Abstract
Although microbial metabolic engineering has traditionally relied on rational and knowledge-driven techniques, significant improvements in strain performance can be further obtained through the use of combinatorial approaches exploiting phenotypic diversification and screening. Here, we demonstrate the combined use of global transcriptional machinery engineering and a high-throughput L-tyrosine screen towards improving L-tyrosine production in Escherichia coli. This methodology succeeded in generating three strains from two separate mutagenesis libraries (rpoA and rpoD) exhibiting up to a 114% increase in L-tyrosine titer over a rationally engineered parental strain with an already high capacity for production. Subsequent strain characterization through transcriptional analysis and whole genome sequencing allowed complete phenotype reconstruction from well-defined mutations and point to important roles for both the acid stress resistance pathway and the stringent response of E. coli in imparting this phenotype. As such, this study presents one of the first examples in which cell-wide measurements have helped to elucidate the genetic and biochemical underpinnings of an engineered cellular property, leading to the total restoration of metabolite overproduction from specific chromosomal mutations.
Genetic modifications identified by the rational analysis of pathway stoichiometry, kinetics, and regulation (such as gene deletions/overexpressions and pathway deregulation) have been routinely used in metabolic engineering applications with remarkable success. Despite this, however, many important multigenic phenotypes still remain inaccessible through these methods. Even when pathways are well elucidated, unexpected disconnects between genotypes and predicted phenotypes often hamper the engineering of highly interconnected biological systems. Combinatorial metabolic engineering has emerged in recent years to address this issue using the concept and tools of phenotypic diversification and screening (1). Unlike traditional rational approaches, these methods require very little a priori knowledge of the metabolic network’s properties and interactions and instead depend on the type of diversity generated and the efficiency of the high-throughput assay used. One such technique, global transcription machinery engineering (gTME), has been shown to be particularly effective in introducing phenotypic diversity by reprogramming the cellular transcriptome (2). In Escherichia coli, mutating two plasmid-encoded components of the RNA polymerase—rpoD, encoding the principal sigma factor σ70, and rpoA, encoding the α subunit—led to improvements in a number of cellular traits, including ethanol tolerance, butanol tolerance, lycopene overproduction, and hyaluronic acid production (3, 4). Both polymerase subunits play natural roles in dictating rates of transcription through their interactions with the –10 and –35 regions of the promoter (rpoD) and upstream-promoter (UP) elements, activators, and repressors (rpoA) (Fig. S1), thus making them important knobs for modulating global transcriptional profiles and, by extension, for generating complex phenotypes.
Interest in developing a robust microbial production process for the amino acid L-tyrosine has remained high due to its use in a myriad of pharmaceutical and industrial applications. Apart from its common use as a dietary supplement (5, 6), L-tyrosine also serves as an important precursor for the Parkinson’s disease drug 3, 4-dihydroxy-L-phenylalanine (L-DOPA) (7), as well as melanin-based UV absorbers, cation exchangers, drug carriers, and amorphous semiconductors (8). In addition, both p-coumaric acid and p-hydroxystyrene can be synthesized directly from L-tyrosine and appear frequently as key components in several polymers, adhesives and coatings, pharmaceuticals, biocosmetics, and flavonoid product precursors (9–11). Work to-date on L-tyrosine and, more broadly, aromatic amino acid (AAA) production in E. coli has focused primarily on rational metabolic engineering strategies to alter regulation and increase pathway flux (12–15). As such, the use of less targeted combinatorial approaches such as gTME presents an as-yet untapped potential for optimizing this important cellular property.
In a recent publication, we described the development of a high-throughput screen for L-tyrosine production based on the accumulation of the dark pigment melanin (16). Here we present the combined application of this assay with gTME for improving L-tyrosine production over a rationally engineered parental strain. We successfully isolated three strains from two plasmid-based mutagenesis libraries (rpoA, rpoD) which exhibited significant increases in both L-tyrosine yields and titers. Unusually, the observed phenotypes were found to be dependent on both the plasmid-encoded rpoA/rpoD gene and unidentified mutations within the bacterial chromosome. Characterization of these isolates using cell-wide assays such as transcriptional analysis and whole genome sequencing allowed us to identify the exact origins for these improvements and, more significantly, reconstruct the phenotype through the introduction of specific chromosomal mutations. Evaluation of this reconstructed strain in large-scale fermentations yielded L-tyrosine titers of up to 13.8 g/l after 36 h at very high volumetric productivities.
Results
Rational Engineering of a Parental Strain.
