Abstract
Aggregation of p53 is initiated by first-order processes that generate an aggregation-prone state with parallel pathways of major or partial unfolding. Here, we elaborate the mechanism and explore its consequences, beginning with the core domain and extending to the full-length p53 mutant Y220C. Production of large light-scattering particles was slower than formation of the Thioflavin T-binding state and simultaneous depletion of monomer. EDTA removes Zn2+ to generate apo-p53, which aggregated faster than holo-p53. Apo-Y220C also aggregated by both partial and major unfolding. Apo-p53 was not an obligatory intermediate in the aggregation of holo-p53, but affords a parallel pathway that may be relevant to oncogenic mutants with impaired Zn2+ binding. Full-length tetrameric Y220C formed the Thioflavin T-binding state with similar rate constants to those of core domain, consistent with a unimolecular initiation that is unaffected by neighboring subunits, but very slowly formed small light-scattering particles. Apo-Y220C and aggregated holo-Y220C had little, if any, seeding effect on the initial polymerization of holo-Y220C (measured by Thioflavin T binding), consistent with initiation being a unimolecular process. But apo-Y220C and aggregated holo-Y220C accelerated somewhat the subsequent formation of light-scattering particles from holo-protein, implying coaggregation. The implications for cancer cells containing wild-type and unstable mutant alleles are that aggregation of wild-type p53 (or homologs) might not be seeded by aggregated mutant, but it could coaggregate with p53 or other cellular proteins that have undergone the first steps of aggregation and speed up the formation of microscopically observable aggregates.
Keywords: kinetics, amyloid, folding, misfolding
The kinetics of aggregation of the core domain of the oncogenic p53 mutant Y220C is most unusual, as it does not appear to follow the conventional nucleation-growth mechanism. (1) The aggregation of p53 differs from that most frequently studied for other proteins (2–4): the kinetics is deceptively simple, fitting a simple sequential scheme of A → B → C with two first-order rate steps; the aggregate is amorphous, and not well-formed amyloid fibrils; and aggregation is fast and easily studied by continuous spectroscopic measurements over minutes, rather than hours or days. (1) We suggest that there is rate-determining first-order formation of an aggregation prone intermediate followed by fast polymerization events prior to very slow formation of light scattering large aggregates. That is, there is not rate-determining formation of an oligomeric nucleus, but initiation is unimolecular.
Here, we explore the consequences of the suggested kinetic mechanism of aggregation of the core domain of Y220C, which might be a paradigm for amorphous aggregation, and extend the studies to full-length protein p53 (Flp53Y220C) with the Y220C mutation. Full-length p53 is a multidomain tetrameric protein with two folded domains: the core domain extending from residues 94–312 and the tetramerization domain (Fig. S1). (5) The core domain is flanked by intrinsically disordered sequences and generally behaves the same in the tetramer as when expressed as individual monomers, including sharing the same melting temperatures that are affected the same way by mutation. (6) The presence of neighboring subunits could affect aggregation kinetics (7).
We investigate the role of the apo-protein and the loss of Zn2+ ions. Zn2+ is important for the structural stability of p53 (8). Loss of Zn2+ on chelation by EDTA gives apo-p53, which is destabilized by 3.2 kcal/mol (9). The isolated apo-protein has some residual native NMR signals, but is reported to aggregate much faster than does the holo-protein and nucleates its aggregation (10). The unstable R175H mutant more rapidly loses its crucial Zn2+ ligand and is proposed to nucleate the aggregation of wild-type p53 core domain in vitro, accounting for the phenomenon of negative dominance (10). We propose an expanded mechanism that involves the first-order formation of an aggregation competent state that rapidly polymerizes to give an oligomer that binds Thioflavin T (ThT), which then aggregates further to form large light-scattering particles. A consequence of the mechanism, which might have biological relevance, is that there is not an initial seeding of the aggregation of p53 by the apo-protein but a later aggregation to form larger particles.
Results
Basic Kinetics.
Depletion of monomer from solution.
Aggregation, as monitored by ThT fluorescence, proceeds according to equation 3 of the accompanying paper (1):
| [1] |
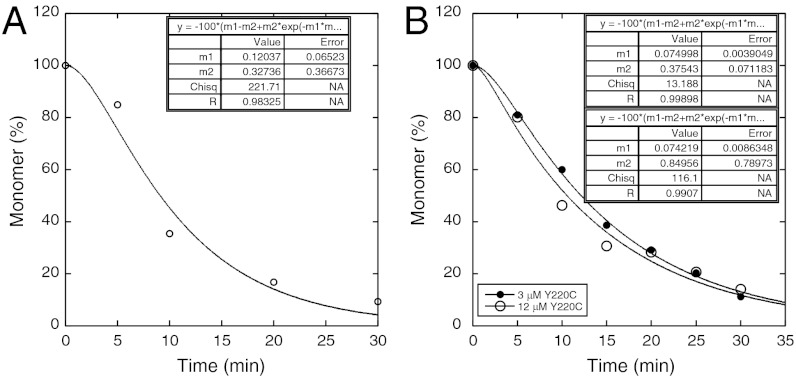
(where Ft is the intensity of ThT fluorescence at time t, and m, the amplitude, = [A]0f, where f is the specific fluorescence of ThT bound to the aggregate) with rate constants of 0.066 ± 0.002 and 0.36 ± 0.01 min-1, the first step being the slower, and k3 close to 0. (1) We monitored the depletion of monomeric protein, At + Bt, from solution by sampling the supernatant after centrifuging at different times, t, followed by measurement by either quantitative gel-electrophoresis or gel-filtration (Fig. S2). Loss of monomeric protein also followed lag kinetics, as expected since Ct = A0 - At–Bt, and so dCt/dt = -d(At + Bt)/dt (Fig. 1). The rate constants for the best data set (3 μM protein in Fig. 1B) were 0.38 ± 0.08 and 0.075 ± 0.004 min-1, in agreement with the ThT data, and the others were within experimental error. The rate constants did not materially change between 3 and 12 μM protein. Accordingly, the rate of loss of monomer equates to the rate of growth of the ThT-binding polymer. Crosslinking the samples prior to centrifugation did not noticeably alter the time course (Fig. S2). Loss of monomer is concomitant with the growth of the ThT-binding species.
