Abstract
The Cuatro Ciénegas Basin (CCB) is an oasis in the desert of Mexico characterized by low phosphorus availability and by its great diversity of microbial mats. We compared the metagenomes of two aquatic microbial mats from the CCB with different nutrient limitations. We observed that the red mat was P-limited and dominated by Pseudomonas, while the green mat was N-limited and had higher species richness, with Proteobacteria and Cyanobacteria as the most abundant phyla. From their gene content, we deduced that both mats were very metabolically diverse despite their use of different strategies to cope with their respective environments. The red mat was found to be mostly heterotrophic, while the green mat was more autotrophic. The red mat had a higher number of transporters in general, including transporters of cellobiose and osmoprotectants. We suggest that generalists with plastic genomes dominate the red mat, while specialists with minimal genomes dominate the green mat. Nutrient limitation was a common scenario on the early planet; despite this, biogeochemical cycles were performed, and as a result the planet changed. The metagenomes of microbial mats from the CCB show the different strategies a community can use to cope with oligotrophy and persist. Key Words: Microbial mats—Metagenomics—Metabolism. Astrobiology 12, 648–658.
1. Introduction
Microbial mats are self-sustaining communities with the capacity to perform all major biogeochemical cycles. Microbial mats are characterized by stratification of the microbial populations into distinct layers and are thought to be the earliest biological communities on Earth, as suggested by fossil stromatolites dated to 3.4 billion years ago (Des Marais, 1990; Tice and Lowe, 2004). Mats have inhabited Earth for many years and are found in many different environments, which highlights their great plasticity in adapting to different conditions (Paerl et al., 2000; Kunin et al., 2008; Lau et al., 2009). Their wide range of metabolic capabilities (see Bender and Phillips, 2004) makes them systems of particular biotechnological importance. The geographical distribution of modern microbial mats is currently restricted to only a few aquatic systems, one of which is the Cuatro Ciénegas Basin (CCB), an oasis in the Chihuahuan Desert of Mexico.
The CCB is composed of a system of springs, pools, and streams that form an inverse archipelago in which each pool is an island (Souza et al., 2008). The pools of the CCB exhibit the lowest phosphorous concentration reported in continental waters (Elser et al., 2005). Geological data show that the valley and its hydrological systems have a unique ancient history (Souza et al., 2006; Szynkiewicz et al., 2009). It has been proposed that aquatic communities in the CCB diverged from their marine ancestors in the Jurassic period, when the CCB was under the ocean (Souza et al., 2006; Moreno-Letelier et al., 2011). Paleo-pollen data show that no new soil has settled in the basin floor, and as a consequence no stores of nutrients have been built up (Minckley and Jackson, 2007). New phosphorus (P) arrives by wind transport and dust deposition, and is quickly occluded by the abundant calcium associated with limestone parent material. This long history of nutrient deprivation has resulted in adaptations, such as bacteria with reduced genomes or cell membranes where sulfolipids substitute for phospholipids (Elser et al., 2005; Alcaraz et al., 2008; Desnues et al., 2008; Souza et al., 2008). Paradoxically, this extremely unbalanced ecosystem has a large diversity of microbes, endemic fishes, snails, and zooplankton, and represents one of the most biodiverse sites by area in North America and the most important hot spot of endemicity at all taxa levels (Tatusov et al., 2001; Souza et al., 2006; Carson et al., 2008; Cerritos et al., 2008; Desnues et al., 2008; Escalante et al., 2008, 2009; Breitbart et al., 2009; The Nature Conservancy, 2010; Wilson and Sherman, 2010).
It is still unknown how a nutrient-deprived ecosystem sustained solely by microbial mats can host such complex and diverse communities. To understand the ecological particularities of CCB microbial communities, we describe and compare in the present study the metagenomes of two aquatic microbial mats that live under different conditions within the oasis. A red mat was sampled from a shallow, highly variable desiccation pond with a very low C:N:P ratio (15820:157:1). The second mat, which we will call the green mat, was sampled from a permanent pool with a different C:N:P ratio (51:2:1) and a constant temperature. The two metagenomes show markedly different community structures and display different strategies for contending with oligotrophy.
2. Materials and Methods
2.1. Sampling site
Microbial mats and water samples were collected in July 2008 in the CCB. Sampling site coordinates were 26°52′17″N, 102°01′11.3″W for the red pond in the ejido Los Venados and 26°49′24.4″N, 102°00′53.2″W for the green pool in the Pozas Azules Ranch (PRONATURA). For each microbial mat, four temperature measurements were recorded daily for more than 6 months with a UA-002-64 data logger (Onset, MA, USA).
2.2. Physicochemical analysis
Two hundred milliliters of water were filtered through a 0.45 μm Millipore filter. All carbon (C) forms were determined with a total carbon analyzer (UIC Mod. CM5012; Chicago, IL, USA), and nitrogen (N) and phosphorus (P) forms were determined by colorimetric methods with use of a Bran-Luebbe Auto Analyzer III (Norderstedt, Germany). Total C and inorganic C were determined by combustion and coulometric detection (Huffman, 1977), respectively. Total organic C was calculated as the difference between total C and inorganic C. Total N and P were determined after acid digestion. P was determined by a molybdate-based colorimetric method after reduction with ascorbic acid (Murphy and Riley, 1962), and N was assayed by a macro-Kjeldahl method with colorimetric determination (Bremner and Mulvaney, 1982). The stoichiometric C:N:P ratio was calculated based on the mass (mg/L) of total organic carbon, total organic nitrogen, and total organic phosphorous. Water analyses were performed in CIECO/UNAM.
2.3. DNA isolation and sequencing
The microbial mat was collected with sterile equipment. DNA extraction was performed as described previously (Zhou et al., 1997; Breitbart et al., 2009) by using freeze/thaw, CTAB, and phenol-chloroform extraction. Samples were then further purified by electrodialysis as described by Rodriguez-Mejia et al. (2008). Total DNA was amplified with Genomiphi polymerase (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer's instructions; random hexamers were used as primers. Ten independent 4 h reactions were carried out and then pooled before sequencing to reduce amplification bias. The resulting DNA was purified on silica columns (Qiagen) and concentrated by ethanol precipitation. Approximately 10 μg DNA was sequenced with pyrosequencing technology (454 FLX Roche Diagnostics, IN, USA) at Cinvestav-LANGEBIO, Irapuato, Mexico.
2.4. Data analysis
Each read from the data set was annotated with BlastN and BlastX (Altschul et al., 1990) with a cutoff e-value of 10−5 with the following databases and annotation systems: Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2008), the SEED (Overbeek et al., 2005), NCBI's NR, and the Clusters of Orthologous Groups (COGs) (Szynkiewicz et al., 2009).
