Abstract
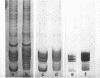
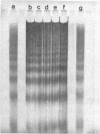
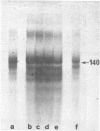
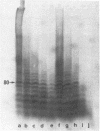
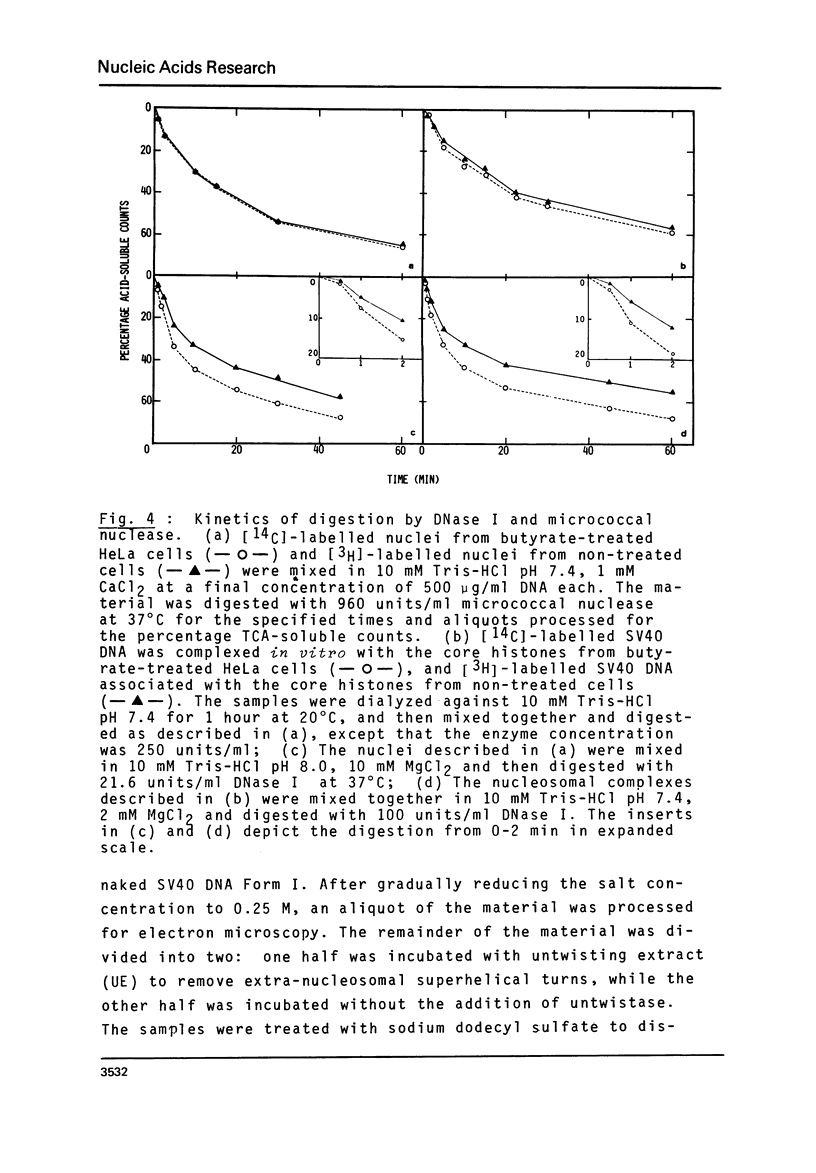
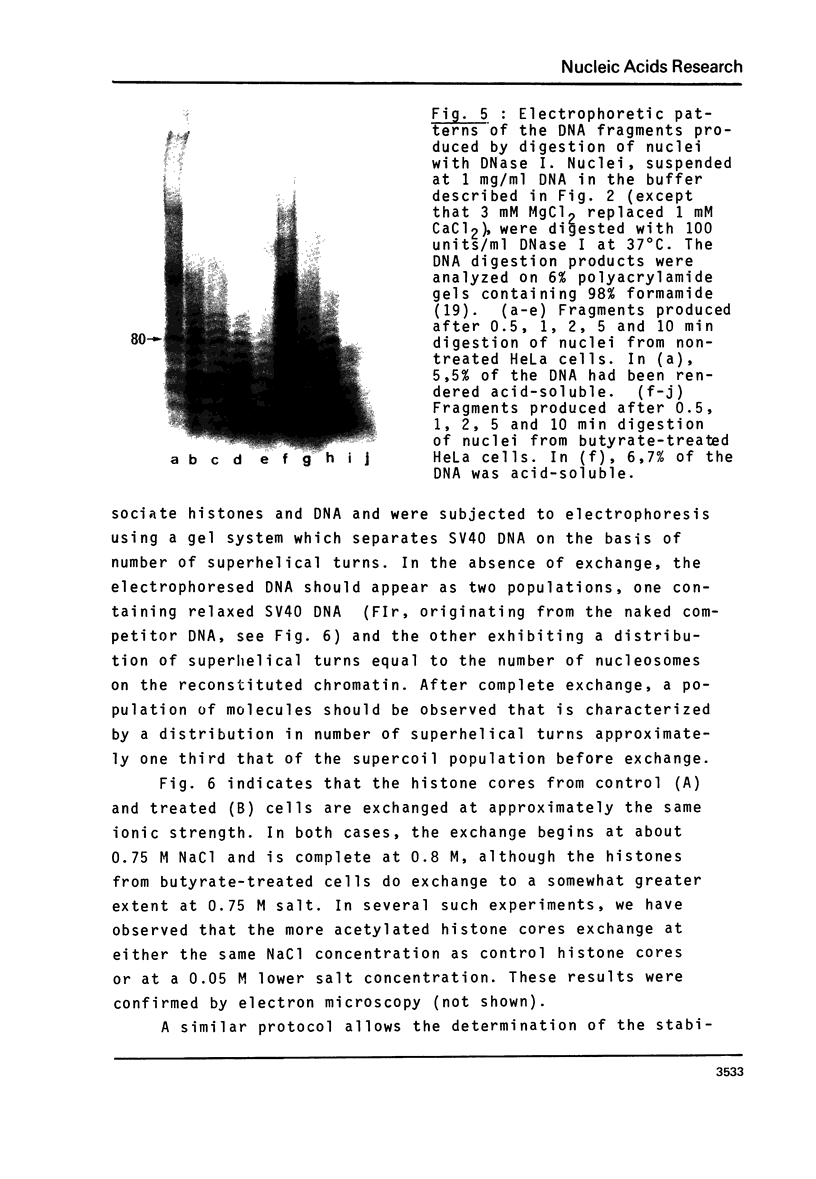
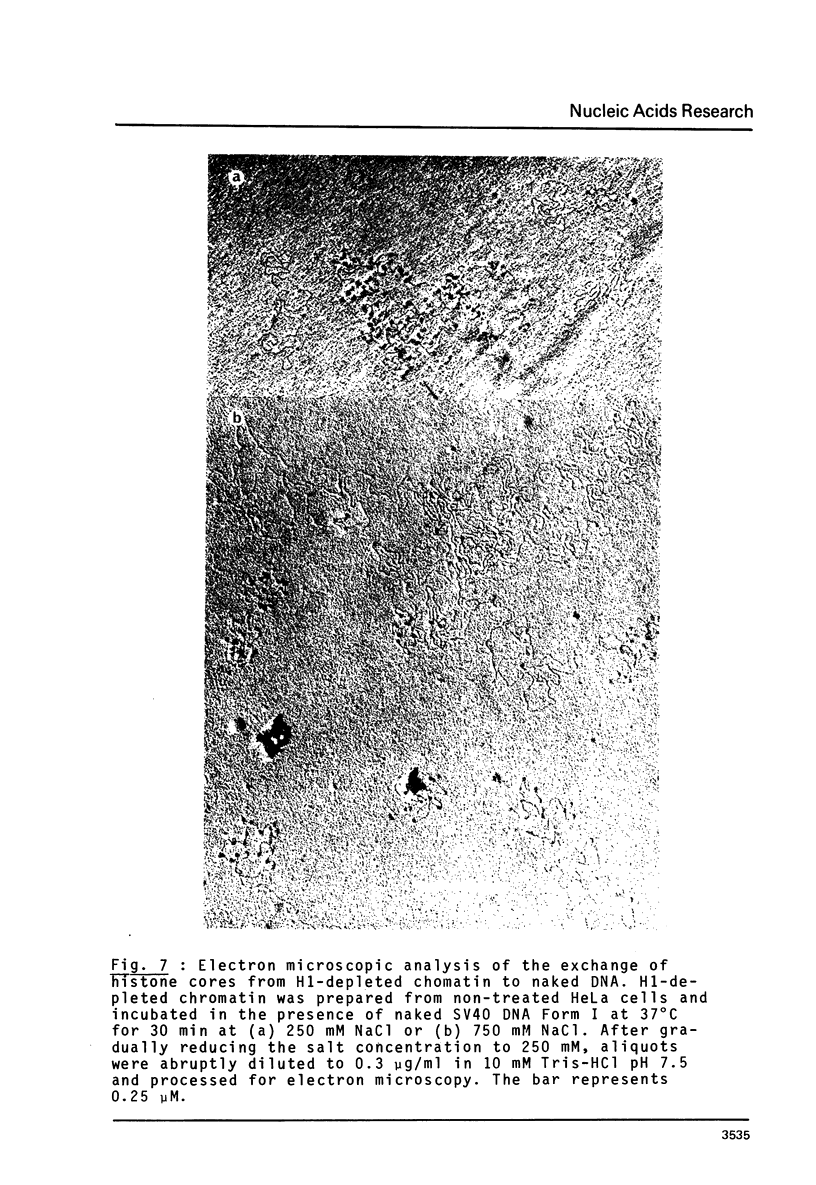
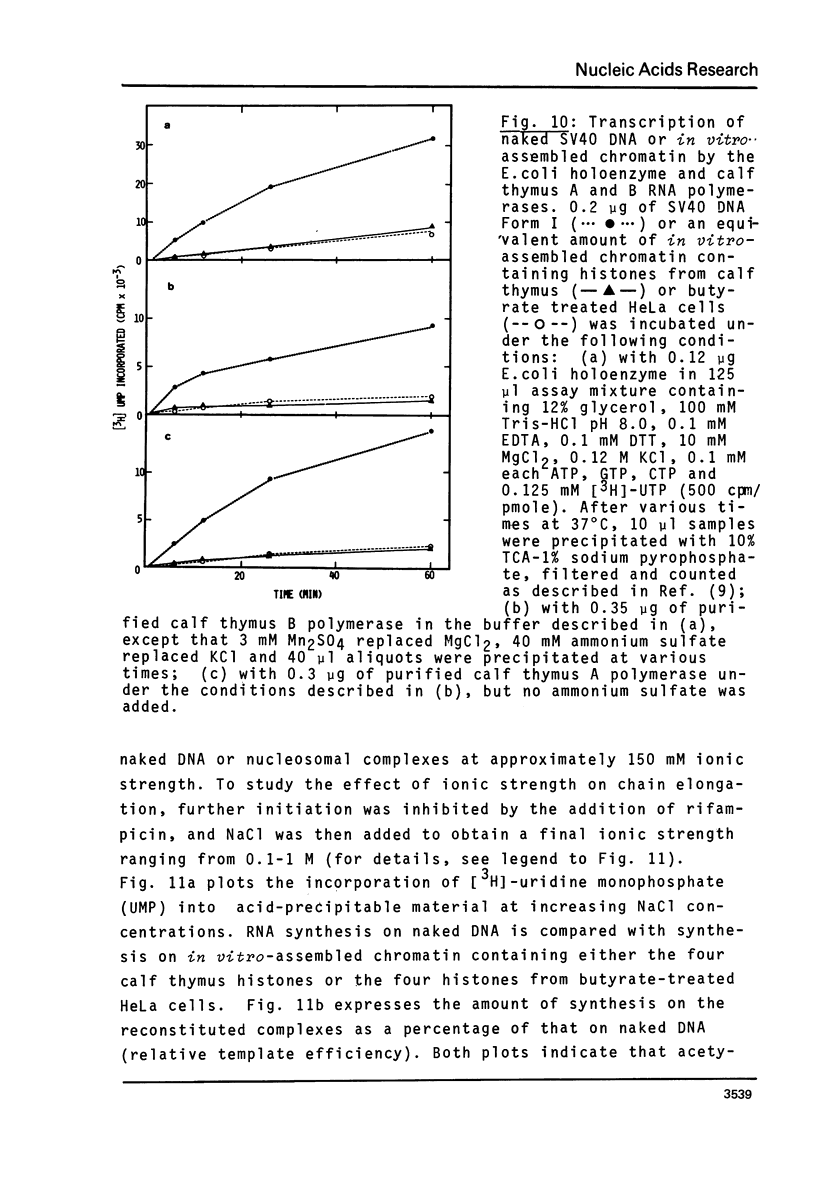
n-Butyrate treatment of growing Hela cells produces a dramatic increase in the levels of histone acetylation. We have exploited this system to study the effect of histone acetylation on chromatin structure. Chromatin containing highly acetylated histones is more rapidly digested to acid-soluble material by DNase I, but not by micrococcal nuclease. The same pattern of nuclease sensitivity was exhibited by in vitro-assembled chromatin consisting of SV40 DNA Form I and the 2 M salt-extracted core histones from butyrate-treated cells. Using