312 [DOI] [PubMed] [Google Scholar]
- 11. Davey GP, Ward LJH, Brown JCS. 1995. Characterisation of four Leuconostoc bacteriophages isolated from dairy fermentations. FEMS Microbiol. Lett. 128:21–25 [Google Scholar]
- 12. Deveau H, Labrie SJ, Chopin M-C, Moineau S. 2006. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72:4338–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Elliott JA, Facklam RR. 1993. Identification of Leuconostoc spp. by analysis of soluble whole-cell protein patterns. J. Clin. Microbiol. 31:1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Facklam R, Elliott JA. 1995. Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin. Microbiol. Rev. 8:479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119 doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hyman P, Abedon ST. 2009. Practical methods for determining phage growth parameters. Methods Mol. Biol. 501:175–202 [DOI] [PubMed] [Google Scholar]
- 17. Jang S, Hwang M, Chang H-I. 2010. Complete genome sequence of ΦMH1, a Leuconostoc temperate phage. Arch. Virol. 155:1883–1885 [DOI] [PubMed] [Google Scholar]
- 18. Kleppen HP, Bang T, Nes IF, Holo H. 2011. Bacteriophages in milk fermentations: diversity fluctuations of normal and failed fermentations. Int. Dairy J. 21:592–600 [Google Scholar]
- 19. Lu Z, Altermann E, Breidt F, Kozyavkin S. 2010. Sequence analysis of Leuconostoc mesenteroides bacteriophage Φ1-A4 isolated from an industrial vegetable fermentation. Appl. Environ. Microbiol. 76:1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Madera C, Monjardin C, Suarez JE. 2004. Milk contamination and resistance to processing conditions determine the fate of Lactococcus lactis bacteriophages in dairies. Appl. Environ. Microbiol. 70:7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marchler-Bauer A, et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mc Grath S, Fitzgerald GF, van Sinderen D. 2007. Bacteriophages in dairy products: pros and cons. Biotechnol. J. 2:450–455 [DOI] [PubMed] [Google Scholar]
- 25. Mehmeti I, MJönsson Fergestad EM, Mathiesen G, Nes IF, Holo H. 2011. Transcriptome, proteome, and metabolite analyses of a lactate dehydrogenase-negative mutant of Enterococcus faecalis V583. Appl. Environ. Microbiol. 77:2406–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moineau S, Lévesque C. 2005. Control of bacteriophages in industrial fermentations, p 286–296 In Kutter E, Sulakvelidze A. (ed), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL [Google Scholar]
- 27. Olsen KN, Brockmann E, Molin S. 2007. Quantification of Leuconostoc populations in mixed dairy starter cultures using fluorescence in situ hybridization. J. Appl. Microbiol. 103:855–863 [DOI] [PubMed] [Google Scholar]
- 28. Rousseau GM, Moineau S. 2009. Evolution of Lactococcus lactis phages within a cheese factory. Appl. Environ. Microbiol. 75:5336–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shin C, Sato Y. 1979. Isolation of Leuconostoc bacteriophages from dairy products. Jpn. J. Zootechnol. Sci. 50:419–422 [Google Scholar]
- 30. Stirling AC, Whittenbury R. 1963. Sources of the lactic acid bacteria occurring in silage. J. Appl. Microbiol. 26:86–90 [Google Scholar]
- 31. Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verreault D, et al. 2011. Detection of airborne lactococcal bacteriophages in cheese manufacturing plants. Appl. Environ. Microbiol. 77:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weigel C, Seitz H. 2006. Bacteriophage replication modules. FEMS Microbiol. Rev. 30:321–381 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.