Abstract
Prion diseases are fatal neurodegenerative disorders that include bovine spongiform encephalopathy (BSE) and scrapie in animals and Creutzfeldt-Jakob disease (CJD) in humans. They are characterized by long incubation periods, variation in which is determined by many factors including genetic background. In some cases it is possible that incubation time may be directly correlated to the level of gene expression. To test this hypothesis, we combined incubation time data from five different inbred lines of mice with quantitative gene expression profiling in normal brains and identified five genes with expression levels that correlate with incubation time. One of these genes, Hspa13 (Stch), is a member of the Hsp70 family of ATPase heat shock proteins, which have been previously implicated in prion propagation. To test whether Hspa13 plays a causal role in determining the incubation period, we tested two overexpressing mouse models. The Tc1 human chromosome 21 (Hsa21) transchromosomic mouse model of Down syndrome is trisomic for many Hsa21 genes including Hspa13 and following Chandler/Rocky Mountain Laboratory (RML) prion inoculation, shows a 4% reduction in incubation time. Furthermore, a transgenic model with eightfold overexpression of mouse Hspa13 exhibited highly significant reductions in incubation time of 16, 15, and 7% following infection with Chandler/RML, ME7, and MRC2 prion strains, respectively. These data further implicate Hsp70-like molecular chaperones in protein misfolding disorders such as prion disease.
Prion diseases or transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative diseases affecting several mammalian species and include scrapie in sheep and goats, chronic wasting disease (CWD) in deer and elk, bovine spongiform encephalopathy (BSE) in cattle, and Creutzfeldt-Jakob disease (CJD) in humans (1). They are characterized by the conversion of endogenous host PrPc to an abnormal form, PrPSc, resulting in distinctive neuropathology that includes PrPSc deposition, vacuolation, gliosis, and neuronal loss.
All prion diseases, regardless of species or etiology, exhibit a long, clinically silent incubation period. The duration of this latent phase is determined by many factors including prion strain, dose, route of infection, and genetic background. In humans and mice the main genetic determinant has been shown to be the prion protein gene (PRNP). In humans, a methionine to valine polymorphism at codon 129 of PrP has been strongly associated with disease susceptibility (2–6), and in mice, incubation time has also been linked to polymorphisms in Prnp where Prnpa (108-Leu, 189-Thr) and Prnpb (108-Phe, 189-Val) are associated short and long incubation times, respectively (7–10).
Although the central role of PRNP in prion disease susceptibility is undisputed, it is acknowledged that other genes also contribute to the observed variation. In mice, this is exemplified by the comparison of incubation times from a range of Prnpa inbred lines showing a difference of over 100 d between the shortest and longest incubation time strains (11). Many approaches have been taken to identify susceptibility genes including a genome-wide association study (GWAS) for variant CJD (vCJD) (6), and in mice, several studies have carried out quantitative trait loci mapping in both two-way crosses and a heterogeneous cross (12–14). In these studies, the underlying functional polymorphism is unknown but could be an amino acid change within the coding region of a protein or splicing variant or may occur within noncoding sequences such as untranslated regions, promoters, or other regulatory regions thereby influencing the pattern or level of expression. Although incubation time is a polygenic trait, it is possible that in some cases, there may be a direct correlation between incubation time and individual gene expression level. To test this hypothesis and look for genes that show a correlation between incubation time and expression level, we carried out a microarray gene expression study to determine the expression profile of transcripts from normal adult brains of five inbred lines of mice with different incubation times. Five genes showed a reproducible correlation with incubation time. One of these genes, Hspa13 (Stch), is a member of the Hsp70 family of ATPase “stress” or “heat shock” proteins (15). Molecular chaperones have been previously implicated in prion disease, suggesting that Hspa13 is a promising candidate gene for a causal role in incubation time (16, 17). To investigate this possibility, we examined two Hspa13 overexpressing mouse models, both of which showed a significant decrease in incubation time. These results reinforce the importance of molecular chaperones and the protein folding machinery in prion disease.
Results
GeneChip Expression Study.
To look for genes whose expression level in the brain correlates with incubation time, we selected five inbred lines of mice with previously characterized different incubation times for the mouse adapted scrapie prion strain, Chandler/Rocky Mountain Laboratory (RML) (18). The strains selected were NZW/OlaHsd (108 ± 1), SJL/OlaHsd (122 ± 1), FVB/NHsd (131 ± 1), SWR/OlaHsd (135 ± 1), and C57BL/6JOlaHsd (143 ± 1) all of which are Prnpa (18). cRNA was prepared from the whole brain of 6-wk-old uninfected mice (n = 5) and hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 arrays that contain probes representing over 39,000 transcripts. After scaling and normalization, we identified probe sets that showed statistically significant differential gene expression (M > 0.5 or < −0.5, equivalent to 1.4-fold difference) between the shortest (NZW) and longest (C57BL/6) incubation time strains. A false discovery rate (FDR) of 0.01 was applied to take into account the large number of statistical tests carried out. This step reduced the number of genes to 420. These genes are differentially expressed between the phenotypic extremes but may not show a correlation across all five inbred strains; therefore, we calculated the square of the correlation coefficient (R2) for each probe set using the mean normalized array signal and the mean incubation time for each strain. These were ranked by R2 value. The top 5% (n = 21) of the distribution had R2 values >9.0. Random permutations (n = 1,000) of the data produced only four R2 values >9.0, suggesting that we have achieved a significant enrichment of hits over what would be expected by chance alone (Table S1).