We first sought to construct a high-performing parental strain through the introduction of rational modifications based on known properties of the AAA biosynthetic pathway (Fig. S2). In particular, prior work in our lab (17) helped us identify several strategies for overcoming key kinetic and regulatory limitations in the pathway: (i) deletion of tyrR to circumvent transcriptional regulation, (ii) deletion of pheA to eliminate the loss of AAA pathway intermediates to competing reactions, and (iii) overexpression of feedback resistant derivatives of 3‐deoxy‐D‐arabinoheptulosonate‐7‐phosphate (DAHP) synthase (aroGfbr) and chorismate mutase/prephenate dehydrogenase (tyrAfbr) to increase flux through the AAA pathway. To avoid potential issues associated with marker and plasmid origin incompatibilities and to eliminate the need for antibiotics and inducers, the tyrAfbr-aroGfbr operon was placed under constitutive control and integrated into the bacterial chromosome. Expression levels of these genes were optimized through the use of varied strength promoters (R, Y, W, B, PL) from a synthetic constitutive promoter library (Table S1) (18). However, because the L-tyrosine titers of these single integration strains still fell short of their plasmid-based counterparts (Fig. 1A), we also explored the integration of a second copy of tyrAfbr-aroGfbr under the promoters W, B, and PL. From this set of strains, P2, which contains two PL-driven copies of tyrAfbr-aroGfbr and achieves a titer of 418 mg/l L-tyrosine after 24 h, was selected as the starting strain for the construction of our gTME-derived libraries (Fig. 1B).
Fig. 1.
L-tyrosine production of rationally engineered strains after 24 h. (A) L-tyrosine production of E. coli K-12 ΔpheA ΔtyrR strains containing plasmid-based overexpression of the tyrAfbr-aroGfbr operon, a single integrated copy, or two chromosomal copies. Plasmid-based copies of tyrAfbraroGfbr under the control of the synthetic constitutive promoters (R, Y, W, B, PL) were provided on pZE-kanFRT-tyrAfbraroGfbr. (B) L-tyrosine production of wild-type E. coli K-12, a double deletion mutant (E. coli K-12 ΔpheA ΔtyrR), and parental strain P2.
Isolation and Sequence Analysis of gTME-derived Mutants.
In a recent publication, we described a high throughput screen for L-tyrosine production based on the tyrosinase-mediated conversion of L-tyrosine to the dark and diffusible pigment melanin (16). When implemented in an agar plate format, high L-tyrosine overproducers could be identified by the extent of colony pigmentation. We applied this simple visual assay to screen two separate gTME libraries based on the mutagenesis of the RNA polymerase subunits, rpoA and rpoD. Libraries consisting of 105–106 viable colonies were created by transforming plasmid-encoded rpoA and rpoD mutants into the rationally engineered parental strain P2. Our search led to the isolation of three strains (rpoA14, rpoA27, and rpoD3) exhibiting 91–113% increases in L-tyrosine production over parental P2 (Fig. 2A). Of note, final L-tyrosine concentrations, which ranged between 798 and 893 mg/L for the three isolates, are reported after 48 h due to the slower growth rates of these strains; however, growth and L-tyrosine production profiles show that levels reach their peaks after just 28 h (Fig. S3).
Fig. 2.
Properties of mutants isolated from gTME libraries. (A) L-tyrosine titers of isolated mutants after 48 h. (B) Sequence analysis of plasmid-encoded mutant rpoA and rpoD. Numbers above wild-type (WT) rpoD-encoded σ70 indicate conserved regions of the protein. NSH stands for the “nonstandard helix” portion of the rpoA-encoded α subunit’s C-terminal domain.
Sequence analysis of the mutant rpoA/rpoD plasmids revealed interesting mechanistic insights into transcriptional diversification for two of the three strains. Although the wild-type σ70 protein typically contains four conserved domains (19), the altered rpoD gene in strain rpoD3 encoded a truncated form comprising just the latter part of a D521E-substituted region 3 and region 4 (Fig. 2B). We hypothesize that overexpression of this shortened protein could result in anti-σ factor (such as Rsd) or RNA polymerase holoenzyme titration events which could affect transcriptional preferences. Sequencing of the mutant rpoA plasmid from strain rpoA14 revealed the presence of two amino acid substitutions (V257F, L281P) within the C-terminal domain of the α subunit (αCTD), in close proximity to residues involved with regulatory factor and UP element interactions (Fig. 2B) (20). The appearance of mutations in key secondary structural elements of the αCTD (the “nonstandard helix” and an α-helix, respectively) (21) suggests that structural deviations may shift the relative alignment of amino acid promoter contacts and, as a result, alter the affinity of the RNA polymerase enzyme for some of its targets. This theory is supported by other rpoA mutations and strains uncovered in our lab (4). Surprisingly, no changes were found in the promoter sequence or coding region of rpoA from strain rpoA27 (Fig. 2B), which suggests that plasmid-based overexpression of even the wild-type gene can lead to phenotypically-advantageous transcriptional modifications.