Fig. 1.
Soluble p53 monomer remaining in supernatant during aggregation of Y220C at 37 °C. (A) Monomer peak from gel filtration assay. (B) SDS-PAGE stained by Sypro Orange for aggregation initiated from 3 μM (Left) and 12 μM (Right) Y220C. Monomer depletion curves obtained by (C), gel filtration assay, and (D), SDS-PAGE assay fitted to Eq. 1.
Effects of EDTA on aggregation.
p53 binds Zn2+ ions strongly. (11) In the absence of EDTA, Zn2+ ions remained bound to the aggregate: the supernatant from centrifuging the solution after 24 h contained < 2.5% of the Zn2+ ions and the aggregate retained 100 ± 10%. The tris(2-carboxyethyl)phosphine hydrochloride (TCEP), used as an antioxidant in the buffers, is a weak Zn2+ chelator, but altering the concentration of TCEP from 0.5 to 1.5 mM had no effect on rates.
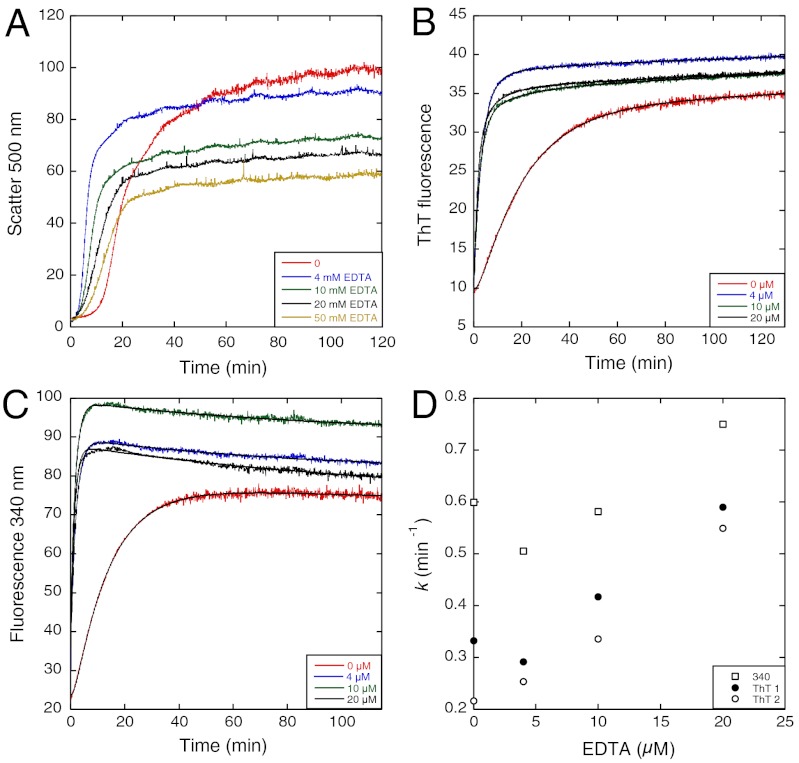
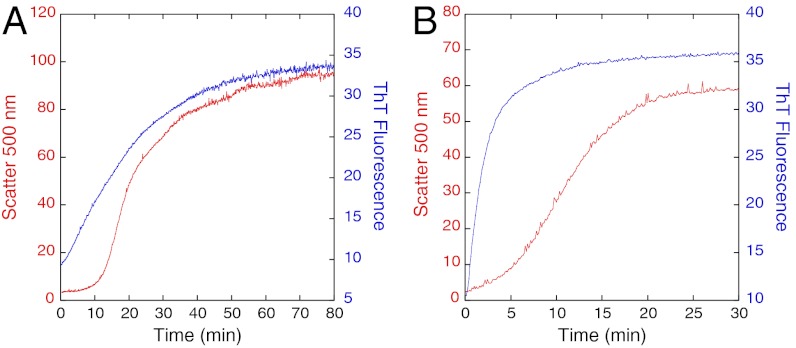
The addition of 4–20 mM EDTA caused the lag phase in light scattering to be significantly reduced (Fig. 2A). The lag in the ThT and 340 nm assays disappeared (Fig. 2 B and C), leaving a major exponential phase with rate constants of approximately 0.3–0.7 min-1, respectively, which were close to the values of the fast phase in the absence of EDTA (Fig. 2D). In the absence of EDTA (Fig. 3A), binding of ThT appears to precede light scattering; at 20 mM EDTA (Fig. 3B), ThT binding was near complete at the early stages of scattering. The effects of EDTA showed clearly that the structural events that lead to the binding of ThT precede the formation of large scattering particles.
Fig. 2.
Effect of EDTA on aggregation kinetics of Y220C at 37 °C. (A) Light-scattering assay shows shorter lag phase in the presence of EDTA. (B) ThT fluorescence and (C) fluorescence at 340 nm indicate faster aggregation with higher concentrations of EDTA. (D) Rate constant values obtained by ThT (duplicate runs) and 340 nm fluorescence versus [EDTA].