The metagenomes of the microbial mats were uploaded and compared against other metagenomes with MG-RAST (Meyer et al., 2008). Whole metagenome data sets were used for functional comparisons; we used a cutoff value of 10−5 and the SEED Subsystems with a minimum identity of 60% in the overall alignment. The data sets employed for the comparisons are publicly available in MG-RAST and have the following accession numbers or names: 4442466.3, 4441363.3, 4440060.4, 4440964.3, 4440963.3, 4440965.3, 4440966.3, 4440967.3, 4440969.3, 4440970.3, 4440968.3, 4440971.3, 4440972.3, 4441576.3, 4441587.3, 4440067.3, 4441124.3, 4441125.3, 4441126.3, 4441127.3, 4441128.3, 4441129.3, 4441130.3, 4441131.3, 4441143.3, 4441144.3, 4441148.3, 4441152.3, 4441153.3, 4441579.3, 4441580.3, 4441582.3, 4441658.3, 4441584.3, 4441590.3, GS001a, GS002, GS003, GS004, GS006, GS009, GS010, GS011, GS012, GS017, GS020, GS038, GS040, GS041, GS042, GS043, GS044, GS045, GS046, GS117b, Guerrero Negro (all samples), PAStromBahamasMic20050722, Río Mesquites Bacteria, and the two metagenomes presented in this work. All abundance data have been normalized to values between 0 and 1. For the statistical analysis, we made use of R version 2.12.1 (x86_64) (R Development Core Team, 2008), and principal component analysis (PCA) was done with the FactoMineR library (Lê et al., 2008). We also conducted a χ2 test for independence across all samples. For the PCA, a normalized matrix, via multiple sample scaling, that contained the abundance of SEED Level 1 functional groups was used as input, as well as a qualitative supplementary variable indicating the sample's origin. PCA clustering was used to determine group formation along with the sample's origin. We chose to cluster the following groups: CC, Guerrero Negro mats, GOS open ocean, and GOS coastal and estuarine samples. One-way ANOVA was performed with the PCA-derived groups, and the significance of this grouping across the gene's functional roles was assessed. A false discovery rate (FDR) (White et al., 2009) p-value was calculated when comparing individual features with 1000 permutations and a significance threshold of 0.05.
3. Results
3.1. Pools
The unstable desiccation red pond (the name of the pond is due to the color of its water) from which the red mat was obtained is characterized by a very low P content (0.6 mg/L total organic phosphorus). The pond's water temperature fluctuates between 10°C and 60°C, with maximum daily fluctuations of 15°C during winter and 45°C in the summer. The water is slightly acidic (pH 5.5), while its conductivity (117.6 μS/cm) reveals a low-salt freshwater similar to that found in the Ice Lake (Minnesota, USA) or the oligotrophic Lakes Superior and Tahoe. This pond also shows an extremely oligotrophic C:N:P ratio of 15820:157:1 (for comparison, the Redfield ratio of 150:15:1 has been found in most aquatic ecosystems).
The permanent green pool is more stable (the water of this pool is green). The temperature was 25°C (±4 standard deviation) across the 6 months in which measurements were taken; the water is also slightly acidic (pH 6.0), and the conductivity is 2.57 μS/cm, which is very low for lentic soft-water bodies and an order of magnitude less than that of the red pond. Even though there are 2 orders of magnitude more P in the green mat than in the red mat, the proportion of N is very low (C:N:P ratio of 51:1.8:1). The green pool is also poor in other elements such as magnesium, sodium, potassium, chloride, and sulfate (the detailed chemical composition of the pools is available as Supplementary Material; Supplementary Data are available online at www.liebertonline.com/ast).
3.2. Metagenomes
The 454 pyrosequencing of the DNA obtained from the red mat yielded 347,728 reads that resulted in a total of 64 Mb of sequence data. The average read length was 226 bp, and the GC content mode was 60%. We identified 991 different ribosomal gene sequences, each of which was assigned to a genus in the SILVA database (Pruesse et al., 2007). The red mat was found to be composed almost exclusively of bacteria (Archaea 0.26%, Eukaryota 1.78%), with the phylum Proteobacteria being predominant, followed by Firmicutes and Cyanobacteria (Table 1). Of the 347,728 total reads, 105,549 were assigned to taxa through the MG-RAST server and use of the RefSeq database (Pruitt et al., 2005; Meyer et al., 2008). Of these assigned reads, 55% belonged to Pseudomonas, which reveals that this gammaproteobacteria was dominant in the mat.
Table 1.
Apparent Taxonomic Distribution of Metagenome Sequences
| Red mat all readsa | Red mat rRNAb | Green mat all readsa | Green mat rRNAb | |
|---|---|---|---|---|
| Bacteria | 97.96 | 93.95 | 95.15 | 92.87 |
| Proteobacteria | 76.52 | 70.33 | 36.27 | 36.48 |
| Cyanobacteria | 11.24 | 1.61 | 18.19 | 7.30 |
| Firmicutes | 4.31 | 10.9 | 12.91 | 19.24 |
| Bacteroidetes | 3.86 | 3.53 | 9.99 | 7.30 |
| Actinobacteria | 0.65 | 1.51 | 2.59 | 1.33 |
| Chloroflexi | 0.45 | 0.1 | 3.75 | 2.16 |
| Planctomycetes | 0.12 | 0 | 1.28 | 1.66 |
| Verrucomicrobia | 0.11 | 0 | 1.75 | 0.33 |
| Chlorobi | 0.1 | 0 | 1.99 | 0.17 |
| Archaea | 0.26 | 0.00 | 2.06 | 0.66 |
| Eukaryota | 1.78 | 6.05 | 2.79 | 6.47 |
| Number of reads | 105,549 | 991 | 94,009 | 603 |
Metagenome affiliation was obtained by using the Metagenome RAST server (Meyer et al., 2008). Only the most common phyla are shown.
Reads identified as LSU and SSU rRNA genes.
The green mat metagenome yielded 427,366 reads with an average size of 202 bp, which resulted in 86 Mb with a GC content mode of 35%. Of these, 94,009 reads were assigned to taxa through MG-RAST (Meyer et al., 2008). In addition, 603 ribosomal genes were identified and assigned to a genus by using the SILVA database (Pruesse et al., 2007). This mat was also composed almost exclusively of bacteria (Archaea 2.06%, Eukaryota 2.79%). In this case, the phyla Proteobacteria and Cyanobacteria were dominant, but no clear dominant taxon was identified (Table 1).
3.3. Red mat metabolic analysis
Functional assignment of the reads was carried out by performing BLAST searches against the KEGG and COGs databases. The COGs annotation (Szynkiewicz et al., 2009) matched 53,485 sequences. These results indicate that 37% of the identified sequences encoded proteins with known metabolic functions. A further 29% were similar to genes involved in cellular processes, while 18% were similar to genes involved in replication, transcription, and translation (Table 2).
Table 2.
Percentage of Metagenome Sequences Similar to Major Metabolism
| COG categorya | Red matb | Green matb | Pc |
|---|---|---|---|
| Information storage and processing | |||
| Translation, ribosomal structure, and biogenesis | 3.89±3.02 | 10.05±1.94 | 2e-300 |
| DNA replication, recombination, and repair | 8.99±3.02 | 10±2.88 | 2e-14 |
| Transcription | 4.77±1.67 | 2.82±2.14 | 1e-92 |
| 17.65 | 22.87 | ||
| Metabolism | |||
| Amino acid transport and metabolism | 10.66±2.85 | 8.78±3.1 | 1e-41 |
| Energy production and conversion | 8.26±2.62 | 7.33±2.77 | 1e-14 |
| Carbohydrate transport and metabolism | 5.41±2.31 | 5.6±2.28 | 0.968 |
| Coenzyme metabolism | 3.88±2.14 | 4.74±1.94 | 2e-20 |
| Nucleotide transport and metabolism | 2.43±1.81 | 3.33±1.55 | 4e-31 |
| Lipid metabolism | 4.52±1.79 | 3.28±2.09 | 2e-42 |
| Secondary metabolites biosynthesis | 1.84±1.07 | 1.14±1.35 | 2e-35 |
| 37 | 34.21 | ||
| Cellular processes | |||
| Cell envelope biogenesis | 5.26±2.8 | 8.48±2.25 | 2e-130 |
| Post-translational modification | 3.56±2.27 | 5.36±1.86 | 1e-71 |
| Inorganic ion transport and metabolism | 7.99±2.03 | 4.26±2.73 | 7e-174 |
| Signal transduction mechanisms | 5.85±2.02 | 4.23±2.36 | 2e-54 |
| Defense | 2.4±1.59 | 2.56±1.54 | 0.989 |
| Cell division and segregation | 0.73±1.28 | 1.65±0.86 | 2e-68 |
| Cell motility | 2.11±1.25 | 1.56±1.44 | 2e-19 |
| Secretion | 0.97±1.15 | 1.33±0.98 | 5e-14 |
| 28.86 | 29.41 | ||
| Poorly characterized | |||
| general function | 9.48±2.81 | 8.51±2.95 | 6e-14 |
| unknown | 6.94±2.17 | 4.89±2.56 | 3e-71 |
| 16.42 | 13.41 | ||
Szynkiewicz et al., 2009.