Requirement for and Interchangeability of Mutant Plasmids and Backgrounds.
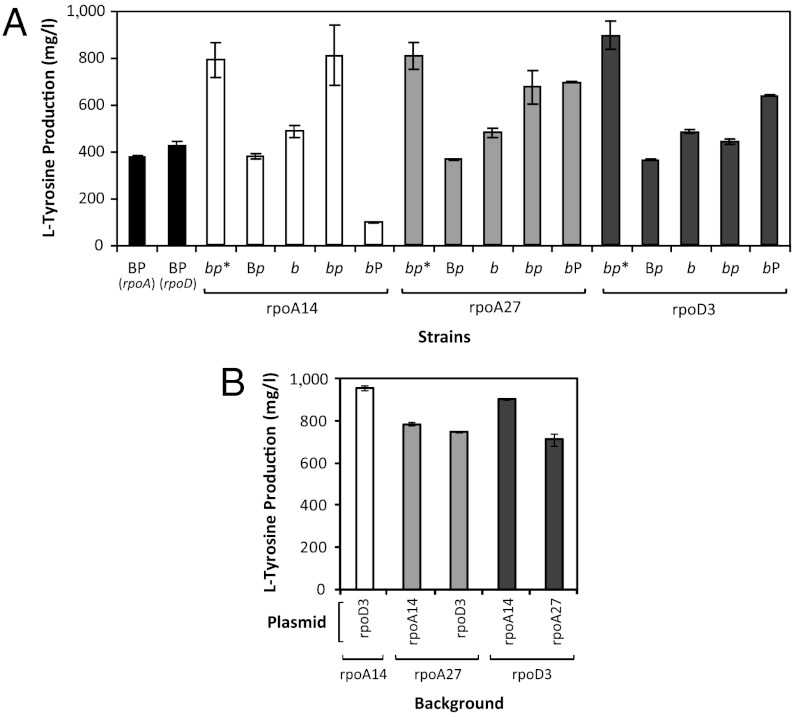
The recovery of a wild-type rpoA plasmid from rpoA27, which by itself has not been shown to be beneficial for L-tyrosine production, introduced the possibility that unidentified chromosomal mutations in the isolated strains may either partially or fully contribute to the observed phenotypes. To explore these scenarios, we constructed and tested a series of strains containing either the wild-type or mutated rpoA/rpoD plasmids in the backgrounds of the parental strain P2 or the plasmid-cured isolates (henceforth referred to as mutant or modified chromosomal backgrounds). We also assessed the individual contributions of the mutant rpoA/rpoD plasmids and mutant backgrounds on cellular phenotype. Unusually, we found that the L-tyrosine production properties of all three gTME strains were uniquely dependent on the presence of both the rpoA/rpoD plasmid and a modified chromosomal background (Fig. 3A), thus highlighting a possible synergism between the plasmid-encoded components and other altered proteins residing on the chromosome. Of note, with the exception of strain rpoA27, the presence of a wild-type or mutant rpoA/rpoD plasmid did not lead to an enhanced frequency of mutation when compared to the parental strain P2 (Fig. S4). We therefore assume that the high rate of chromosomal variation in our isolates arose out of a requirement for these specific background mutations to increase L-tyrosine production and, as a result, be identified by our screen.
Fig. 3.
Effect of background-plasmid combinations on L-tyrosine production (48 h). (A) Comparison of strains containing wild-type or mutant rpoA/rpoD plasmids and/or wild-type (P2) or modified chromosomal backgrounds. B = background, P = plasmid, wild-type = uppercase, mutant = italicized lowercase. PA and PD represent the wild-type rpoA and rpoD plasmids, respectively; bp* represents the original strains isolated from the mutant libraries; bp indicates a cured background strain that has been retransformed with the mutant plasmid. (B) Several combinations of mutant rpoA and rpoD plasmids and mutant backgrounds result in similarly high L-tyrosine titers. Results for the rpoA27 plasmid (equivalent to pHACM-rpoAWT) in an rpoA14 background were not included, as this strain has previously been shown to have low L-tyrosine titers (Fig. 3A).