Fig. 3.
Effects of EDTA on light scattering and ThT fluorescence kinetics of 3 μM Y220C at 37 °C. (A) In the absence of EDTA, ThT appears to precede growth of light scattering. (B) Addition of 20 mM EDTA shows clearly ThT binding preceding light scattering.
ApoY220Cp53.
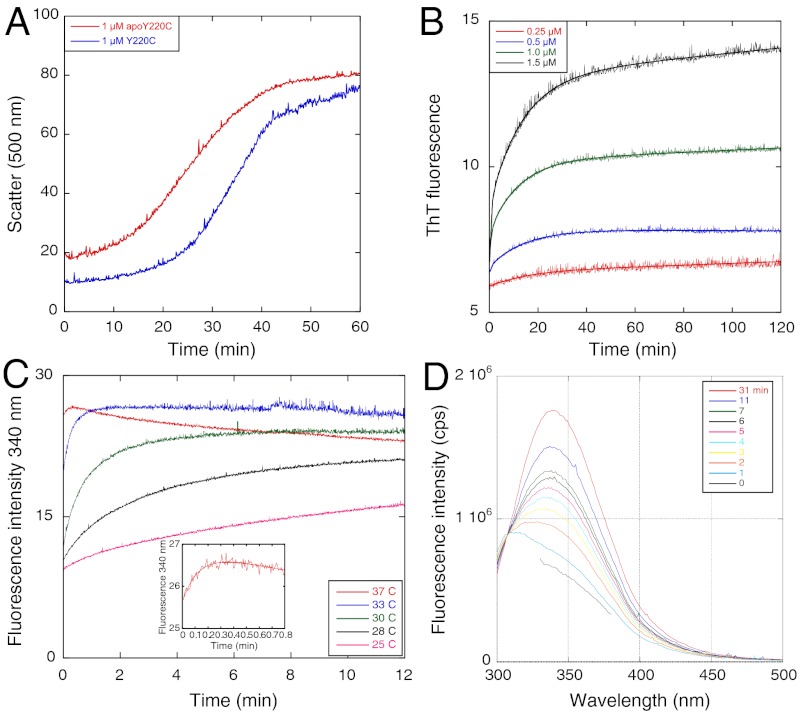
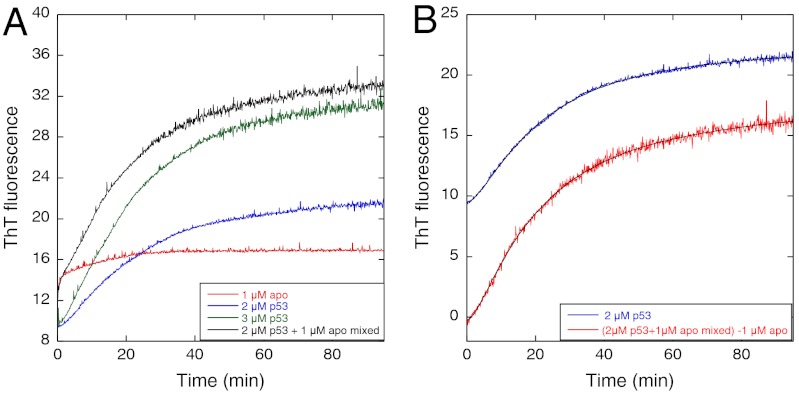
Given the previous suggestion of apop53 being an intermediate in the aggregation of p53 (10) and the acceleration of aggregation by EDTA, we prepared apoY220C to compare its aggregation kinetics with that of the holo-protein and the effects of EDTA. A solution of 1 μM apoY220C had a shorter lag but similar time course to Y220C, measured by light scattering at 37 °C (Fig. 4A). ThT binding preceded significant light scattering (compare 1 μM protein in Fig. 4 A and B). The ThT-monitored kinetics of aggregation of apop53 fitted a double exponential increase (with a small linear drift term, Eq. 2, where m are arbitrary amplitudes, Fig. 4B)
| [2] |
with high precision, rather than Eq. 1. The rate constants for 0.5–1.5 μM protein were 0.079 ± 0.005 min-1 and 1.5 ± 0.2 min-1, the amplitude of the fast phase increasing from 16–36% of the total change as concentration increased from 0.5–1.5 μM, and was undetectable at 0.25 μM. At 33 °C, there were similar rate constants with about 50% amplitude for the fast phase, independent of concentration. The corresponding rate constants for the holo-protein are 0.064 ± 0.0016 min-1 and 0.32 ± 0.03 at 3 μM protein, for kinetics that is sequential rather than biphasic.
Fig. 4.
Aggregation kinetics of apop53 Y220C. (A) Light scattering shows much shorter lag phase for apo than for holoY220C at 37 °C. (B) Aggregation of apoY220C at different concentrations monitored by ThT fluorescence at 37 °C. (C) Aggregation of apoY220C at different temperatures monitored by fluorescence at 340 nm. (D) Time-resolved fluorescence spectra of apo at 25 °C.
The kinetics monitored by fluorescence at 340 nm became uncoupled from and faster than that monitored by ThT (Fig. 4 B and C). At 37 °C, the half-life was 6 s, which increased with decreasing temperature. Spectra recorded at 25 °C showed a move to a maximum emission at 340 nm, with an isoemission wavelength of 307 nm.