The given error is the standard error from a FDR analysis.
The P-values were calculated with a two-tailed t test.
By using the KEGG database (Kanehisa et al., 2008), 43,112 sequences were assigned. The sequences were found to correspond to 3228 KEGG unique features (KO, KEGG Orthology) and 211 metabolic pathways. The most abundant pathways were the ABC transporters (8.83%), purine metabolism (6.43%), and the two-component systems (4.59%) (Table 3). Pseudomonas genomes have 111–123 KEGG pathways represented per genome. All the metabolic pathways present in the completely sequenced Pseudomonas genomes were found in the red mat metagenome, in addition to many other pathways such as photosynthesis, glycan biosynthesis, and linoleic acid metabolism. The red mat metagenome also contained pathways for the synthesis and degradation of secondary metabolites that are not present in any known Pseudomonas genome.
Table 3.
Percentage of Red Mat Sequences Showing Homology to Genes Associated with KEGG Pathways (Kanehisa et al., 2008)
| KEGG pathway [path id] | % |
|---|---|
| ABC transporters [PATH:ko02010] | 8.83 |
| Purine metabolism [PATH:ko00230] | 6.43 |
| Two-component system [PATH:ko02020] | 4.59 |
| Oxidative phosphorylation [PATH:ko00190] | 3.83 |
| Glycolysis/Gluconeogenesis [PATH:ko00010] | 3.40 |
| Glycine, serine, and threonine metabolism [PATH:ko00260] | 2.75 |
| Porphyrin and chlorophyll metabolism [PATH:ko00860] | 2.59 |
| Alanine, aspartate, and glutamate metabolism [PATH:ko00250] | 2.30 |
| Arginine and proline metabolism [PATH:ko00330] | 2.29 |
| Aminoacyl-tRNA biosynthesis [PATH:ko00970] | 2.16 |
| Pyrimidine metabolism [PATH:ko00240] | 2.07 |
| Fatty acid metabolism [PATH:ko00071] | 2.07 |
| Cysteine and methionine metabolism [PATH:ko00270] | 1.97 |
| Fructose and mannose metabolism [PATH:ko00051] | 1.87 |
| Citrate cycle (TCA cycle) [PATH:ko00020] | 1.85 |
| Ribosome [PATH:ko03010] | 1.81 |
| Fatty acid biosynthesis [PATH:ko00061] | 1.64 |
| Valine, leucine, and isoleucine biosynthesis [PATH:ko00290] | 1.52 |
| Pentose phosphate pathway [PATH:ko00030] | 1.51 |
| Starch and sucrose metabolism [PATH:ko00500] | 1.39 |
The most frequent paths are shown. The full table is available as Supplementary Data, Table S1 (www.liebertonline.com/ast).
Similarly to Pseudomonas, the red mat metagenome also exhibited many genes involved in metabolic pathways for the degradation of organic compounds. We found genes involved in the degradation of toxic compounds such as toluene, xylene, ethylbenzene, naphthalene, styrene, DDT (insecticide), atrazine (herbicide), dichlorobenzene, hexachlorocyclohexane, tetrachloroethane, dichloroethane, chloroacrylate fluorine, and fluorobenzoate. We also found pathways for producing antibiotics such as tetracycline, penicillin, streptomycin, novomycin, ansamycin, and vancomycin, as well as genes such as those coding for beta-lactamases and metallo-lactamases that indicate ways to resist those antibiotics.
3.4. Green mat metabolic analysis
The green mat metagenome was analyzed in the same way as the red mat, and 63,364 reads were assigned and classified by using the COGs database (Szynkiewicz et al., 2009). Of these reads, 34% corresponded to genes involved in metabolism, 29% to genes involved in cellular processes, and 23% to genes involved in information storage and processing (Table 2). By using the KEGG database (Kanehisa et al., 2008), 106,841 reads were classified by sequence analysis. These corresponded to 228 pathways, 19 of which are not represented in the red mat. Among the pathways absent in the red mat are those involved in signaling pathways and posttranslational modifications in eukaryotes. The most frequent pathways in the green mat are those involved in purine metabolism (6.54%), ABC transport (5.12%), and aminoacyl-tRNA biosynthesis (4.34%) (Table 4).
Table 4.
Percentage of Green Mat Sequences Showing Homology to Genes Associated with KEGG Pathways (Kanehisa et al., 2008)
| KEGG pathway [path id] | % |
|---|---|
| Purine metabolism [PATH:ko00230] | 6.54 |
| ABC transporters [PATH:ko02010] | 5.12 |
| Aminoacyl-tRNA biosynthesis [PATH:ko00970] | 4.34 |
| Oxidative phosphorylation [PATH:ko00190] | 3.89 |
| Glycolysis/Gluconeogenesis [PATH:ko00010] | 3.31 |
| Nucleotide excision repair [PATH:ko03420] | 2.43 |
| Two-component system [PATH:ko02020] | 2.43 |
| Valine, leucine, and isoleucine biosynthesis [PATH:ko00290] | 2.35 |
| Porphyrin and chlorophyll metabolism [PATH:ko00860] | 2.34 |
| Pyrimidine metabolism [PATH:ko00240] | 2.26 |
| Alanine, aspartate, and glutamate metabolism [PATH:ko00250] | 2.12 |
| Ribosome [PATH:ko03010] | 1.95 |
| Glycine, serine, and threonine metabolism [PATH:ko00260] | 1.83 |
| Fructose and mannose metabolism [PATH:ko00051] | 1.72 |
| Peptidoglycan biosynthesis [PATH:ko00550] | 1.61 |
| Protein export [PATH:ko03060] | 1.60 |
| Citrate cycle (TCA cycle) [PATH:ko00020] | 1.58 |
| Homologous recombination [PATH:ko03440] | 1.50 |
| Pentose phosphate pathway [PATH:ko00030] | 1.24 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis [PATH:ko00400] | 1.22 |
The most frequent paths are shown. The full table is available as Supplementary Data, Table S1 (www.liebertonline.com/ast).
3.5. Metagenome comparison
By analyzing the frequency distributions of the COGs categories, we observed that categories involved in translation and cellular envelope biogenesis are more abundant in the green mat, while genes involved in transcriptional regulation and metabolism of inorganic ions are more frequent in the red mat (Table 2). We explored whether gene function differences correlated with genome size. To this end, the effective genome sizes were calculated as described by Raes et al. (2007), and we obtained a genome average size of 3.69 Mb for the red mat and 1.27 Mb for the green mat.