Verification of Microarray Data.
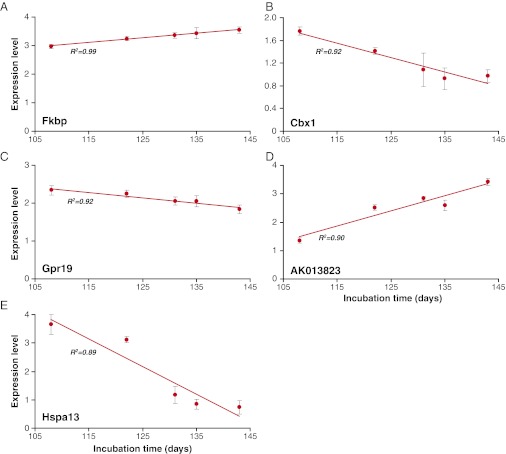
To verify the correlations observed in the GeneChip study, we looked at the expression of the top 30 genes by quantitative reverse transcriptase PCR (qRT-PCR) using SYBR green. Initially, the same RNA as was used for the arrays was tested (n = 5) and subsequently the qRT-PCR was repeated in an additional independent panel of samples from each mouse strain (n = 5). Eight of 25 of these genes replicated the microarray data; therefore, they were tested again using 5′Fam/3′Tamra dual-labeled probes to increase the specificity. Correlations were confirmed for five genes Fkbp9, Cbx1, Gpr19, AK013823, and Hspa13 (Stch) (Fig. 1 and Fig. S1).
Fig. 1.
Correlation of gene expression level with incubation time. Data are taken from GeneChip Mouse Genome 430 2.0 arrays (Affymetrix) hybridized with cRNA prepared from the whole brains from each of five mouse inbred lines: NZW/OlaHsd, SJL/OlaHsd, FVB/NHsd, SWR/OlaHsd, and C57BL/6JOlaHsd (n = 5 per strain). Mean incubation time of each mouse strain inoculated with the Chandler/RML prion strain is measured in days and is shown on the x axis. Mean expression level for each probe is represented on the y axis (±SD). Expression level data are shown following scaling and normalization using the robust multichip average (RMA) method. R2 represents the square of the Pearson product moment correlation coefficient. (A) Fkbp9, (B) Cbx1, (C) Gpr19, (D) AK013823, and (E) Hspa13 (Stch).
Mouse Models.
Tc1 mice.
Although the correlation between gene expression and incubation time was verified and our analysis suggests that we have enriched for genuine associations, it is still possible that these correlations could occur by chance, and so we addressed this possibility by testing prion incubation time in mouse models. Hspa13 is a member of the Hsp70 family of ATPase molecular chaperones that have been previously implicated in prion propagation, suggesting that Hspa13 is an excellent candidate for modifying prion disease incubation time (16, 17). For Hspa13, a shorter incubation time is associated with an increased level of expression (Fig. 1); therefore, we looked at overexpression models. Hspa13 is located on mouse chromosome 16 (Mmu16), with its human homolog on human chromosome 21 (Hsa21); thus, it is likely to be overexpressed in the Tc(Hsa21)1TybEmcf (Tc1) transchromosomic mouse model of Down syndrome (19). The Tc1 mouse contains a normal mouse diploid genome and an additional freely segregating almost complete Hsa21. To confirm the presence of the human HSPA13 gene we sequenced the exons and intron–exon boundaries using human specific primers and we also sequenced the human cDNA transcript to ensure that no rearrangements had occurred. Human and mouse-specific qRT-PCR was used to confirm that the level of endogenous mouse Hspa13 in Tc1 mice was the same as wild-type litter mate controls and that human HSPA13 was expressed in the Tc1 mice. The level of human HSPA13 expression was similar to that in a pool of human brain RNA (n = 23), which is consistent with the duplication of HSPA13 in the Tc1 mouse (Fig. S2 A and B). We also confirmed the presence of human HSPA13 protein in Tc1 mice on a Western blot using an antihuman HSPA13 rabbit polyclonal antibody (ProteinTech Group) (Fig. S2D).