Gel electrophoresis studies were consistent with the uncoupling of 340-nm changes and ThT binding (Fig. S3). Incubation of apoY220C at 37 °C followed by low speed centrifugation of samples (Fig. S3B) showed that loss of soluble protein, as measured by protein appearing at the monomer position on SDS gels, occurred at 0.076 ± 0.005 min-1 (Fig. S3B, single run), close to the value of 0.079 ± 0.005 min-1 for the slow phase in the binding of ThT (Fig. 4). But crosslinking prior to centrifugation showed that monomeric protein was rapidly lost with a rate constant of 1 min-1 or greater (Fig. S3A). Clearly, there are soluble oligomers present at early time that are not removed by low-speed centrifugation and are dissociated by SDS, but are fixed by crosslinking. This was not observed for the holoY220C (Fig. 1 and Fig. S2), where loss of monomeric protein corresponds to its incorporation into insoluble aggregate and where 340-nm fluorescence changes are coupled to ThT binding. It is most likely that the changes in 340-nm fluorescence correspond to the formation of small oligomers, whereas ThT binds tightly to larger, more irreversibly formed, larger polymers. The formation of small oligomers is sufficiently faster from apoY220C that they accumulate and can be detected.
Seeding of Aggregation by apoY220C.
The apoY220C has slow phases for binding of ThT and for light scattering that suggest that it could be on the pathway for aggregation of Y220C. We investigated the apoprotein’s being a potential intermediate by examining the effects of its seeding the aggregation of the holoprotein. These experiments were performed at very high concentrations of apoY220C, well above the quantities usually required for seeding, and are more a measure of coreaction.
Seeding of light-scattering particles.
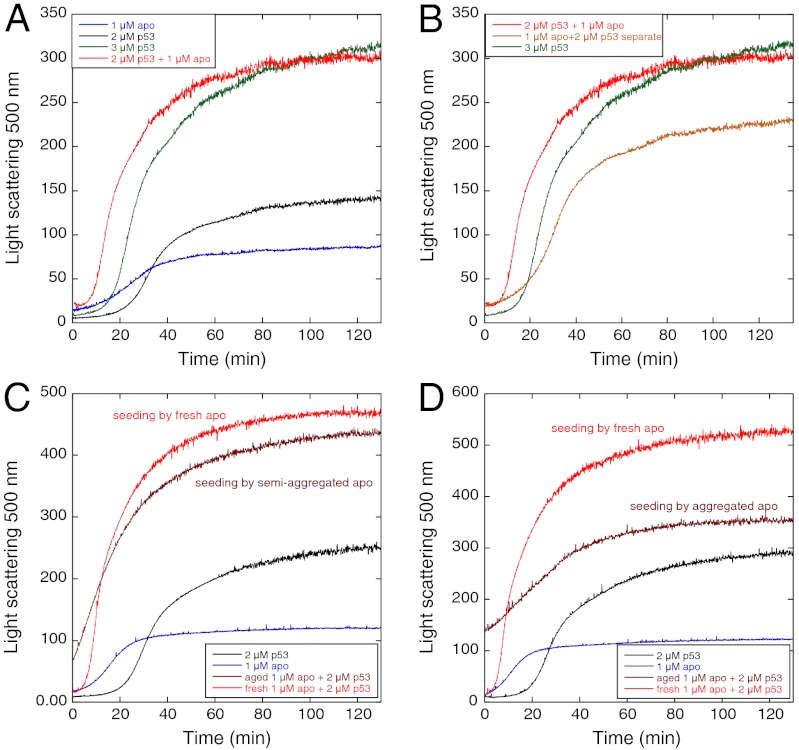
The aggregation of Y220C, as monitored by light scattering at 37 °C, was seeded by apoY220C (Fig. 5). Adding 2 μM Y220C to freshly prepared 1 μM apo had a similar time course to that for the aggregation of 3 μM Y220C, but with a shorter lag time (Fig. 5B). Further, the aggregation was faster and gave a higher value of light-scattering intensity than the sum of the scattering intensities of 2 μM Y220C and 1 μM apo monitored separately, suggesting that the two, together, coaggregate (Fig. 5B). Aggregated apo seeded the aggregation, but with different kinetics. Adding Y220C to apo that had been incubated at 37 °C for 23 min (and was not fully aggregated) eliminated the lag phase (Fig. 5C). Adding Y220C to apo that had been incubated at 37 °C for 150 min (which was fully aggregated) gave qualitatively different kinetics and smaller scattering particles (Fig. 5D).
Fig. 5.
Seeding effect of apo to holoY220C by light scattering at 37 °C. (A) Addition of fresh apo reduced the lag phase for aggregation of Y220C. (B) Aggregation of 1 μM apo and 2 μM holoY220C mixture was much faster than 3 μM Y220C and the sum of separate signals of 1 μM apo and 2 μM holoY220C. Addition of (C), 23-min aged apo,and (D), 150-min aged apo eliminated the lag phase.
ThT binding.
High concentrations of apoY220C had only minor, if any, effects on the kinetics of aggregation of Y220C, as monitored by ThT kinetics (Figs. 6 and 7A). The change in fluorescence of a mixture of 2 μM Y220C and 1 μM apoY220C was similar to that of 3 μM Y220C. Further, the time course of fluorescence of the mixture of 2 μM Y220C and 1 μM apoY220C, from which that of 1 μM apoY220C was subtracted (Fig. 6B), had near identical lag kinetics to that of 2 μM Y220C alone (0.048 ± 0.001 and 0.31 ± 0.02 min-1 for 2 μM Y220C versus 0.050 ± 0.002 and 0.38 ± 0.03 min-1 on addition of 1 μM apo). Thus, as far as ThT binding is concerned, apo and holo proteins act independently.
Fig. 6.