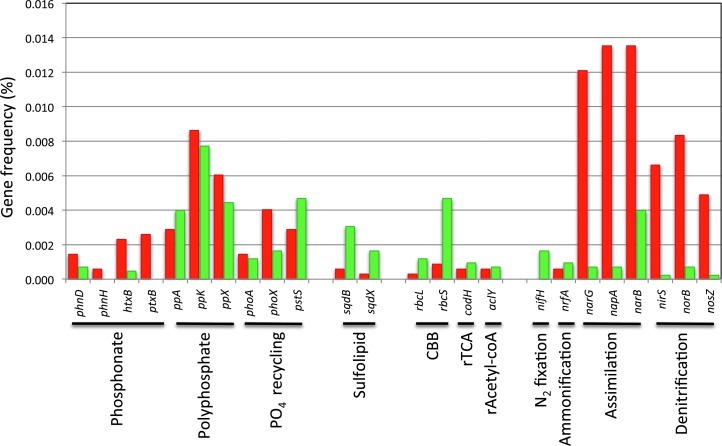
Specific genes were searched for as proxies or indicators of particular biogeochemical cycles. We looked for genes involved in phosphonate utilization (phnD, phnH, htxB, and ptxB), polyphosphate metabolism (ppA, ppK, and ppX), and phosphate recycling (phoA, phoX, and pstS). All these genes were found in both mats, with the exception being genes for phosphite and phosphonate utilization (ptxB and phnH) that were not found in the green mat. Sulfolipid biosynthesis genes sqdB and sqdX, in contrast, were more notable in the green mat. The genes rbcL, rbcS, codH, and aclY, used as markers for major pathways of carbon fixation, were all present in both mats.
Nitrogen analysis was performed by examining the genes nifH, nrfA, narG, napA, narB, nirS, norB, and nosZ. Nitrogen assimilatory pathways were observed to be more abundant in the green mat, with a marked preference for assimilatory nitrate reductases (narB) over their respiratory counterparts (narG, napA, nirS). There is a remarkable preference for iron-containing cytochrome cd1 nitrate reductase (nirS) because no copper-containing dissimilatory nitrate reductase (nirK) was found in either mat. Nitrogen fixation genes were detected only in the green mat (Fig. 1).
FIG. 1.
Frequency of reads corresponding to gene markers for biochemical cycles. Red mat is shown in red bars while green mat is shown in green bars. Genes correspond to the following proteins: phnD and ptxB (phosphonate transporters), phnH and htxB (C-P lyase), ppA (pyrophosphatase), ppK (polyphosphatase kinase), ppX (exopolyphosphatase), phoA and phoX (alkaline phosphatases), pstS (phosphate transporter), rbcL and rbcS (RuBisCO), codH (CO dehydrogenase), aclY (citrate lyase), nifH (nitrogenase reductase), nrfA (nitrite reductase), narG (nitrate reductase), napA (nitrate reductase), narB (nitrate reductase), nirS (nitrite reductase), norB (nitric oxide reductase), and nosZ (nitrous oxide reductase). Color images available online at www.liebertpub.com/ast
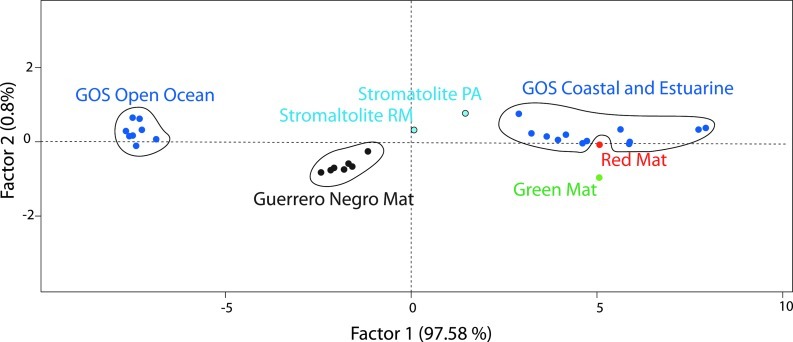
We next compared these two CCB mats against other aquatic microbiomes whose metagenomes are known. We used metagenomes from two stromatolites from the CCB (also isolated from phosphorus-deprived oligotrophic conditions), the hypersaline microbial mat of Guerrero Negro—for which each layer was studied independently—and the Global Ocean Sampling (GOS) Expedition (Rusch et al., 2007; Kunin et al., 2008; Breitbart et al., 2009). We used the SEED subsystems to compare the annotations for each of the analyzed metagenomes by PCA. Three large clusters were formed: (1) GOS open ocean, (2) Guerrero Negro, and (3) GOS coastal and estuarine. The red mat clustered together with the GOS coastal water samples, and the green mat was also close to this cluster. The nearest neighbor of stromatolite PA was stromatolite RM, although these two clustered apart from the rest of the samples. Interestingly, when analyzing differences in gene functions by clustering the groups from the CCB, marine environments, and layers of the Guerrero Negro hypersaline mat, the only significant difference found by PCA (SEED level 1) was in genes of the photosynthesis category (FDR p=0.0374). Within this category, the most differences were found in genes involved in electron transport and photophosphorylation that were more abundant (FDR p=0.0096) in the metagenomes from coastal waters and the CCB than in the metagenomes from the open ocean. Light-harvesting complex-related genes were also more abundant (FDR p=0.0111) in the basal group of the PCA (Fig. 2).
FIG. 2.
Functional comparison of metagenomes from aquatic microbiomes by PCA analysis of SEED Subsystems level 1 (Meyer et al., 2008). Stromatolites PA and RM (Breitbart et al., 2009), Global Ocean Sampling (GOS) (Rusch et al., 2007), Guerrero Negro hypersaline mat (Kunin et al., 2008). The correlation of each variable with factor 1 is available as Supplementary Material, Table S2. Color images available online at www.liebertonline.com/ast
4. Discussion
In this work, we compared the metagenomes of two oligotrophic microbial mats, one a P-limited red mat from a desiccation pond and the other a N-limited green mat from a permanent pool at the CCB. The two mats were collected from locations that are less than 10 km apart, and their pools are immersed in a calcareous environment. Oligotrophic environments are defined by their low nutrient availability; oligotrophic continental water bodies are limited by P (Correll, 1999), while oceans are limited by P, N, and Fe (Mills et al., 2004). In the latter, primary producers such as cyanobacteria can fix atmospheric nitrogen, using nitrogenase. This is not the case with phosphorus because it lacks volatile atmospheric compounds and thus enters into aquatic systems mainly through deposition on surface waters. The C:N:P ratio in both pools is well below the Redfield ratio (106:16:1) required in the ocean to allow phytoplankton growth and sustain ecosystems (Redfield, 1934; Souza et al., 2007).
We found that microbial communities in both the red and green mats are dominated by known heterotrophic taxa, while other previously reported phosphorus-limited microbial communities, such as the Mediterranean Ocean and the Río Mesquites oncolite (stromatolite RM), are dominated by autotrophs (Krom et al., 1991; Breitbart et al., 2009). The oceanic N:P ratio is regulated by nitrogen-fixing organisms. When the N:P ratio decreases, diazotrophs acquire an adaptive advantage due to their ability to obtain new nitrogen. As the ratio increases, rapidly growing heterotrophic organisms displace the high-energy-requiring diazotrophs (Tyrrell, 1999). This fine homeostatic mechanism seems to be disrupted in the CCB, where phosphorus is rapidly mineralized and most of the available phosphorus-containing compounds exist within standing organic biomass. This sequestration of phosphorus explains the abundance of heterotrophic taxa.