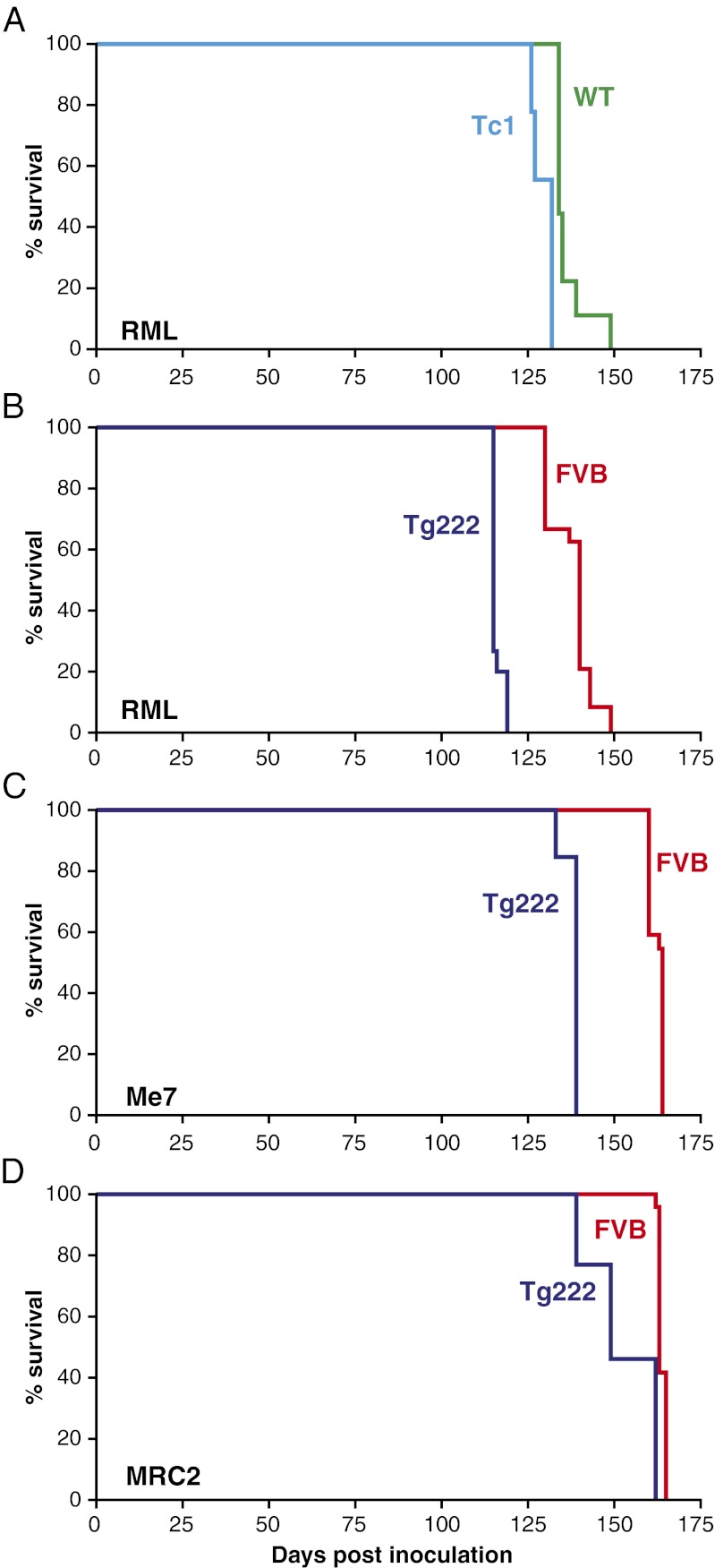
Tc1 (n = 9) and wild-type litter mate controls (n = 9) were inoculated intracerebrally with the mouse adapted scrapie prion strain Chandler/RML. Tc1 mice showed a 4% reduction in incubation time (130 ± 1) relative to wild type (136 ± 2) and this was highly significant (P < 0.001, Kaplan–Meier log-rank survival) (Table 1 and Fig. 2A).
Table 1.
Prion disease incubation times in Hspa13 overexpressing mouse models
| Mouse strain | Prion strain | Incubation time ± SEM (n) | P value |
| Tc1 | Chandler/RML | 130 ± 1 (9) | <0.001 |
| Wild type | 136 ± 2 (9) | ||
| Tg222 | Chandler/RML | 116 ± 0 (15) | <0.001 |
| FVB | 138 ± 1 (24) | ||
| Tg222 | ME7 | 138 ± 1 (13) | <0.001 |
| FVB | 162 ± 0 (22) | ||
| Tg222 | MRC2 | 153 ± 3 (13) | <0.001 |
| FVB | 164 ± 0 (24) |
Data were analyzed using the Kaplan–Meier log-rank survival test.
Fig. 2.
Kaplan–Meier survival curves. Data are shown as percentage of animals surviving (y axis) plotted against the number of days postinoculation (x axis). (A) Transmission of Chandler/RML prion strain to Tc1 and wild-type litter mate controls. (B) Transmission of Chandler/RML prion strain to Tg222 (Hspa13 overexpressor) and FVB/NHsd controls. (C) Transmission of ME7 prion strain to Tg222 and FVB/NHsd controls. (D) Transmission of MRC2 mouse-adapted BSE prion strain to Tg222 and FVB/NHsd controls. Reduction in mean incubation time of 4, 16, 15, and 7% was seen in A–D, respectively. This reduction in survival was statistically significant for each transmission (P < 0.001, Kaplan–Meier log-rank survival test).
Western blotting showed that there was no difference in the PrPSc strain type produced in Tc1 mice relative to litter mate controls (Fig. S3A). Histological examination of infected mice showed that the overall pattern of PrPSc distribution (Fig. S4), spongiosis, and gliosis were the same for both Tc1 and controls. Mild neuronal loss was seen in the hippocampus of Tc1 mice; however, very little loss was seen in the controls (P = 0.02, Fisher’s exact test). The hippocampal ribbon was five- to six-cells thick in controls and thinner (two to three cells) in Tc1 mice accompanied by reactive astrocytes and more frequent neuronal apoptosis (Fig. S5).
Hspa13 transgenic mice.
Although the Tc1 model overexpresses HSPA13, this is a modest increase in expression (×2) of the human protein and the known mosaicism suggests that this is not present in all neurons (19). More importantly, this model overexpresses many additional human genes from Hsa21, making it difficult to ascribe any difference in incubation time directly to HSPA13 overexpression. We therefore generated a transgenic mouse overexpressing only mouse Hspa13 under the control of the Syrian hamster Prnp promoter (Tg222). By qRT-PCR, homozygous Tg222 mice showed approximately eightfold increased Hspa13 mRNA expression relative to wild-type FVB mice (Fig. S2C). Due to the absence of a usable antimouse Hspa13 antibody, we were unable to confirm the overexpression of Hspa13 protein.