Effects of apoY220C on holoY220C monitored by ThT fluorescence at 37 °C. (A) Aggregation of 1 μM apo and 2 μM Y220C mixture shows similar trace to that of 3 μM Y220C. (B) Signal of 1 μM apo and 2 μM Y220C combined mixture minus that of 1 μM apo has lag phase similar to that of 2 μM Y220C.
Fig. 7.
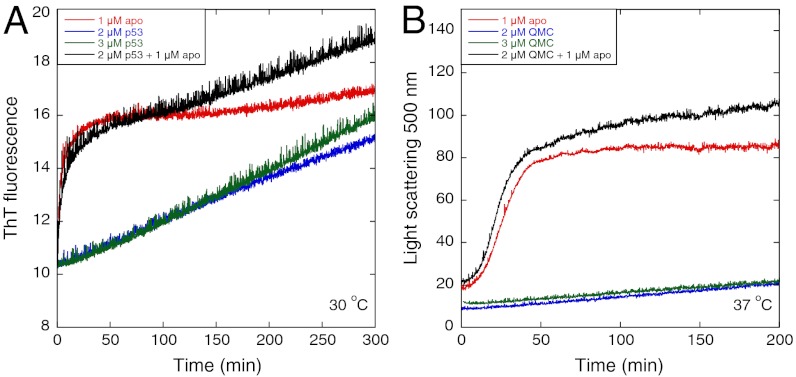
Seeding effect of apoY220C on p53 variants under stable conditions. (A) apoY220C showed little seeding effect on Y220C at 30 °C. (B) apoY220C had little seeding effect on stabilized quadruple-mutant core domain at 37 °C.
Undetectable seeding of p53 variants under more stable conditions.
Consistent with apoY220C not seeding the initial events in aggregation, we found that under conditions where Y220C is relatively stable (30 °C, Fig. 7A), apoY220C did not seed its aggregation. Similarly, the aggregation of a p53 mutant with Tm of 50 °C was not seeded at 37 °C (Fig. 7B). In both cases, the time courses for the mixture of apo and holo protein were the sum of the separate time courses.
Inhibition of Aggregation of apoY220C by Ligands.
Addition of the ligand 5174, which binds to Y220C, (1, 12) did not inhibit the rate of increase of ThT fluorescence at 37 °C. At 30 °C, the rate constants for fluorescence increase at 340 nm were inhibited by 5174, with a dissociation constant of 17.2 ± 1.6 μM, similar to the value of 16 μM for dissociation from the holo-protein (1) at 30 °C (Fig. S4). The ThT fluorescence increased clearly biphasically. The fast phase was close in rate constant to that for the 340 nm fluorescence, at 0.43 min-1, which was also inhibited by 5174 with a KD of 20 ± 2 μM. A slower phase of 45% of the total amplitude at 0.05 min-1 was not inhibited.
Competition Between Inhibition by Ligand Binding and EDTA Acceleration.
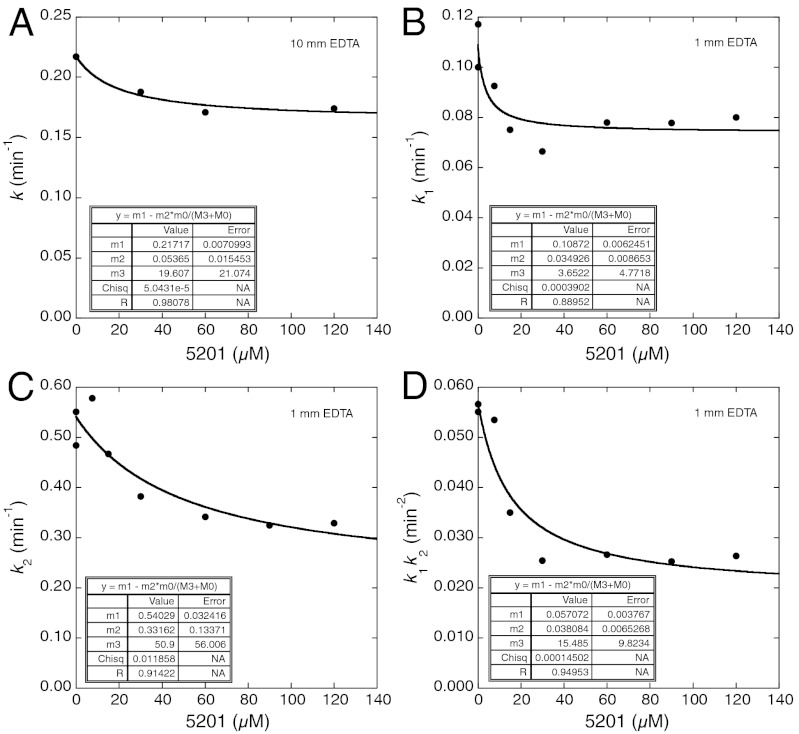
The dissociation constant of 5201 from Y220C in the absence of EDTA is 8 μM. (1) The single phase in the presence of 10 mM EDTA was only partly inhibited, with KD of approximately 20 μM (Fig. 8A). The ligand-free protein reacted with a rate constant of 0.22 min-1 and the ligand-bound at 0.16 min-1. The presence of 1 mM EDTA was not sufficient to cause the changeover from lag to single exponential kinetics, but increased k1 and k2 by about 2-fold. They were inhibited by 5201 approximately 40% and approximately 30%, respectively (Fig. 8 B and C). k1k2 was inhibited approximately 60% with a KD of 16 ± 10 μM (Fig. 8D). Again, there were parallel pathways of ligand-free and ligand-bound protein.
Fig. 8.