As previously mentioned, the pond where the red mat was sampled is small and has large fluctuations in temperature and water volume, with consequent changes in conductivity. The red mat is strongly dominated by the genus Pseudomonas, as 55% of the red mat sequence reads reveal identity to this genus. Pseudomonas are characterized by a large genomic and metabolic plasticity that allows them to survive in many environmental conditions (Mathee et al., 2008; Kümmerli et al., 2009; Klockgether et al., 2011). In contrast, the pool from which the green mat was harvested has much lower temperature fluctuations, an extreme N limitation instead of a P limitation, and a large microbial diversity. When assigning a genus to each read, we observed a greater richness in the green mat (801 genera in the green mat vs. 698 in the red mat). Likewise, we observe a greater evenness in the green mat; Cyanothece is the most abundant genus in the green mat, with a representation of 4.5%. Both richness and evenness indicate that the diversity of the green mat is much greater than the diversity of the red carpet, as is confirmed by the Simpson's index (D), which is 0.0087 for the green mat and 0.2823 for the red mat.
Surprisingly, despite the differences in ecology, nutrient limitation, and species richness and diversity, both mats can perform a wide array of metabolic functions and have almost the same diversity of COGs (i.e., 3125 COGs in the red mat versus 3025 COGs in the green mat). In this work, we studied gene presence, and it should be noted that further gene expression analysis is needed for validation of the metabolic capacities of the mats. The number of metabolic pathways detected in each mat is quite similar (211 for the red mat and 228 for the green mat) (Supplementary Fig. S1). Nevertheless, large differences were observed within the relative frequencies in which different pathways are represented. In the red mat, we observed a higher frequency of ABC transporters and two-component system pathways (with red-mat-to-green-mat ratios of 1.7 and 1.9, respectively; Tables 3 and 4). ABC transporters and two-component system genes are particularly important for organisms to be able to survive in a variety of environmental conditions.
Biogeochemical cycles are of special interest because these pathways indicate how a community incorporates essential elements for later reuse by various mat components. Microbial mats are known to display complete biogeochemical cycles (Canfield and Des Marais, 1993). Here, we determined whether a given biogeochemical cycle was present by analyzing the data for the presence of key proxy or marker genes within the cycle's various pathways.
Many different mechanisms exist to cope with low phosphorus availability, such as using alternative phosphorus sources, using polyphosphates as storage compounds, or employing a highly effective phosphate-recycling mechanism. The use of alternative phosphorus sources (phosphonates, phosphites, and hypophosphites) is revealed by the presence of the high affinity transporters phnD and ptxB as well as by C-P lyase genes phnH and htxB (White and Metcalf, 2004). The genes ppA, ppK, and ppX are also induced under phosphate-limiting conditions; these genes are involved in polyphosphate metabolism and code for pyrophosphatase, polyphosphatase kinase, and exopolyphosphatase, respectively. Polyphosphate acts as a reservoir of intracellular phosphate, a strategy that seems to be particularly important for motility and biofilms (Brown and Kornberg, 2004). Extracellular phosphates are recycled by the overexpression of alkaline phosphatases phoA and phoX as well as by the high-affinity phosphate transporter pstS (Scanlan et al., 1993; Suzuki et al., 2004; Zaheer et al., 2009). All three of the strategies described above are utilized by both mats; however, the use of alternative sources of phosphates appeared to be more important for the red mat because ptxB and phnH were not found in the green mat (Fig. 1). Also exclusive to the red mat was the presence of coding genes for phosphate-binding DING proteins, which may be another resource more typically used by Pseudomonas to deal with phosphate starvation (Berna et al., 2009). We detected in both mats the sqdB and sqdX genes involved in sulfolipid biosynthesis, which is another mechanism to contend with limited phosphate (van Mooy et al., 2004: Alcaraz et al., 2008). Interestingly, despite the low N:P ratio of the green mat, seqB and sqdX abundance is 6 times higher than in the red mat, which suggests that sulfolipid biosynthesis is not induced only by P limitation as expected (Fig. 1).
Autotrophic primary production appears to be more important in the green mat, because the gene frequency of carbon-fixation pathways in the green mat is greater than in the red mat. This is especially true for RuBisCO, which has a frequency 5 times greater in the green mat than in the red mat (Fig. 1). This is consistent with a higher proportion of Cyanobacteria detected in the green mat (Table 1). However, both mats also contain genes for the reductive acetyl-CoA pathway and the reductive tricarboxylic acid (rTCA) cycle, two alternative carbon fixation pathways that are present in a variety of microorganisms (Hugler et al., 2011). The reductive acetyl-CoA pathway is twofold more common in the green mat, as indicated by the frequency of the CO dehydrogenase gene codH. This pathway is present in both archaea and bacteria under reductive and anaerobic conditions (Berg et al., 2010). Citrate lyase is one of the few enzymes unique to the rTCA cycle (Wahlund and Tabita, 1997), a pathway exclusive to anaerobic and microaerophilic bacteria that also uses many of the enzymes involved in the TCA cycle. Citrate lyase (aclY) was observed to be 1.5 times more frequent in the green than in the red mat. The other three known pathways for carbon fixation were not detected in any of the mats. It is noteworthy that the distribution of these routes is much more restricted: the 3-hydroxypropionate (3-HP) bicycle is exclusive to Chloroflexaceae, while the 3-hydroxypropionate/4-hydroxybutyrate (3-HP/4-HB) cycle and the dicarboxylate/4-hydroxybutyrate (DC/4-HB) cycle occur only in Crenarchaeota (Hugler et al., 2011).
Nitrogen metabolism was found to be very different between the two mats. Nitrogen fixation genes were only detected in the green mat, where we also observed a low GC content (GC mode of 35%). This is consistent with the extreme N limitation of the site, as a high GC content requires more nitrogen. The low GC content represents an adaptive advantage in environments with low nitrogen availability (Biers et al., 2009). In contrast, nitrogen cycle genes in the red mat are predominantly involved with nitrate (NO3) assimilation and respiration in agreement with a much higher GC content, which peaks at 60%. These observations also suggest that nitrogen is not a limiting nutrient in the red mat environment.
Transmembrane transporter analysis revealed other relationships between the organisms and their environment (Patel et al., 2010). A larger amount of transporters were found in the red mat, notably the ATP-hydrolyzing ABC transporter family (8.83% in the red mat vs. 5.12% in the green mat). The red mat also has cellobiose transporters, which suggests that it has the capacity to degrade cellulose as a carbon source. In the red mat, the frequency of genes coding for transporters of the osmoprotectants choline and betaine is more than 10 times that of the green mat. This reinforces the observation that the red mat experiences stressful conditions during periods of desiccation. In contrast, there are 8 times more transporters involved in the assembly of Fe-S clusters in the green mat, which confirms the importance of photosynthesis and nitrogen fixation for this mat.
We observed yet more contrasts between the two mats through the analysis of those genes with greater differences in their relative frequencies of appearance (Table 2). In the green mat, there are several genes involved in synthesis and degradation of the cell wall. In particular, carboxypepetidase genes are twice as frequent in the green mat as in the red mat. In contrast, the red mat has 3 times more outer membrane-related genes, such as porin genes. The high frequency of outer membrane proteins in the red mat is consistent with the particularities of Pseudomonas, as they have several of these proteins that help them respond to environmental changes (Remans et al., 2010).