this very defined system, it was possible to demonstrate that acetylated nucleosomes do not have a greatly diminished stability. Stability was measured in terms of exhange of histone cores onto competing naked DNA or sliding of histone cores along ligated naked DNA. Finally, it was shown that acetylated nucleosomes are efficient inhibitors of in vitro RNA synthesis by the E. coli holoenzyme as well as by the mammalian polymerases A and B.
Full text
PDF





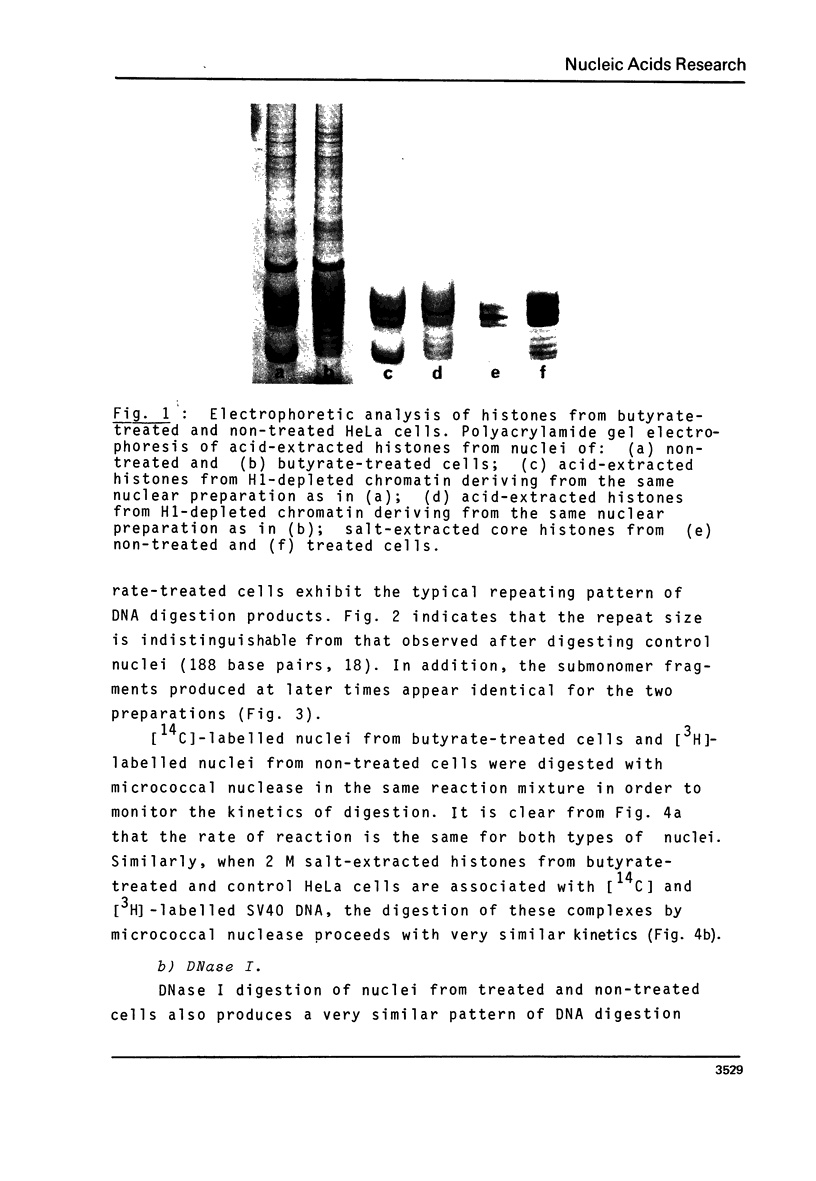













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALLFREY V. G., FAULKNER R., MIRSKY A. E. ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proc Natl Acad Sci U S A. 1964 May;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Oudet P., Germond J. E., Chambon P. Subunit structure of simian-virus-40 minichromosome. Eur J Biochem. 1976 Nov 15;70(2):543–553. doi: 10.1111/j.1432-1033.1976.tb11046.x. [DOI] [PubMed] [Google Scholar]