Even more surprisingly, we discovered that transforming the recovered rpoA/rpoD plasmids into each of the three mutant chromosomal backgrounds led to several unique strains with enhanced L-tyrosine production levels. As shown in Fig. 3B, plasmid and chromosomal mutations possess the unique property of complete interchangeability, with each newly generated strain yielding L-tyrosine titers that either closely approached or even exceeded values for the original isolates. These results are clearly atypical, given the recovery of these plasmids from two functionally divergent transcriptional libraries as well as the low likelihood of having recovered the same chromosomal mutation(s) in all three isolates.
Transcriptional Analysis Reveals Roles for the Acid Stress and Stringent Responses in E. coli.
To elucidate the biochemical significance of these plasmid and background mutations, we conducted a full transcriptional analysis of the parental strain P2 and the three gTME isolates, rpoD3, rpoA14, and rpoA27. Although several hundred genes were found to be differentially expressed, specific patterns of pathway up- and down-regulation offered notable insights into the underlying changes in these strains.
An analysis of the upregulated genes in the plasmid-bearing strains returned only a handful of loci for rpoD3 and rpoA27 (and none for rpoA14), with a significant number of these presumed hits possessing functions related to acid stress resistance (Table 1). Indeed, several of these genes were found to reside within the same transcriptional network, a complex regulon with at least three separate layers of control exerted by the proteins encoded by gadE, ydeO, and evgA (in ascending hierarchical order) (22).
Table 1.
List of upregulated genes related to acid resistance
|
rpoD3 |
rpoA27 |
|||||
| Gene name |
Gene ID |
Function |
Log fold change |
Q value*, % |
Log fold change |
Q value, % |
| b3517 | gadA | glutamate decarboxylase | 2.236 | 0 | 1.974 | 11.05 |
| b1493 | gadB | glutamate decarboxylase | 2.699 | 0 | 2.515 | 0 |
| b1492 | gadC | glutamate:γ-aminobutyrate antiporter | 2.027 | 0 | - | - |
| b3512 | gadE | transcriptional activator | 2.137 | 0 | 2.160 | 0 |
| b3510 | hdeA | acid stress chaperone | 1.950 | 0 | - | - |
| b3509 | hdeB | acid stress chaperone | 2.115 | 0 | - | - |
| b3511 | hdeD | acid resistance membrane protein | 1.707 | 0 | - | - |
| b3506 | slp | starvation lipoprotein | 1.609 | 10.17 | 1.897 | 5.67 |
*Q values represent the lowest false discovery rate at which that gene is called significant. It is like the well-known P value, but adapted to multiple testing situations.
Although hundreds of downregulated genes were uncovered by our analysis, overlaying the transcriptional data on top of a preassembled network (using Ecocyc’s Pathway Tools Omics Viewer) revealed interesting global reductions in several pathways, including ribosomal protein and RNA formation, fatty acid elongation, de novo purine/pyrimidine biosynthesis, and DNA replication (Table 2). Taken altogether, these transcriptional patterns implicated a stringent response in E. coli, a metabolic state triggered by amino acid starvation and other conditions of nutritional stress (23–25). This cellular shift from proliferation and growth to maintenance and stress survival (including, notably, acid stress resistance) is mediated by the small nucleotide (p)ppGpp, a regulator whose levels are jointly controlled by the relA-encoded ppGpp synthase and the spoT-encoded ppGpp synthase/hydrolase.
Table 2.
List of downregulated pathways in rpoD3, rpoA14, and rpoA27
| Pathway | rpoD3 | rpoA14 | rpoA27 |
| Arginine synthesis | ✓ | ✓ | ✓ |
| Isoleucine synthesis | ✓ | ✓ | ✓ |
| Leucine/valine synthesis | ✓ | ✓ | ✓ |
| Histidine synthesis | ✓ | ✓ | |
| Tryptophan synthesis | ✓ | ✓ | |
| Lysine synthesis | ✓ | ✓ | |
| Glutamate synthesis | ✓ | ✓ | ✓ |
| De novo purine/pyrimidine biosynthesis | ✓ | ||
| DNA replication | ✓ | ✓ | |
| Ribosomal proteins and RNA | ✓ | ✓ | ✓ |
| Fatty acid elongation | ✓ |
Because several of these cellular changes were observed in all three gTME mutants, we suspected that the acid resistance pathway and/or the stringent response may in fact play important roles in L-tyrosine overproduction. If this were the case, then altering the expression of known regulators of these pathways may have interesting implications for engineering this particular phenotype. To test this hypothesis, we constitutively overexpressed four regulators—gadE, ydeO, evgA, and a truncated form of relA (26)—to ascertain their effects on L-tyrosine production.