The nucleotide excision repair and mismatch repair (MMR) pathways also showed significant differences, as there are more than twice as many genes from these pathways in the green mat than in the red. Nucleotide excision repair is associated with DNA repair following UV damage (Goosen and Moolenaar, 2008). Both pools receive similar high levels of solar radiation, but the green pool is considerably more translucent than the red pool. MMR is usually involved with DNA repair after replication errors; however, the MMR pathway also participates in repairing damage caused by different types of stress (Kunkel and Erie, 2005). The above data suggest that maintenance of genome stability is more important in the green mat than in the red mat, where the plasticity of the Pseudomonas genomes may allow the community to survive the fluctuating environment.
We estimated the average size of the genomes in these metagenomes, using a method that relies on the relative frequency of 35 genes that are found as a single copy in bacterial genomes (Raes et al., 2007). The estimated size of the genomes in the green mat was 3 times smaller (∼1.27 Mb) than that estimated for the genomes in the red mat (∼3.69 Mb), which suggests very different environmental strategies. The differences in genome size are linked to the overrepresentation of some functional categories, such as translation and replication in the green mat. This phenomenon occurs because different functional categories can be lost during genome streamlining in different organisms, while informational pathways are essential in all organisms and hence will be common to all genomes in the sample. Small genomes, such as those inferred to occur in the green mat, have been reported to be the consequence of genome streamlining as a response to low nitrogen and phosphate availability in some oceanic environments (Giovannoni et al., 2005; Lauro et al., 2009). Small genomes suggest an abundance of specialized bacteria that can only survive in very specific microniches, because they lack the required plasticity to survive in other environments.
In contrast, in the red mat we observed larger genomes with an overrepresentation of COGs involved in energy-dependent transport systems, cell motility, and transcriptional regulation, as well as genes involved in signal transduction that are crucial to sense and respond to changing environmental conditions (Konstantinidis and Tiedje, 2004). All these genomic features are characteristic of copiotrophic lifestyles (Lauro et al., 2009), in which organisms are not dependent on carbon and nitrogen fixation and heterotrophic organisms predominate. These features also suggest that, despite the extremely biased Redfield ratio, the microbial community from the red mat is not nutrient limited because it is slowly consuming the already fixed nutrient-rich standing biomass. In contrast, in the oligotrophic green mat community, N limitation results in smaller genomes, an abundance of carbon- and nitrogen-fixing pathways, a high abundance of COGs involved in DNA repair, and a lower abundance of COGs involved in energy-dependent transport systems.
5. Conclusions
Thus we can conclude that, although both mat communities exist under the enormous environmental pressure that is nutrient deprivation, they seem to cope with it in very different ways, which suggests the existence of a wide array of strategies to survive in low-nutrient environments. The green mat represents a highly structured and fractioned niche inhabited by highly specialized bacteria (Diamond, 1975), while the red mat follows a “Red Queen” model, in which the plastic Pseudomonas must continuously change their strategy to maintain their dominance in an ever-changing environment (Table 5).
Table 5.
Summary of the Differences between the Red and Green Mat
| Red mat | Green mat | |
|---|---|---|
| Pool | Shallow pond | Permanent pool |
| Nutrient limitation | Phosphorus | Nitrogen |
| Richness | 698 genera | 801 genera |
| Evenness (most abundant genus) | Pseudomonas 55% | Cyanothece 4.5% |
| Simpson index (D) | 0.2823 | 0.0087 |
| Gene presence | More transporters | More photosynthesis |
| More transcriptional regulators | More DNA repair | |
| Nitrogen fixation | ||
| Average genome size | 3.69 Mb | 1.27 Mb |
| Strategy | Generalist bacteria | Specialized bacteria |
Finally, if we compare the functions inferred for CCB metagenomes with those from the GOS (Rusch et al., 2007) and Guerrero Negro (Kunin et al., 2008) by PCA, we observe that metabolic differences in such dissimilar microbiomes can be explained almost entirely by the photosynthesis category. This suggests that the functional differences between aquatic microbiomes are subtle and should be studied in detail rather than by large functional categories. Moreover, this relatively low functional diversity invites us to think that, in these ancient communities, the function is what is being selected for rather than the species composition. Detailed community ecology and experimental evolution studies are needed in order to explain how the CCB microbial communities, which are dissimilar species assemblies analogous to ancient life on Earth, perform the same functions.
Supplementary Material
Acknowledgments
We thank Rodrigo González Chauvet for extraordinary technical logistics and field assistance. We also thank Africa Islas, Varinia Lopez, Frederique Reverchon, Eria Rebollar, Morena Avitia, Ana Gutierrez for assistance in DNA isolation; Celeste Martinez-Piedragil from CIECO/UNAM for assistance in the chemical analyses; Laura Espinosa from IE/UNAM for laboratory and technical assistance. This work was done with grants CONACyT 057507 and SEMARNAT 2006-C01-23459 to V.S. The manuscript was done while V.S. and L.E.E. were on sabbatical in UCI with support of DGAPA, UNAM, and UC-Mexus-CONACYT. G.B.R. was supported by CONACYT scholarship 196814 and Programa de Posgrado en Ciencias Biomedicas UNAM, and M.P. had a postdoctoral salary from CONACyT 057507.
Author Disclosure Statement
No competing financial interests exist.
Abbreviations
CCB, Cuatro Ciénegas Basin; COGs, Clusters of Orthologous Groups; FDR, false discovery rate; GOS, Global Ocean Sampling; KEGG, Kyoto Encyclopedia of Genes and Genomes; MMR, mismatch repair; PCA, principal component analysis.