To look at the effect of Hspa13 overexpression on prion disease incubation time, Tg222 (homozygous) and FVB wild-type controls were inoculated with two different mouse-adapted scrapie prion strains, Chandler/RML and ME7, and a mouse-adapted BSE strain of prions, MRC2 (20). In Tg222 mice, a significant decrease in incubation time was seen for all three prion strains relative to nontransgenic controls. For Chandler/RML, the onset of symptoms was reduced from wild-type control values of 138 ± 1 to 116 ± 0 (16%) (P < 0.001, Kaplan–Meier log-rank survival). Similar results were seen with ME7, where incubation time decreased from control values of 162 ± 0 to 138 ± 1 (15%) (P < 0.001, Kaplan–Meier log-rank survival). For MRC2, there was a smaller decrease of 7% from control values of 164 ± 0 to 153 ± 3 (P < 0.001, Kaplan–Meier log-rank survival) (Fig. 2 B–D and Table 1).
Western blotting confirmed that the strain types were faithfully propagated in the Tg222 mice (Fig. S3 B–D). Histological comparison of PrPSc deposition, spongiosis, and gliosis showed the characteristic patterns for the prion strain type and showed no significant differences between Tg222 and controls (Figs. S6 and S7). Unlike the Tc1 model there was no increase in hippocampal neuronal death for Chandler/RML-inoculated Tg222 mice. For some individual animals, there was more severe neuronal loss in the CA2 region of the hippocampus for MRC2-inoculated Tg222 animals; however, this loss was not seen across the whole group and was not statistically significant (Fig. S7).
PrPc expression.
PrP expression is a major influence on incubation time in experimental mouse models where short incubation times are seen in PrP overexpressors and hemizygous mice have a prolonged incubation time relative to wild type (21, 22). To determine whether differences in endogenous levels of PrPc expression in both Tc1 and Tg222 mice could explain the reduction in incubation time, we measured total PrPc in 10% (wt/vol) uninfected brain homogenates using a PrP-specific ELISA (23). No difference was seen between Tc1 and wild-type litter mate controls or between Tg222 and FVB wild-type controls (Fig. S8).
Hspa13 knockdown in a cell-based prion curing assay.
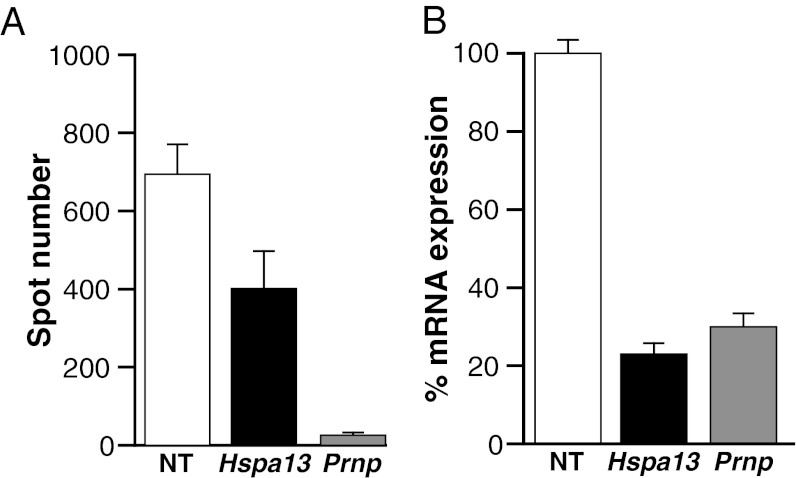
Only one mouse line for Hspa13 overexpression was established; therefore, it is possible that the reduction in incubation time could be the result of disrupting another gene during the random integration of the transgene. To provide additional support for the role of Hspa13 in prion disease, we used a pool of four siRNAs (Accell SMART pool) to transiently knockdown mRNA expression of Hspa13 in chronically prion infected N2a cells (IPKS7 subclone). “Curing” of the cells was determined by counting PrPSc positive cells using an ELISPOT assay compared with cells treated with a nontargeting control siRNA pool (24). Prnp knockdown was used as a positive control (25). As determined by quantitative real-time RT-PCR, mRNA expression was reduced to 23 and 30% for Hspa13 and Prnp, respectively. Hspa13 and Prnp knockdown significantly reduced the spot number to 57.8 and 3.7% of the nontargeting control respectively (P = 0.04, P < 0.0001, t test) (Fig. 3).
Fig. 3.
Treatment of chronically prion-infected PK1 cells with siRNAs. (A) Quantification of “curing” by PrPSc-based ELISPOT assay. (B) Quantification of mRNA by real-time RT-PCR. NT, nontargeting control.
Discussion
Incubation time is a complex trait determined by many genes; therefore, although the expression level of an individual gene may have a profound effect on incubation time, this effect may not be detectable in the overall incubation time of the animal due to opposing effect of other genes on the phenotype. Where the effect in very strong or in the same direction as other effectors, a correlation between incubation time and expression may be seen. This is exemplified experimentally by PrP expression where transgenic mice overexpressing mouse PrP have a very short incubation time relative to wild-type mice and conversely, hemizygous mice have an extended incubation time (21, 22). The principle of correlating endogenous expression levels with phenotype has also been applied successfully to mouse models of anxiety (26).