- Brison O., Kedinger C., Wilhelm J. Enzymatic properties of viral replication complexes isolated from adenovirus type 2-infected HeLa cell nuclei. J Virol. 1977 Nov;24(2):423–435. doi: 10.1128/jvi.24.2.423-435.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissinger F., Chambon P. Animal DNA-dependent RNA polymerases. 2. Purification of calf-thymus AI enzyme. Eur J Biochem. 1972 Jul 13;28(2):277–282. doi: 10.1111/j.1432-1033.1972.tb01911.x. [DOI] [PubMed] [Google Scholar]
- Hagopian H. K., Riggs M. G., Swartz L. A., Ingram V. M. Effect of n-butyrate on DNA synthesis in chick fibroblasts and HeLa cells. Cell. 1977 Nov;12(3):855–860. doi: 10.1016/0092-8674(77)90284-7. [DOI] [PubMed] [Google Scholar]
- Ilyin Y. V., Varshavsky A. Y., Mickelsaar U. N., Georgiev G. P. Studies on deoxyribonucleoprotein structure. Redistribution of proteins in mixtures of deoxyribonucleoproteins, DNA and RNA. Eur J Biochem. 1971 Sep 24;22(2):235–245. doi: 10.1111/j.1432-1033.1971.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Kedinger C., Chambon P. Animal DNA-dependent RNA polymerases. 3. Purification of calf-thymus BI and BII enzymes. Eur J Biochem. 1972 Jul 13;28(2):283–290. doi: 10.1111/j.1432-1033.1972.tb01912.x. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Marushige K. Activation of chromatin by acetylation of histone side chains. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3937–3941. doi: 10.1073/pnas.73.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Riggs M. G., Whittaker R. G., Neumann J. R., Ingram V. M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature. 1977 Aug 4;268(5619):462–464. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978 Apr;13(4):691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Vidali G., Boffa L. C., Bradbury E. M., Allfrey V. G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci U S A. 1978 May;75(5):2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Localization of the sites along nucleosome DNA which interact with NH2-terminal histone regions. J Biol Chem. 1977 Sep 25;252(18):6516–6520. [PubMed] [Google Scholar]
- Wilhelm J., Brison O., Kedinger C., Chambon P. Characterization of adenovirus type 2 transcriptional complexes isolated from infected HeLa cell nuclei. J Virol. 1976 Jul;19(1):61–81. doi: 10.1128/jvi.19.1.61-81.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M. Steroid receptors: elements for modulation of eukaryotic transcription. Annu Rev Biochem. 1976;45:721–746. doi: 10.1146/annurev.bi.45.070176.003445. [DOI] [PubMed] [Google Scholar]