References
- Alcaraz L.D. Olmedo G. Bonilla G. Cerritos R. Hernandez G. Cruz A. Ramirez E. Putonti C. Jimenez B. Martinez E. Lopez V. Arvizu J.L. Ayala F. Razo F. Caballero J. Siefert J. Eguiarte L. Vielle J.P. Martinez O. Souza V. Herrera-Estrella A. Herrera-Estrella L. The genome of Bacillus coahuilensis reveals adaptations essential for survival in the relic of an ancient marine environment. Proc Natl Acad Sci USA. 2008;105:5803–5808. doi: 10.1073/pnas.0800981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F. Gish W. Miller W. Myers E.W. Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bender J. Phillips P. Microbial mats for multiple applications in aquaculture and bioremediation. Bioresour Technol. 2004;94:229–238. doi: 10.1016/j.biortech.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Berg I.A. Ramos-Vera W.H. Petri A. Huber H. Fuchs G. Study of the distribution of autotrophic CO2 fixation cycles in Crenarchaeota. Microbiology. 2010;156:256–269. doi: 10.1099/mic.0.034298-0. [DOI] [PubMed] [Google Scholar]
- Berna A. Scott K. Chabriere E. Bernier F. The DING family of proteins: ubiquitous in eukaryotes, but where are the genes? Bioessays. 2009;31:570–580. doi: 10.1002/bies.200800174. [DOI] [PubMed] [Google Scholar]
- Biers E.J. Sun S.L. Howard E.C. Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Appl Environ Microbiol. 2009;75:2221–2229. doi: 10.1128/AEM.02118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M. Hoare A. Nitti A. Siefert J. Haynes M. Dinsdale E. Edwards R. Souza V. Rohwer F. Hollander D. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Cienegas, Mexico. Environ Microbiol. 2009;11:16–34. doi: 10.1111/j.1462-2920.2008.01725.x. [DOI] [PubMed] [Google Scholar]
- Bremner J.M. Mulvaney C.S. Nitrogen-total. In: Page A.L., editor; Miller R.H., editor; Keeney D.R., editor. Methods of Soil Analysis: Chemical and Microbiological Properties. American Society of Agronomy and Soil Science Society of America; Madison, WI: 1982. pp. 595–624. [Google Scholar]
- Brown M.R.W. Kornberg A. Inorganic polyphosphate in the origin and survival of species. Proc Natl Acad Sci USA. 2004;101:16085–16087. doi: 10.1073/pnas.0406909101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield D.E. Des Marais D.J. Biogeochemical cycles of carbon, sulfur, and free oxygen in a microbial mat. Geochim Cosmochim Acta. 1993;57:3971–3984. doi: 10.1016/0016-7037(93)90347-y. [DOI] [PubMed] [Google Scholar]
- Carson E.W. Elser J.J. Dowling T.E. Importance of exogenous selection in a fish hybrid zone: insights from reciprocal transplant experiments. Copeia. 2008;4:794–800. [Google Scholar]
- Cerritos R. Vinuesa P. Eguiarte L.E. Herrera-Estrella L. Alcaraz-Peraza L.D. Arvizu-Gomez J.L. Olmedo G. Ramirez E. Siefert J.L. Souza V. Bacillus coahuilensis sp. nov., a moderately halophilic species from a desiccation lagoon in the Cuatro Cienegas Valley in Coahuila, Mexico. Int J Syst Evol Microbiol. 2008;58:919–923. doi: 10.1099/ijs.0.64959-0. [DOI] [PubMed] [Google Scholar]
- Correll D.L. Phosphorus: a rate limiting nutrient in surface waters. Poult Sci. 1999;78:674–682. doi: 10.1093/ps/78.5.674. [DOI] [PubMed] [Google Scholar]
- Des Marais D.J. Microbial mats and the early evolution of life. Trends Ecol Evol. 1990;5:140–144. doi: 10.1016/0169-5347(90)90219-4. [DOI] [PubMed] [Google Scholar]
- Desnues C. Rodriguez-Brito B. Rayhawk S. Kelley S. Tran T. Haynes M. Liu H. Furlan M. Wegley L. Chau B. Ruan Y. Hall D. Angly F.E. Edwards R.A. Li L. Thurber R.V. Reid R.P. Siefert J. Souza V. Valentine D.L. Swan B.K. Breitbart M. Rohwer F. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- Diamond J.M. Assembly of species communities. In: Cody M.L., editor; Diamond J.M., editor. Ecology and Evolution of Communities. The Belknap Press of Harvard University Press; Cambridge, MA: 1975. pp. 342–444. [Google Scholar]
- Elser J.J. Schampel J.H. Garcia-Pichel F. Wade B.D. Souza V. Eguiarte L. Escalante A.E. Farmer J.D. Effects of phosphorus enrichment and grazing snails on modern stromatolitic microbial communities. Freshw Biol. 2005;50:1808–1825. [Google Scholar]
- Escalante A.E. Eguiarte L.E. Espinosa-Asuar L. Forney L.J. Noguez A.M. Souza Saldivar V. Diversity of aquatic prokaryotic communities in the Cuatro Cienegas basin. FEMS Microbiol Ecol. 2008;65:50–60. doi: 10.1111/j.1574-6941.2008.00496.x. [DOI] [PubMed] [Google Scholar]
- Escalante A.E. Caballero-Mellado J. Martinez-Aguilar L. Rodriguez-Verdugo A. Gonzalez-Gonzalez A. Toribio-Jimenez J. Souza V. Pseudomonas cuatrocienegasensis sp. nov., isolated from an evaporating lagoon in the Cuatro Cienegas valley in Coahuila, Mexico. Int J Syst Evol Microbiol. 2009;59:1416–1420. doi: 10.1099/ijs.0.006189-0. [DOI] [PubMed] [Google Scholar]
- Giovannoni S.J. Tripp H.J. Givan S. Podar M. Vergin K.L. Baptista D. Bibbs L. Eads J. Richardson T.H. Noordewier M. Rappe M.S. Short J.M. Carrington J.C. Mathur E.J. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Goosen N. Moolenaar G.F. Repair of UV damage in bacteria. DNA Repair (Amst) 2008;7:353–379. doi: 10.1016/j.dnarep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Huffman E.W.D. Performance of a new automatic carbon dioxide coulometer. Microchem J. 1977;22:567–573. [Google Scholar]
- Hugler M. Petersen J.M. Dubilier N. Imhoff J.F. Sievert S.M. Pathways of carbon and energy metabolism of the epibiotic community associated with the deep-sea hydrothermal vent shrimp Rimicaris exoculata. PLoS One. 2011;6:e1601810.1371. doi: 10.1371/journal.pone.0016018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Araki M. Goto S. Hattori M. Hirakawa M. Itoh M. Katayama T. Kawashima S. Okuda S. Tokimatsu T. Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J. Cramer N. Wiehlmann L. Davenport C.F. Tummler B. Pseudomonas aeruginosa genomic structure and diversity. Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K.T. Tiedje J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom M.D. Kress N. Brenner S. Phosphorus limitation of primary productivity in the E. Mediterranean sea. Limnol Oceanogr. 1991;36:424–432. [Google Scholar]
- Kümmerli R. Jiricny N. Clarke L.S. West S.A. Griffin A.S. Phenotypic plasticity of a cooperative behaviour in bacteria. J Evol Biol. 2009;22:589–598. doi: 10.1111/j.1420-9101.2008.01666.x. [DOI] [PubMed] [Google Scholar]
- Kunin V. Raes J. Harris J.K. Spear J.R. Walker J.J. Ivanova N. von Mering C. Bebout B.M. Pace N.R. Bork P. Hugenholtz P. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4 doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T.A. Erie D.A. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lau M.C. Aitchison J.C. Pointing S.B. Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles. 2009;13:139–149. doi: 10.1007/s00792-008-0205-3. [DOI] [PubMed] [Google Scholar]
- Lauro F.M. McDougald D. Thomas T. Williams T.J. Egan S. Rice S. DeMaere M.Z. Ting L. Ertan H. Johnson J. Ferriera S. Lapidus A. Anderson I. Kyrpides N. Munk A.C. Detter C. Han C.S. Brown M.V. Robb F.T. Kjelleberg S. Cavicchioli R. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S. Josse J. Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25:1–18. [Google Scholar]