We have successfully identified five genes whose mRNA expression correlates with incubation time. Although the microarray data have been verified by qRT-PCR, it is possible that this correlation may occur by chance; therefore, functional studies such as we show here for Hspa13 are essential to confirm the relevance of Fkbp9, Gpr19, Cbx1, and AK013823 to prion disease incubation time. In this study we used RNA extracted from whole brains to look at global effects; however, it is possible that this choice may have masked the effect of some genes that are only differentially expressed in specific regions of the brain. Previous genome-wide expression studies in prion disease have focused on differential expression between the normal brain and different stages during disease pathogenesis (27). In this study we chose to use normal brains; therefore, we may not have detected genes that are only differentially expressed during the disease process. By comparing normal brains from different inbred lines of mice, our aim was to identify genes that are differentially expressed in the normal state and the basis of the differential regulation is therefore likely to be genetic.
The microarray data for Hspa13 predicts that overexpression is likely to result in a shortening of the incubation time. For both Tc1 and Tg222 incubations, times for all prion strains tested were significantly reduced. In both cases, the actual reduction is not as great as predicted by the mRNA expression of Hspa13 alone, which suggests that other factors are also influencing the incubation time. We have shown that this effect is not due to any direct up-regulation of PrPc expression.
We have shown that the Tc1 mouse model shows a 4% reduction in incubation time with Chandler/RML prions. Although Tc1 expresses both mouse and human Hspa13, it is a complex model carrying large, often rearranged, regions of Hsa21, thereby expressing both mouse and human versions of several genes (19). Two genes present on Hsa21, APP and SOD1, have previously been implicated in prion disease (28). In mouse models, loss of App and overexpression of human SOD1 have been shown to prolong prion disease incubation time by 13 and 19%, respectively. However, in the Tc1 model, the human APP gene is rearranged and therefore not expressed (29). We have confirmed that human SOD1 as well as mouse Sod1 is expressed in the Tc1 model, which should act to prolong incubation time (Fig. S2E) (19). The mixture of human and mouse Hspa13 in the Tc1 model, the lower level of expression relative to the Tg222 model, mosaicism for Hsa21 and the overexpression of SOD1 may offer some explanation as to why the Tc1 mice inoculated with Chandler/RML show only a 4% reduction in incubation time compared with a 16% reduction in Tg222. It should also be noted that the genetic background of the Tc1 mice is mixed and although wild-type litter mate controls were used, this difference may also introduce additional variation. Similar data have also been observed for trisomy 16—diploid aggregation chimeras (Ts16), which showed an 11% reduction in the onset of scrapie symptoms following challenge with the Chandler scrapie isolate (30). As seen for Tc1, the Ts16 model is trisomic for a large number of additional genes, any of which may influence incubation time. Due to the complexity of the Tc1 and Ts16 models, it is not possible to ascribe specifically the reduction in incubation time to the overexpression of Hspa13.
We have shown that there is significantly more neuronal loss in Tc1 mice relative to litter mate controls following infection with Chandler/RML prions. This loss was not seen in the Tg222, suggesting that this effect is not directly related to Hspa13 overexpression but rather an increased susceptibility in Tc1 neurons as a result of the Hsa21 trisomy. An increase in neuronal death was also seen in individual Tg222 animals inoculated with MRC2; however, this finding was not statistically significant over the whole group.
We have demonstrated that overexpression of Hspa13 in a transgenic mouse model significantly reduces the prion disease incubation period for three different mouse prion strains and we also show that knockdown of Hspa13 mRNA in chronically prion infected N2a cells significantly reduces the PrPSc spot number, thus suggesting that changes in expression of Hspa13 leads to a direct effect on phenotype. Hspa13 (Stch) is a member of the Hsp70 core ATPase family of protein chaperones (15, 31). Hspa13 is truncated relative to Hsp70; however, it has a similar size and activity to proteolytically cleaved N-terminal Hsc70 and Grp78/BiP. Hspa13 is constitutively expressed and is induced by Ca2+ released by ionophore A23187 but not heat shock. As predicted by the absence of a peptide-binding domain, the ATPase activity of Hspa13 is independent of peptide stimulation. Hspa13 has been localized to the luminal side of microsome fractions and its cellular distribution is similar, although not identical, to that of Gpr78/BiP, which is consistent with an endoplasmic reticulum (ER) and other intraorganellar localization (15). Protein misfolding, such as occurs in prion disease, can trigger ER stress and activate the unfolded protein response (UPR) resulting in the up-regulation of ER chaperones such as Gpr78/BiP (32, 33). Toxic peptides of PrP have also been implicated in the alteration of Ca2+ homeostasis thereby contributing to further ER stress (34). Loss of PrP may also contribute to the disruption of Ca2+ homeostasis within neurons (35). Furthermore, overexpression of STCH in HEK293 cells sensitized them to tumor necrosis factor-related apoptosis-related ligand (TRAIL)-induced apoptosis, which was abolished by the stomach cancer-derived del223V-226L STCH mutation, which occurs within the ATP-binding domain (36). TRAIL is not normally expressed in the brain; however, it may be induced under pathological conditions such as Alzheimer’s disease (37). The normal function of Hspa13 is poorly defined; however, its distribution and similarity to Gpr78/BiP implies a role that is sensitive to Ca2+ flux in a membrane-bound compartment, thus suggesting pathways through which it influences prion pathogenesis.