Inhibition of EDTA-accelerated aggregation kinetics of Y220C by ligand 5201 at 37 °C. (A) Inhibition of Y220C aggregation by 5201 in the presence of 10 mM EDTA. (B), (C), and (D) are inhibition of 5201 in the presence of 1 mM EDTA of k1, k2, and k1k2, obtained from ThT binding assays.
Full-Length p53Y220C.
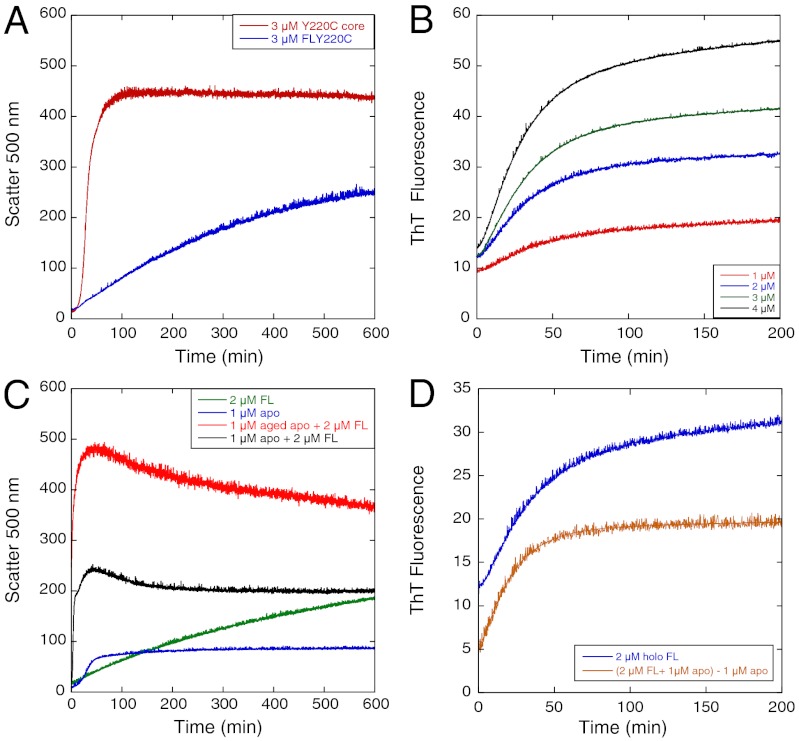
We compared full-length Y220C, which is predominantly tetrameric in the μM concentration range, (13) with the core domain (Fig. 9). A most notable difference is that the formation of light-scattering particles was much slower (Fig. 9A). But the rate constants for the formation of the ThT binding state were reduced by only 50% and were constant between 1–4 μM Flp53Y220C (as monomeric subunit concentrations) at 0.033 ± 0.001 and 0.18 ± 0.02 min-1 (Fig. 9B). The addition of 1 μM apoY220C had only minor effects on the ThT fluorescence curves (Fig. 9D), but greatly accelerated light scattering (Fig. 9C). Electron micrographs after 16 h incubation at 37 °C showed only small particles (Fig. S5), unlike the large amorphous particles seen for core domain (1). Like core domain, the Flp53Y220C forms a state that binds ThT in a first-order process. That state then slowly forms light-scattering particle.
Fig. 9.
Aggregation kinetics of full-length p53 Y220C at 37 °C. (A) Comparison of the aggregation kinetics of Y220C and Flp53Y220C. (B) ThT binding kinetics of Flp53Y220C at different concentrations. (C) Seeding effect of apoY220C on Flp53Y220C monitored by light scattering. (D) Seeding effect of apoY220C on Flp53Y220C monitored by ThT fluorescence.
ThT binding to polymerized Flp53Y220C was inhibited by 5201 (Fig. S6) analogously to that of Y220C. k1 decreased to about 50% of initial value, indicating that the ligand–bound protein was still able to aggregate.
It was proposed that tetrameric p53 aggregates faster than monomeric constructs. (7) But, as previously pointed out, (14) the construct used in that study is a truncated form, lacking the N-terminus, and is significantly destabilized with an exposed hydrophobic patch. Here, the full-length protein is more stable than the core domain.
Discussion
Construction of a Kinetic Mechanism.
The resolution of the kinetic mechanism of aggregation is complicated because there are parallel pathways of aggregation that depend on environmental conditions. A satisfactory kinetic mechanism for the amorphous aggregation of the p53 core domain of Y220C has to account for the series of following results.
The growth of polymer, as measured by binding of ThT, the 340-nm signal and directly measured depletion of monomer from solution follows similar apparent two-step unimolecular lag kinetics. The inhibition of ThT binding by ligands is incomplete, and levels off at finite values at saturating concentrations.
In the accompanying paper (1), we showed that the simplest scheme to account for those basic observations involves Y220C core domain converting by two-apparent first-order sequential rate constants to an aggregation competent state that rapidly aggregates to form a ThT-binding state. That process is not conventional nucleation-growth because there is not rate-determining formation of an oligomeric nucleus. The partial inhibition is explained by there being parallel pathways of aggregation from unfolded and ligand-bound folded states.
Effects of EDTA and the role of apoY220C.