- Letunic I. Yamada T. Kanehisa M. Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem Sci. 2008;33:101–103. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Mathee K. Narasimhan G. Valdes C. Qiu X. Matewish J.M. Koehrsen M. Rokas A. Yandava C.N. Engels R. Zeng E. Olavarietta R. Doud M. Smith R.S. Montgomery P. White J.R. Godfrey P.A. Kodira C. Birren B. Galagan J.E. Lory S. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci USA. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. Paarmann D. D'Souza M. Olson R. Glass E.M. Kubal M. Paczian T. Rodriguez A. Stevens R. Wilke A. Wilkening J. Edwards R.A. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M.M. Ridame C. Davey M. La Roche J. Geider R.J. Iron and phosphorus co-limit nitrogen fixation in the eastern tropical North Atlantic. Nature. 2004;429:292–294. doi: 10.1038/nature02550. [DOI] [PubMed] [Google Scholar]
- Minckley T. Jackson S. Ecological stability in a changing world? Reassessment of the palaeo-environmental history of Cuatrociénegas, Mexico. J Biogeogr. 2007;35:188–190. [Google Scholar]
- Moreno-Letelier A. Olmedo G. Eguiarte L.E. Martinez-Castilla L. Souza V. Parallel evolution and horizontal gene transfer of the pst operon in Firmicutes from oligotrophic environments. Int J Evol Biol. 2011;2011 doi: 10.4061/2011/781642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. Riley J.P. A modified single solution method for the determination of phosphorus in natural water. Anal Chim Acta. 1962;27:31–36. [Google Scholar]
- Overbeek R. Begley T. Butler R.M. Choudhuri J.V. Chuang H.Y. Cohoon M. de Crecy-Lagard V. Diaz N. Disz T. Edwards R. Fonstein M. Frank E.D. Gerdes S. Glass E.M. Goesmann A. Hanson A. Iwata-Reuyl D. Jensen R. Jamshidi N. Krause L. Kubal M. Larsen N. Linke B. McHardy A.C. Meyer F. Neuweger H. Olsen G. Olson R. Osterman A. Portnoy V. Pusch G.D. Rodionov D.A. Ruckert C. Steiner J. Stevens R. Thiele I. Vassieva O. Ye Y. Zagnitko O. Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl H.W. Pinckney J.L. Steppe T.F. Cyanobacterial-bacterial mat consortia: examining the functional unit of microbial survival and growth in extreme environments. Environ Microbiol. 2000;2:11–26. doi: 10.1046/j.1462-2920.2000.00071.x. [DOI] [PubMed] [Google Scholar]
- Patel P.V. Gianoulis T.A. Bjornson R.D. Yip K.Y. Engelman D.M. Gerstein M.B. Analysis of membrane proteins in metagenomics: networks of correlated environmental features and protein families. Genome Res. 2010;20:960–971. doi: 10.1101/gr.102814.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E. Quast C. Knittel K. Fuchs B.M. Ludwig W. Peplies J. Glockner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D. Tatusova T. Maglott D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2008. R: a language and environment for statistical computing. [Google Scholar]
- Raes J. Korbel J.O. Lercher M.J. von Mering C. Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield A.C. On the proportions of organic derivations in sea water and their relation to the composition of plankton. In: Daniel R.J., editor. James Johnstone Memorial Volume. University Press of Liverpool; Liverpool: 1934. pp. 177–192. [Google Scholar]
- Remans K. Vercammen K. Bodilis J. Cornelis P. Genome-wide analysis and literature-based survey of lipoproteins in Pseudomonas aeruginosa. Microbiology. 2010;156:2597–2607. doi: 10.1099/mic.0.040659-0. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mejia J.L. Martinez-Anaya C. Folch-Mallol J.L. Dantan-Gonzalez E. A two-step electrodialysis method for DNA purification from polluted metallic environmental samples. Electrophoresis. 2008;29:3239–3244. doi: 10.1002/elps.200700829. [DOI] [PubMed] [Google Scholar]
- Rusch D.B. Halpern A.L. Sutton G. Heidelberg K.B. Williamson S. Yooseph S. Wu D. Eisen J.A. Hoffman J.M. Remington K. Beeson K. Tran B. Smith H. Baden-Tillson H. Stewart C. Thorpe J. Freeman J. Andrews-Pfannkoch C. Venter J.E. Li K. Kravitz S. Heidelberg J.F. Utterback T. Rogers Y.H. Falcon L.I. Souza V. Bonilla-Rosso G. Eguiarte L.E. Karl D.M. Sathyendranath S. Platt T. Bermingham E. Gallardo V. Tamayo-Castillo G. Ferrari M.R. Strausberg R.L. Nealson K. Friedman R. Frazier M. Venter J.C. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan D.J. Mann N.H. Carr N.G. The response of the picoplanktonic marine cyanobacterium Synechococcus species Wh7803 to phosphate starvation involves a protein homologous to the periplasmic phosphate-binding protein of Escherichia coli. Mol Microbiol. 1993;10:181–191. doi: 10.1111/j.1365-2958.1993.tb00914.x. [DOI] [PubMed] [Google Scholar]
- Souza V. Espinosa-Asuar L. Escalante A.E. Eguiarte L.E. Farmer J. Forney L. Lloret L. Rodriguez-Martinez J.M. Soberon X. Dirzo R. Elser J.J. An endangered oasis of aquatic microbial biodiversity in the Chihuahuan Desert. Proc Natl Acad Sci USA. 2006;103:6565–6570. doi: 10.1073/pnas.0601434103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza V. Falcón L.I. Elser J.J. Eguiarte L.E. Protecting a window into the ancient Earth: towards a Precambrian park at Cuatro Cienegas, Mexico. The Citizen's Page, Evolutionary Ecology Research. 2007. http://www.evolutionary-ecology.com/citizen/citizen.html http://www.evolutionary-ecology.com/citizen/citizen.html
- Souza V. Eguiarte L.E. Siefert J. Elser J.J. Microbial endemism: does phosphorus limitation enhance speciation? Nat Rev Microbiol. 2008;6:559–564. doi: 10.1038/nrmicro1917. [DOI] [PubMed] [Google Scholar]
- Suzuki S. Ferjani A. Suzuki I. Murata N. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in synechocystis. J Biol Chem. 2004;279:13234–13240. doi: 10.1074/jbc.M313358200. [DOI] [PubMed] [Google Scholar]
- Szynkiewicz A. Ewing R.C. Moore C.H. Glamoclija M. Bustos D. Pratt L.M. Origin of terrestrial gypsum dunes—implications for martian gypsum-rich dunes of Olympia Undae. Geomorphology. 2009;121:69–83. [Google Scholar]
- Tatusov R.L. Natale D.A. Garkavtsev I.V. Tatusova T.A. Shankavaram U.T. Rao B.S. Kiryutin B. Galperin M.Y. Fedorova N.D. Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Nature Conservancy. The Nature Conservancy. 2010. http://www.nature.org http://www.nature.org
- Tice M.M. Lowe D.R. Photosynthetic microbial mats in the 3,416-Myr-old ocean. Nature. 2004;431:549–552. doi: 10.1038/nature02888. [DOI] [PubMed] [Google Scholar]
- Tyrrell T. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature. 1999;400:525–531. [Google Scholar]
- van Mooy B.A.S. Devol A.H. Keil R.G. Quantifying H-3-thymidine incorporation rates by a phylogenetically defined group of marine planktonic bacteria (Bacteriodetes phylum) Environ Microbiol. 2004;6:1061–1069. doi: 10.1111/j.1462-2920.2004.00636.x. [DOI] [PubMed] [Google Scholar]
- Wahlund T.M. Tabita F.R. The reductive tricarboxylic acid cycle of carbon dioxide assimilation: initial studies and purification of ATP-citrate lyase from the green sulfur bacterium Chlorobium tepidum. J Bacteriol. 1997;179:4859–4867. doi: 10.1128/jb.179.15.4859-4867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.K. Metcalf W.W. Two C-P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J Bacteriol. 2004;186:4730–4739. doi: 10.1128/JB.186.14.4730-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.R. Nagarajan N. Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.G. Sherman P.W. Anciently asexual bdelloid rotifers escape lethal fungal parasites by drying up and blowing away. Science. 2010;327:574–576. doi: 10.1126/science.1179252. [DOI] [PubMed] [Google Scholar]
- Zaheer R. Morton R. Proudfoot M. Yakunin A. Finan T.M. Genetic and biochemical properties of an alkaline phosphatase PhoX family protein found in many bacteria. Environ Microbiol. 2009;11:1572–1587. doi: 10.1111/j.1462-2920.2009.01885.x. [DOI] [PubMed] [Google Scholar]
- Zhou J. Davey M.E. Figueras J.B. Rivkina E. Gilichinsky D. Tiedje J.M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.