In conclusion, we have shown that the endogenous expression level of five genes in mouse brain is correlated with prion disease incubation time, and for Hspa13, this finding has been confirmed by the demonstration that overexpression in a mouse model significantly reduces the incubation time with three different prion strains and that mRNA knockdown in a cell-based prion curing assay significantly reduces the level of PrPSc. The precise role of Hspa13 in prion disease remains to be established but may involve the UPR or TRAIL-induced apoptosis. Further evaluation of the role of Hspa13 should increase our understanding of these pathways in prion disease.
Methods
Mice.
All inbred lines were obtained from Harlan. The 129S8Nimr;C57BL/6J-Tc(Hsa21)1TybEmcf (Tc1) mice (19) were obtained from E.M.C.F. and V.L.J.T. (UCL Institute of Neurology and MRC National Institute for Medical Research, London).
Expression Arrays.
RNA for expression arrays was prepared from 6-wk-old mouse brains (n = 5) from NZW/OlaHsd, SJL/OlaHsd, FVB/NHsd, SWR/OlaHsd, and C57BL/6JOlaHsd using a Maxi RNeasy kit (Qiagen). This preparation was followed with an additional DNase I digest and clean up step using a Mini RNeasy kit (Qiagen). Biotinylated cRNA was generated using a One Cycle Target Labeling Assay kit (Affymetrix) and hybridized to GeneChip Mouse Genome 430 2.0 arrays (Affymetrix) by UCL Scientific Support Service using a GeneChip Scanner 3000 (Affymetrix). All kits were used according to the manufacturer’s instructions. Array data were scaled and normalized using the robust multichip average (RMA) method. Microarray data have been deposited in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under accession no. GSE38205. Probes with negative values or absent calls were excluded from the analysis. Differential expression between the shortest incubation time (NZW/OlaHsd) and longest incubation time (C57BL/6JOlaHsd) mice was calculated using Limma (38) applying a FDR of 0.01 and filtering for an M value (log2 fold difference) of >0.5 or < −0.5. The square of the Pearson product moment correlation coefficient (R2) between the mean incubation time for each mouse strain and the mean normalized array signal for each probe was calculated in Excel (Microsoft). Estimates of enrichment beyond chance were obtained by permuting the data 1,000 times and recalculating the R2 values.
Generation of Tg222 Transgenic Mice.
The ORF of mouse Hspa13 was amplified from C57BL/6 mouse brain cDNA using Pfu DNA polymerase (Promega) with a forward (5′-ATGTCGACGGACTGATGGCCGGAGAGAT-3′) and reverse (5′-AACTCGAGGCACTCCCTCAGTTCAGTTGA-3′) primer containing a SalI and XhoI restriction site, respectively (underlined). The PCR was carried out using a PTC-225 (Bio-Rad) thermal cycler as follows: 95 °C for 2 min; 95 °C for 60 s, 60 °C for 30 s, 73 °C for 2 min for 40 cycles; and 73 °C for 5 min. The resulting product was gel purified (Gel Elution kit; Qiagen), blunt end ligated into SmaI-digested pSP72 vector (Promega), and sequenced to confirm that no errors had been incorporated. The Hspa13 ORF was excised with a SalI/XhoI double digest, gel purified, and ligated into the cosmid vector SHaCosTt cut with SalI (39). Preparation of high-quality DNA and insert was as previously described (40) and was microinjected into FVB/NHsd eggs (41). Eggs were cultured to the two-cell stage and surgically transferred to F1 (CBA × C57BL/6) recipient females. A single founder (Tg222) was identified by real-time PCR screening of DNA extracted from tail biopsies using primers and probes specific to the SHaCosTt vector (forward primer 5′-GGGAGAGATGGTTAGGACACAAA-3′, reverse primer 5′-GGGCATGACCTAATTGTGACTTT-3′ and probe 5′Fam-TCACGGCGCTTGGCGTTTCTTC-Tamra) on a 7500 fast real-time PCR system (Applied Biosystems) using rodent GAPDH endogenous control (Applied Biosystems) and Taqman Gene Expression Master Mix (Applied Biosystems). Reactions were carried out in a total volume of 15 μL using 6 pmoles of each primer and 3 pmoles of probe and 20 ng of DNA. Cycling conditions were as follows: 95 °C for 10 min; 95 °C for 15 s, 60 °C for 60 s for 35 cycles. Mice were bred to homozygosity and screened using the quantitative PCR described above.
Prion Strains.
Inocula were made from the brains of terminally sick mice as 1% (wt/vol) homogenates in PBS. The Chandler/RML prion strain was originally obtained from A. Aguzzi (Institute of Neuropathology, University of Zurich, Zurich, Switzerland) and amplified by passage in CD-1 Swiss mice (I9900). ME7 was obtained from the Institute for Animal Health, United Kingdom and further passaged in C57BL/6JOlaHsd mice (I9459). The MRC2 prion strain (I9468) is derived from a pool of five brainstems from naturally occurring cases of BSE (Veterinary Laboratories Agency, United Kingdom) passaged twice in C57BL6/JOlaHsd and once in SJL/OlaHsd mice (20).
Prion Inoculation and Phenotyping.