While the overexpression of all four genes in the parental strain P2 typically yielded only minimal increases in titer, a completely different cellular response was observed upon transfer of the evgA and relA plasmids into the modified backgrounds (Fig. 4). Indeed, strains containing one of the mutant chromosomal backgrounds and either evgA or relA exhibited L-tyrosine titers that closely matched those of the original isolates. Testing both weak and strong constitutive expression also revealed gene- and background-specific optima for each combination. Given such remarkable phenotypic sensitivity to promoter strength, it seems plausible that additional fine-tuning of evgA and relA expression may yield even greater titers than those observed here.
Fig. 4.
Overexpression of evgA and relA in parental and mutant backgrounds. The genes evgA and a truncated form of relA were individually overexpressed in four different backgrounds—the parental strain P2 and the cured backgrounds of rpoA14, rpoA27, and rpoD3. Two promoter strengths were tested for each gene in order to ascertain the effects of both low- and high-level constitutive expression on cellular phenotype. All strong promoters were PL. Weak promoters for evgA and relA were AA and II, respectively (Table S1). L-tyrosine titers were measured after 48 h cultivation in 50 ml MOPS minimal medium.
Whole Genome Sequencing Identifies Beneficial Substitutions in hisH and purF.
Although specific cellular pathways were found to be important for L-tyrosine overproduction, the identities of chromosomal alterations necessary for the desired phenotype remained unknown. We therefore conducted whole genome sequencing on parental P2 and the three gTME strains to parse out the identities of these mutation(s). Auspiciously, our analysis revealed only a single base pair (bp) change in each of the three gTME strains—an L82R substitution in the hisH-encoded imidazole glycerol phosphate (IGP) synthase subunit of strain rpoA14, a V5G shift in the purF-encoded amidophosphoribosyl transferase gene of strain rpoD3, and a T → C nucleotide substitution 17 bp upstream of the purF gene in strain rpoA27 (Fig. S5 and Table S2). Furthermore, reintroduction of these specific mutations into a clean parental P2 background resulted in high L-tyrosine titers when strains were also supplied with their respective rpoA or rpoD plasmids (reconstructed strains were named rpoA14R, rpoA27R, and rpoD3R) (Fig. S6). These results clearly demonstrate the importance of each hisH and purF mutation in establishing an L-tyrosine overproduction phenotype. More significantly, the identification and validation of these chromosomal mutations have led us to the construction of a completely genetically defined strain, rpoA14R, which possesses a titer of 902 mg/l L-tyrosine and a yield of 0.18 g L-tyrosine/g glucose in 50-ml shake flask cultures. To put these numbers into perspective, this yield on glucose is more than 150% greater than a classically improved L-phenylalanine auxotroph (DPD4195) currently used for the industrial production of L-tyrosine (cultivated under similar conditions) (27) and, when excluding glucose diverted to biomass, represents 85% of the maximum theoretical yield.
Performance of rpoA14R in Large-Scale Bioreactors.
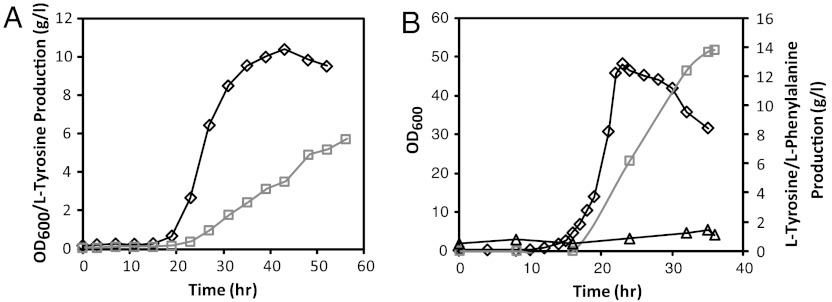
To evaluate the performance of rpoA14R in larger-scale fermentations, we ran 2-l bioreactors using two different minimal medium formulations—MOPS minimal medium (Teknova) (28) and R medium (29). When cells were cultivated in MOPS minimal medium, strain characteristics remained comparable to those previously observed in 50-ml shake flask cultures, with yields on glucose of 0.204 g L-tyrosine/g glucose and a specific growth rate of 0.275 h-1. Unfortunately, however, cell growth was severely hampered (OD600 of 10), leading to a titer of just 5.7 g/l L-tyrosine after 56 h (Fig. 5A). Switching to another defined synthetic medium (R medium) led to much higher maximum productivities (2.1 g L-tyrosine/l/h), titers (13.8 g/l after 36 h), and growth rates (0.405 h-1) but with a lower overall yield (0.120 g L-tyrosine/g glucose) (Fig. 5B). Future fermentation optimization should therefore aim at identifying limiting nutrients in MOPS minimal medium to extend the growth and production phase of the culture or, alternatively, modifying the composition of R medium to boost L-tyrosine yields on glucose.