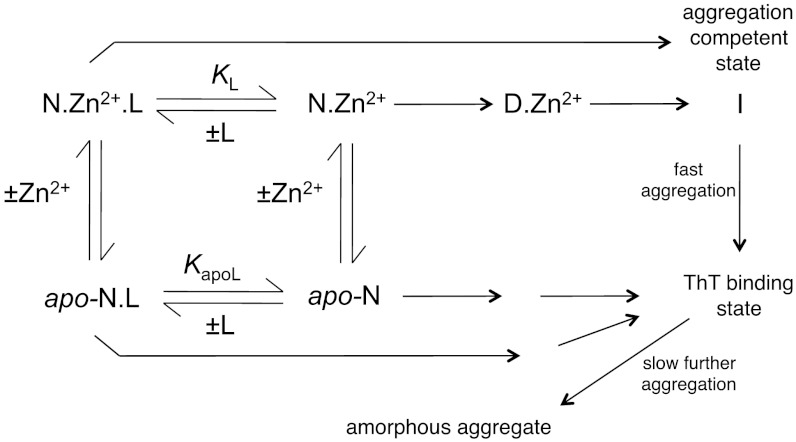
The two first-order rate constants for the ThT-monitored kinetics are 0.066 ± 0.002 and 0.36 ± 0.01 min-1 in the absence of EDTA. The addition of EDTA abolishes the slow phase to give mono-phasic kinetics of approximately 0.3 min-1. The simplest explanation at first sight would be that the slow step in aggregation involves the loss of Zn2+, which is enhanced by EDTA, perhaps by displacing an unfavorable equilibrium between the holoprotein and an apoprotein. Independently prepared apoY220C does indeed aggregate faster than Y220C, with biphasic rather than lag kinetics. But in the absence of EDTA, Zn2+ remains bound to the aggregate. It is possible that Zn2+ dissociates prior to aggregation and rebinds to an apo state. The weak chelator TCEP does not affect the rate, and so the simplest explanation is that there are two pathways for aggregation: in the absence of a Zn2+-chelator, the major route involves the holo-protein, and in the presence of the chelator, the mechanism changes to an apo-protein route.
The 340-nm signal on the initial events of apo-protein aggregation appears much faster than and is decoupled from ThT binding (Fig. 4). apo-protein forms soluble oligomers, with the 340-nm signal, that progress to the ThT binding state (Fig. S3). Presumably, the formation of the initial small oligomers from holo-protein is slower than their rate of incorporation into the ThT-binding large polymers and so 340-nm events and ThT-binding seem synchronous.
ApoY220C seeds light scattering of Y220C but not ThT-binding.
The formation of large light-scattering particles, as in Fig. 5, is a structural event that occurs after the formation of the ThT-binding state; the difference in time scale is seen most clearly in Fig. 3. The simplest explanation is that apo- and holo-proteins undergo the first stages of aggregation separately and then coaggregate to form large particles. The preformed aggregates do not seed the initial stages of aggregation. That conclusion is consistent with the first-order kinetics of aggregation.
Inhibition by ligands of ThT kinetics of apoY220C is only partial as is their effect with EDTA on Y220C.
Partial inhibition of apoY220C aggregation shows that, like for the inhibition of the holo-protein, there are parallel pathways for initiation of aggregation. One route involves the unfolding of the binding site, which is inhibited by 5201 (Fig. 8) in the EDTA-catalyzed reactions. The other retains the bound 5201, as suggested by the inhibition of the aggregation of apoY220C by 5174 (Fig. S4).
Roberts and colleagues (15, 16) have formulated a general Lumry–Eyring nucleated polymerization model to describe aggregation kinetics, which has some overlapping steps with Scheme 1. A key assumption in their analysis that dynamics of monomer conformational transitions is rapid compared with the rate-limiting steps in aggregation (i.e., the monomers are in a pre-equilibrium) does not apply to Scheme 1 because some conformational transitions between monomers are slow and were measured.
Scheme 1.
Aggregation of p53 Y220C with or without zinc. N.Zn2+.L, native holo-Y220C bound to ligand; N.Zn2+, native holo-Y220C; D.Zn2+, denatured holo-Y220C; I, aggregation-competent intermediate; apo-N.L, native apo-Y220C bound to ligand; apo-N, native apo-Y220C.
The results above are accommodated most simply by Scheme 1.
Structural Events Initiating Aggregation.
Xu et al (17) have demonstrated that residues 251–257 in β-strand 9 of the β-sandwich of p53 are amyloidogenic. These are buried in the protein and will be exposed on major unfolding. They are covered by residues around 94–100. Residue 94 is usually assumed to be the start of the core domain, but we have noted that extending the domain boundary to include Trp91 increases the stability of the core domain and lessens its propensity to aggregate. (14) Fraying of that N-terminal region could expose the amyloidogenic sequence and be the route for aggregation without major unfolding.
Relevance of Biophysical to Cellular Studies.
The preliminary studies on full-length protein showed the same unimolecular initiation of aggregation as the isolated core domain. Unimolecular rate determining initiation indicates that the dominant negative effect whereby an aggregation prone mutant seeds aggregation of wild-type protein is a late event in aggregation. It raises the possibility that the observed coaggregation of p53 with p63 and p73 (17) might follow rather than initiate early aggregation events, especially as the formation of large light-scattering particles is very slow. The apo-protein is not an obligatory intermediate in the aggregation of Y220C in the absence of a strong Zn2+ chelator. But mutants that have impaired Zn2+-binding, such as R175H that is the third most frequent oncogenic mutation, (18, 19) are likely to aggregate by the faster apo route in the cell.
There is a plasticity of mechanism of aggregation of p53, with parallel pathways and the effects of binding of Zn2+ ions. It is possible that there could be further pathways of aggregation, including classical nucleation-growth, which will be induced by different experimental conditions or mutations.
Methods
Protein Expression and Purification.