Mice were anesthetized with isofluorane/O2 and inoculated intracerebrally into the right parietal lobe with 30 μL of a 1% prion-infected brain homogenate as previously described (13). Mice were examined daily for neurological signs of prion disease and were culled once a definitive diagnosis had been made or earlier if showing signs of distress. Diagnostic criteria for clinical prion disease were as previously described (42). Incubation time was calculated retrospectively and defined as the number of days from inoculation to the onset of clinical signs. All procedures were conducted in accordance with institutional, United Kingdom, and international regulations and standards on animal welfare and conform to the Animal Research: Reporting in Vivo Experiments (ARRIVE) guidelines (43). Experiments were approved by the MRC Prion Unit Ethics Committee and carried out under UK Home Office license PPL 70/6454.
Immunohistochemistry.
Mouse brains were fixed in 10% (vol/vol) buffered formal saline (BFS) and prion-infected tissue was treated by incubation in 98% formic acid for 1 h to remove infectivity. Tissues were paraffin wax embedded, sectioned, and stained as previously described (42). Sections were stained with hematoxylin and eosin (H&E) for general examination and determination of spongiosis and neuronal loss. Prion deposition was visualized with anti-PrP monoclonal antibody ICSM35 (D-Gen) and gliosis was determined with an anti-glial fibrillary acid protein (anti-GFAP) antibody (Dako).
Cell-Based Prion Curing Assay.
Chronically prion-infected N2a cells (IPKS7 subclone) were grown for 1 d in OPTIMEM (Life Technologies) supplemented with 10% (vol/vol) FCS and 1% penicillin/streptomycin (44). The medium was then changed to Accell siRNA delivery media (Thermo Fisher) supplemented with 1.5% (vol/vol) FCS and 1 μM Accell SMART pool siRNA (Thermo Fisher) and cells were allowed to grow for 3 d (n = 6 per siRNA). Cells were split 1:6 and the Accell media and reagents reapplied. This was repeated after a further 3 d of growth. Following the third application of siRNA, cells were grown for 3 d and ∼25,000 cells plated for quantification of PrPSc positive cells by ELISPOT assay (24). The remainder of the cells were used for quantitative RT-PCR using a Cell-to-CT kit (Life Technologies) according to the manufacturer’s instructions. mRNA knockdown was quantified using Prnp and Hspa13 Taqman Gene Expression assays (Life Technologies) with GAPDH endogenous control (Life Technologies).
Statistical Analysis.
Statistical tests were carried out using GraphPad InStat and SPSS (IBM). The Kaplan–Meier log-rank test was used to analyze survival data.
Supplementary Material
Acknowledgments
We are grateful to Jackie Linehan and her staff for histology, Huda Al-Doujaily and Jonathan Wadsworth for preparation of inocula, Michelle Smidak for preparation of cosmid DNA for microinjection, Olivia Sheppard for genotyping Tc1 mice, the Biological Services Facility staff for animal care, and Ray Young for preparation of figures. Hybridization and scanning of GeneChips were carried out by University College London Scientific Support Services. V.L.J.T. was supported by the Medical Research Council (Programme U117527252). This work was funded by the Medical Research Council, United Kingdom.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The array data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38205).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208917109/-/DCSupplemental.
References
- 1.Collinge J. Prion diseases of humans and animals: Their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Collinge J, Palmer MS, Dryden AJ. Genetic predisposition to iatrogenic Creutzfeldt-Jakob disease. Lancet. 1991;337:1441–1442. doi: 10.1016/0140-6736(91)93128-v. [DOI] [PubMed] [Google Scholar]
- 3.Mead S, et al. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science. 2003;300:640–643. doi: 10.1126/science.1083320. [DOI] [PubMed] [Google Scholar]
- 4.Palmer MS, Dryden AJ, Hughes JT, Collinge J. Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature. 1991;352:340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 5.Collinge J. Molecular neurology of prion disease. J Neurol Neurosurg Psychiatry. 2005;76:906–919. doi: 10.1136/jnnp.2004.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mead S, et al. Genetic risk factors for variant Creutzfeldt-Jakob disease: A genome-wide association study. Lancet Neurol. 2009;8:57–66. doi: 10.1016/S1474-4422(08)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore RC, et al. Mice with gene targetted prion protein alterations show that Prnp, Sinc and Prni are congruent. Nat Genet. 1998;18:118–125. doi: 10.1038/ng0298-118. [DOI] [PubMed] [Google Scholar]
- 8.Westaway D, et al. Distinct prion proteins in short and long scrapie incubation period mice. Cell. 1987;51:651–662. doi: 10.1016/0092-8674(87)90134-6. [DOI] [PubMed] [Google Scholar]
- 9.Carlson GA, et al. Genetics and polymorphism of the mouse prion gene complex: Control of scrapie incubation time. Mol Cell Biol. 1988;8:5528–5540. doi: 10.1128/mcb.8.12.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson GA, Ebeling C, Torchia M, Westaway D, Prusiner SB. Delimiting the location of the scrapie prion incubation time gene on chromosome 2 of the mouse. Genetics. 1993;133:979–988. doi: 10.1093/genetics/133.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd SE, et al. Identification and characterization of a novel mouse prion gene allele. Mamm Genome. 2004;15:383–389. doi: 10.1007/s00335-004-3041-5. [DOI] [PubMed] [Google Scholar]