Fig. 5.
Performance of rpoA14R in 2–l reactors. Two different medium formulations were tested: (A) MOPS minimal medium and (B) R medium. Measurements for cell density (OD600) (◊), L-tyrosine concentrations (□), and L-phenylalanine concentrations (△) are reported.
Discussion
In this study, we have demonstrated the clear potential of utilizing a combinatorial metabolic engineering approach for optimizing cellular phenotype. This methodology led to the identification of rpoA/rpoD mutants capable of enhancing L-tyrosine production over a rationally engineered parental, albeit in a mutated genetic background. An additional unique feature of this work is the successful identification of the specific chromosomal mutations and global regulators that, in combination, allowed the complete recovery of the L-tyrosine overproduction phenotype.
Screening two libraries constructed through the mutagenesis of rpoA and rpoD led to significant increases in L-tyrosine production in three strains, with titers of 798–893 mg/l L-tyrosine and yields of 0.16–0.18 g L-tyrosine/g glucose after 48 h. This performance more than doubles the previous levels observed in the parental strain P2. It is important to note that the capabilities of these gTME-derived strains far surpass not only those of rationally engineered constructions but also those of strains previously recovered from a random knockout library (16). This capacity to reach formerly inaccessible phenotypes therefore distinguishes gTME from other promising and more established combinatorial engineering methods. Its advantages likely stem from its unique ability to reprogram the transcriptome, alter the entire cellular milieu, and as a result, generate an enormous amount of phenotypic diversity.
An analysis of our transcriptional data revealed that the overexpression of one of two transcriptional regulators—evgA or relA—could completely supplant the phenotypic requirement for either a mutant rpoA or rpoD gene. This interchangeability provides compelling evidence regarding the specific biochemical mechanisms induced by these mutant transcriptional components and clearly implicates both acid resistance and the stringent response as important pathways for eliciting L-tyrosine overproduction in E. coli. However, because (p)ppGpp levels are known to directly or indirectly govern the onset of both responses (23–25), it is plausible that these similar phenotypic outcomes are in fact linked by a common mode of action. Although we still lack a complete mechanistic understanding of how alterations in these global cellular processes can lead to increases in L-tyrosine titers, these results provide a solid footing for future investigations aimed at deciphering the underlying mechanism.
Whole genome sequencing of our strains was required to identify spontaneous chromosomal alterations found to be essential for a L-tyrosine overproduction phenotype. Although large-scale sequencing projects oftentimes return more mutations than can be functionally characterized (30), only a single base-pair change was identified in each of our three gTME strains. These individual mutations were isolated to the genes hisH and purF, two loci encoding proteins possessing striking similarities in both function and structure. Each belongs to the class of glutamine amidotransferases, enzymes which utilize glutamine as a source of an amide nitrogen with the concomitant production of glutamate. Remarkably, both of the internal substitutions recovered during our analysis (V5G in rpoD3 and L82R in rpoA14) map to helical structures directly adjacent to two catalytic cysteine residues with important roles in glutamine binding (31–33). It is therefore possible that these chromosomal mutations may exert their effects by altering the substrate affinities, and hence, activities of these enzymes. Such a result would then point to relative glutamine/glutamate levels as a possible control point for L-tyrosine production, a theory with plausible grounds given the requirement for glutamate during the final transamination step in the L-tyrosine pathway. Data from our transcriptional analysis lends credence to this idea, as glutamate is also used as a node for maintaining intracellular pH levels in the upregulated acid resistance pathway (22).