The T-p53-Y220C core domain protein (Y220C) was expressed and purified as described. (1) T-p53Y220C full-length p53 was similarly expressed and purified, except that the final gel filtration buffer was composed of 25 mM sodium phosphate, pH 7.2, 300 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and 10% glycerol. Apo T-p53-Y220C core domain (apoY220C) was produced based on the reported method (10) with modifications. Briefly, T-p53-Y220C was treated by 1/33 volume of 10% acetic acid and 1/100 volume of 0.5 M EDTA, incubated 20 min on ice, then 1.5 volume of 0.5 M bis-tris propane (pH 6.8) was quickly added to initiate the refolding of p53. ApoY220C was allowed to stand for 1 h to refold, and then purified by HiLoad 26/60 Superdex 75 column or Superdex 75 10/300 GL (GE Healthcare) to exchange buffer (to 25 mM sodium phosphate, pH 7.2, 150 mM NaCl, 1 mM TCEP) and remove unfolded/aggregated protein. Residual zinc in apoY220C was checked by 4-(2-pyridylazo)resorcinol) (200 μM) after addition of 625 μM methyl methanethiosulfonate (Sigma) to force release of zinc. Residual zinc was undetectable.
Kinetics of Denaturation and Aggregation and Electron Microscopy.
Light scattering, Thioflavin T assays, time-resolved fluorescence spectra, and electron microscopy were all carried out as described at 37 °C in 25 mM sodium phosphate, pH 7.2, 150 mM NaCl and 1 mM TCEP. (1) For experiments with Flp53Y220C, the final buffer contains 1% glycerol. Concentration of zinc was measured using 4-[2-pyridylazo)resorcinol].(20) Aliquots of Y220C aggregated at 37 °C for different times were centrifuged at 13,000 rpm for 30 min to remove the aggregate before assaying.
Measurement of Soluble Protein by Gel Filtration and SDS-PAGE.
Y220C, 3 μM and 12 μM, were incubated at 37 °C in 25 mM sodium phosphate, pH 7.2, 150 mM NaCl, and 1 mM TCEP. Samples were taken at different time points, kept on ice, and then centrifuged at 13,000 rpm for 30 min (4 °C) to remove aggregate. The supernatant was injected into Superdex 200 10/300 GL column (GE Healthcare), and monomer quantified by area integration of its peak. No oligomer peak could be detected. In addition, the supernatants were also run on a 4–12% denaturating NuPAGE Bis-Tris gel (Invitrogen), and experiments were repeated with 1 μM apoY220C. The gel was stained for 1 h with SYPRO®Orange protein gel stain (Invitrogen) in 7.5% (v/v) acetic acid, washed for 1 min in 7.5% (v/v) acetic acid, and scanned on a Typhoon TRIO Variable Mode Imager (GE Healthcare). Bands were quantified with ImageQuant TL software (GE Healthcare). The monomer depletion kinetics was also checked by quantifying the monomer in the gel of samples reacted with 20-fold excess of crosslinker, BS3 (Thermo Scientific), at 4 °C for 2 h.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211557109/-/DCSupplemental.
References
- 1.Wilcken R, Wang G, Boeckler FM, Fersht AR. Kinetic mechanism of p53 oncogenic mutant aggregation and its inhibition. Proc Natl Acad Sci USA. 2012;109:13584–13589. doi: 10.1073/pnas.1211550109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris AM, Watzky MA, Finke RG. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim Biophys Acta. 2009;1794:375–397. doi: 10.1016/j.bbapap.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 6.Ang HC, Joerger AC, Mayer S, Fersht AR. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J Biol Chem. 2006;281:21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- 7.Lubin DJ, Butler JS, Loh SN. Folding of tetrameric p53: Oligomerization and tumorigenic mutations induce misfolding and loss of function. J Mol Biol. 2010;395:705–716. doi: 10.1016/j.jmb.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Hainaut P, Milner J. A structural role for metal ions in the “wild-type” conformation of the tumor suppressor protein p53. Cancer Res. 1993;53:1739–1742. [PubMed] [Google Scholar]
- 9.Bullock AN, Henckel J, Fersht AR. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: Definition of mutant states for rescue in cancer therapy. Oncogene. 2000;19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 10.Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- 11.Bullock AN, et al. Thermodynamic stability of wild-type and mutant p53 core domain. Proc Natl Acad Sci USA. 1997;94:14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcken R, et al. Halogen-enriched fragment libraries as leads for drug rescue of mutant p53. J Am Chem Soc. 2012;134:6810–6818. doi: 10.1021/ja301056a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajagopalan S, Huang F, Fersht AR. Single-Molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res. 2011;39:2294–2303. doi: 10.1093/nar/gkq800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natan E, et al. Interaction of the p53 DNA-binding domain with its n-terminal extension modulates the stability of the p53 tetramer. J Mol Biol. 2011;409:358–368. doi: 10.1016/j.jmb.2011.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews JM, Roberts CJ. A Lumry–Eyring nucleated polymerization model of protein aggregation kinetics: 1. Aggregation with pre-equilibrated unfolding. J Phys Chem B. 2007;111:7897–7913. doi: 10.1021/jp070212j. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Roberts CJ. Lumry–Eyring nucleated-polymerization model of protein aggregation kinetics. 2. Competing growth via condensation and chain polymerization. J Phys Chem B. 2009;113:7020–7032. doi: 10.1021/jp8083088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 18.Palecek E, Ostatna V, Cernocka H, Joerger AC, Fersht AR. Electrocatalytic monitoring of metal binding and mutation-induced conformational changes in p53 at picomole level. J Am Chem Soc. 2011;133:7190–7196. doi: 10.1021/ja201006s. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-Specific p53 Mutant Reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt JB, Neece SH, Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.