- 12.Manolakou K, et al. Genetic and environmental factors modify bovine spongiform encephalopathy incubation period in mice. Proc Natl Acad Sci USA. 2001;98:7402–7407. doi: 10.1073/pnas.121172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd SE, et al. Identification of multiple quantitative trait loci linked to prion disease incubation period in mice. Proc Natl Acad Sci USA. 2001;98:6279–6283. doi: 10.1073/pnas.101130398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd SE, et al. HECTD2 is associated with susceptibility to mouse and human prion disease. PLoS Genet. 2009;5:e1000383. doi: 10.1371/journal.pgen.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otterson GA, et al. Stch encodes the ‘ATPase core’ of a microsomal stress 70 protein. EMBO J. 1994;13:1216–1225. doi: 10.1002/j.1460-2075.1994.tb06371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Funez P, et al. In vivo generation of neurotoxic prion protein: Role for hsp70 in accumulation of misfolded isoforms. PLoS Genet. 2009;5:e1000507. doi: 10.1371/journal.pgen.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma D, Masison DC. Hsp70 structure, function, regulation and influence on yeast prions. Protein Pept Lett. 2009;16:571–581. doi: 10.2174/092986609788490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd S, Collinge J. Genetic susceptibility to prion diseases in humans and mice. Curr Genomics. 2005;6:1–11. [Google Scholar]
- 19.O’Doherty A, et al. An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science. 2005;309:2033–2037. doi: 10.1126/science.1114535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd SE, et al. Characterization of two distinct prion strains derived from bovine spongiform encephalopathy transmissions to inbred mice. J Gen Virol. 2004;85:2471–2478. doi: 10.1099/vir.0.79889-0. [DOI] [PubMed] [Google Scholar]
- 21.Fischer M, et al. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 22.Büeler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 23.Wadsworth JD, et al. Phenotypic heterogeneity in inherited prion disease (P102L) is associated with differential propagation of protease-resistant wild-type and mutant prion protein. Brain. 2006;129:1557–1569. doi: 10.1093/brain/awl076. [DOI] [PubMed] [Google Scholar]
- 24.Klöhn PC, Stoltze L, Flechsig E, Enari M, Weissmann C. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc Natl Acad Sci USA. 2003;100:11666–11671. doi: 10.1073/pnas.1834432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White MD, et al. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci USA. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hovatta I, et al. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature. 2005;438:662–666. doi: 10.1038/nature04250. [DOI] [PubMed] [Google Scholar]
- 27.Hwang D, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamgüney G, et al. Genes contributing to prion pathogenesis. J Gen Virol. 2008;89:1777–1788. doi: 10.1099/vir.0.2008/001255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheppard O, et al. Altered regulation of tau phosphorylation in a mouse model of Down syndrome aging. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2011.06.025. 33:828e31–828e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epstein CJ, Foster DB, DeArmond SJ, Prusiner SB. Acceleration of scrapie in trisomy 16----diploid aggregation chimeras. Ann Neurol. 1991;29:95–97. doi: 10.1002/ana.410290117. [DOI] [PubMed] [Google Scholar]
- 31.Otterson GA, Kaye FJ. A ‘core ATPase’, Hsp70-like structure is conserved in human, rat, and C. elegans STCH proteins. Gene. 1997;199:287–292. doi: 10.1016/s0378-1119(97)00383-1. [DOI] [PubMed] [Google Scholar]
- 32.Hetz CA, Soto C. Stressing out the ER: A role of the unfolded protein response in prion-related disorders. Curr Mol Med. 2006;6:37–43. doi: 10.2174/156652406775574578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyle KM, et al. Unfolded proteins and endoplasmic reticulum stress in neurodegenerative disorders. J Cell Mol Med. 2011;15:2025–2039. doi: 10.1111/j.1582-4934.2011.01374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreiro E, Resende R, Costa R, Oliveira CR, Pereira CM. An endoplasmic-reticulum-specific apoptotic pathway is involved in prion and amyloid-beta peptides neurotoxicity. Neurobiol Dis. 2006;23:669–678. doi: 10.1016/j.nbd.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Fuhrmann M, et al. Loss of the cellular prion protein affects the Ca2+ homeostasis in hippocampal CA1 neurons. J Neurochem. 2006;98:1876–1885. doi: 10.1111/j.1471-4159.2006.04011.x. [DOI] [PubMed] [Google Scholar]
- 36.Yamagata N, Furuno K, Sonoda M, Sugimura H, Yamamoto K. Stomach cancer-derived del223V-226L mutation of the STCH gene causes loss of sensitization to TRAIL-mediated apoptosis. Biochem Biophys Res Commun. 2008;376:499–503. doi: 10.1016/j.bbrc.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Umberti D, et al. TRAIL is expressed in the brain cells of Alzheimer's disease patients. Neuroreport. 2003;15:579–581. doi: 10.1097/00001756-200403220-00002. [DOI] [PubMed] [Google Scholar]
- 38.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004 doi: 10.2202/1544-6115.1027. 3:Article 3. [DOI] [PubMed] [Google Scholar]
- 39.Scott M, et al. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 40.Asante EA, et al. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J. 2002;21:6358–6366. doi: 10.1093/emboj/cdf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1994. [Google Scholar]
- 42.O’Shea M, et al. Investigation of mcp1 as a quantitative trait gene for prion disease incubation time in mouse. Genetics. 2008;180:559–566. doi: 10.1534/genetics.108.090894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enari M, Flechsig E, Weissmann C. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc Natl Acad Sci USA. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.