Insights from our transcriptional analysis and whole genome sequencing studies allowed us to apply an inverse metabolic engineering paradigm to construct a robust, genetically defined strain for L-tyrosine overproduction. On the shake flask level, our reconstructed strain rpoA14R is capable of outperforming the industrial strain DPD4195 with respect to both yields and titers (27). Early 2-l-scale bioreactor experiments point to the potential for scale-up and have demonstrated improved performance beyond other E. coli strains rationally engineered for this purpose (E. coli T2: final titer of 9.7 g/l L-tyrosine and yield of 0.102 g L-tyrosine/g glucose) (17). However, these studies also illustrate the need for additional media optimization to achieve maximum titers, yields, and productivities. Because previous publications have already established impressive gains through scale-up and process optimization (up to 55 g/l L-tyrosine for DPD4195 on a 200 L scale) (27, 34), we remain confident that similar strides can be taken to transform our engineered strains into competitive industrial performers.
Methods
Cultivation Conditions.
L-tyrosine production experiments were performed at 37 °C with 225 rpm orbital shaking in 50-ml MOPS minimal medium (Teknova) (28) cultures supplemented with 5 g/l glucose and an additional 4 g/l NH4Cl. All liquid cultivations were conducted in at least triplicates. When appropriate, antibiotics were added in the following concentrations: 34 μg/ml chloramphenicol for maintenance of pHACM-derived plasmids, 50 μg/ml spectinomycin for maintenance of pCL1920 ∷ tyrAfbraroGfbr, and 20 μg/ml kanamycin for maintenance of pZE-derived plasmids. Isopropyl-β-D-thiogalactopyranoside (IPTG) (EMD Chemicals) was added at a concentration of 2 mM for the induction of pCL1920 ∷ tyrAfbraroGfbr. For L-phenylalanine auxotrophs (ΔpheA), L-phenylalanine (Sigma) was supplied at a concentration of 0.35 mM.
Generation and Screening of gTME Libraries.
RpoA and rpoD plasmid libraries were generated as described previously (3, 4). Briefly, fragment mutagenesis was performed on wild-type rpoA or rpoD using the GenemorphII Random Mutagenesis Kit (Stratagene) to induce low (0–4.5 mutations/kb), medium (4.5–9 mutations/kb), and high (9–16 mutations/kb) frequencies of mutation. Following digestion and ligation, preparations of the resulting pHACM-rpoA/rpoD plasmid libraries were then transformed into parental strain P2 to generate rpoA and rpoD mutant strain libraries. Approximately 7.5 × 105 and 3.1 × 106 viable colonies (for rpoA and rpoD, respectively) were screened using a melanin-based high-throughput assay for L-tyrosine production (16).
Plasmid Curing and Retransformation.
To investigate the individual contributions of plasmid-based and chromosomal-based mutations, isolated mutant strains were subcultured in LB medium to promote loss of its corresponding pHACM-derived plasmid. Following three to four rounds of reinoculation, strains were streaked out on LB-agar and LB-chloramphenicol plates to check for the loss of chloramphenicol resistance. Routine chemical transformation protocols were utilized in the construction of strains containing a combination of wild-type or mutant backgrounds and plasmids.
Transcriptional Analysis.
Strains P2, rpoA14, rpoA27, and rpoD3 were grown in 50 ml MOPS minimal medium (Teknova) (28) to an OD600 of approximately 0.4. Triplicates samples of RNA (on three separate days) were then extracted using the QIAGEN RNeasy Mini Kit according to the manufacturer’s protocol. Microarray services were provided by the David Koch Institute for Integrative Cancer Research Microarray Technologies Core Facility using the GeneChip E. coli Genome 2.0 Arrays (Affymetrix). Arrays were run in triplicate with biological replicates to allow for statistical confidence in differential gene expression. Microarray data was deposited in the Gene Expression Omnibus database under accession number GSE21652. Expression profile deviations between mutants and parental were identified by a Significance Analysis of Microarrays (SAM) analysis using SAM 3.0 (35).
Whole Genome Sequencing and SNP Detection.
Whole genome sequencing for strains P2, rpoA14, rpoA27, and rpoD3 was performed using the Illumina/Solexa Genome Analyzer System. Briefly, genomic DNA from all four strains was extracted using the Wizard Genomic DNA Purification Kit (Promega). Samples were then fragmented and prepared for paired-end sequencing using the Paired-End DNA Sample Prep Kit (Illumina). Samples were analyzed and processed at the Massachusetts Institute of Technology Biopolymers Laboratory, with duplicate lanes used for each strain. Sequence alignment and analysis, including the detection of insertions, deletions, and SNPs, was performed by the David Koch Institute Bioinformatics Facility. All sequence deviations from P2 were additionally validated by Sanger sequencing.
Fed‐batch Fermentations of rpoA14R.
RpoA14R was cultured in 5 ml LB medium until the mid‐exponential phase, then transferred into 2–50 ml MOPS minimal or R medium flask cultures at a starting OD600 of 0.1. These were cultivated at 37 °C with 225 rpm orbital shaking until the mid‐exponential phase and were subsequently used as the inoculum for a 1.5–l culture (5% by volume). Fed‐batch fermentations for rpoA14R were performed in 2–l glass vessels using the BioFlo110 modular fermentor system (New Brunswick Scientific). Two separate experiments were conducted using MOPS minimal medium and R medium (29) supplemented with 0.5 g/l L‐phenylalanine and 68 μg/ml chloramphenicol. Fermentations were performed at 37 °C with the pH automatically adjusted to 7.0 using a 30% solution of ammonium hydroxide. Dissolved oxygen (pO2) was maintained at ≥25% by agitation speed (100–1,000 rpm) and gas mix control (air/oxygen). Foam formation was controlled by the addition of Tego Antifoam KS911 (Evonik Goldschmidt). Operations were controlled and recorded with the BioCommand Lite Data Logging software (New Brunswick Scientific).
Analytical Methods.
The L-tyrosine content of cell-free culture supernatants was analyzed via HPLC with a Waters 2690 Separations module and a Waters 996 Photodiode Array detector set to a wavelength of 278 nm. Samples were separated on a Waters Resolve C18 column with 0.1% (vol/vol) trifluoroacetic acid (TFA) in water (solvent A) and 0.1% (vol/vol) TFA in acetonitrile (solvent B) as the mobile phase. The following gradient was used at a flow rate of 1 ml/ min: 0 min, 95% solvent A +5% solvent B; 8 min, 20% solvent A +80% solvent B; 10 min, 80% solvent A +20% solvent B; 11 min, 95% solvent A +5% solvent B. Cell densities of cultures were determined by measuring their absorbance at 600 nm with an Ultrospec 2100 pro UV/Visible spectrophotometer (Amersham Biosciences).
Supplementary Material
ACKNOWLEDGMENTS.
We thank C. Whitaker of the Koch Institute Bioinformatics and Computing Core Facility for his help in analyzing our genome sequencing data. This work was supported by a National Science Foundation Graduate Fellowship (CNSS), NSF Grant Number CBET-0730238, and the Singapore-MIT Alliance (SMA).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE21652).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206346109/-/DCSupplemental.
References
- 1.Santos CNS, Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol. 2008;12:168–176. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 2.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 3.Alper H, Stephanopoulos G. Global transcription machinery engineering: A new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Klein-Marcuschamer D, Santos CN, Yu H, Stephanopoulos G. Mutagenesis of the bacterial RNA polymerase alpha subunit for improvement of complex phenotypes. Appl Environ Microbiol. 2009;75:2705–2711. doi: 10.1128/AEM.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deijen JB, Orlebeke JF. Effect of tyrosine on cognitive function and blood pressure under stress. Brain Res Bull. 1994;33:319–323. doi: 10.1016/0361-9230(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 6.Van Spronsen FJ, van Rijn M, Bekhof J, Koch R, Smit PG. Phenylketonuria: Tyrosine supplementation in phenylalanine-restricted diets. Am J Clin Nutr. 2001;73:153–157. doi: 10.1093/ajcn/73.2.153. [DOI] [PubMed] [Google Scholar]
- 7.Rajput A, Rajput AH. Parkinson’s disease management strategies. Expert Rev Neurother. 2006;6:91–99. doi: 10.1586/14737175.6.1.91. [DOI] [PubMed] [Google Scholar]
- 8.Bell AA, Wheeler MH. Biosynthesis and functions of fungal melanins. Annu Rev Phytopathol. 1986;24:411–451. [Google Scholar]
- 9.Leonard E, Lim KH, Saw PN, Koffas MA. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi WW, et al. Functional expression of prokaryotic and eukaryotic genes in Escherichia coli for conversion of glucose to p-hydroxystyrene. Metab Eng. 2007;9:268–276. doi: 10.1016/j.ymben.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Sariaslani FS. Development of a combined biological and chemical process for production of industrial aromatics from renewable resources. Annu Rev Microbiol. 2007;61:51–69. doi: 10.1146/annurev.micro.61.080706.093248. [DOI] [PubMed] [Google Scholar]
- 12.Gosset G. Production of aromatic compounds in bacteria. Curr Opin Biotechnol. 2009;20:651–658. doi: 10.1016/j.copbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Lütke-Eversloh T, Santos CNS, Stephanopoulos G. Perspectives of biotechnological production of L-tyrosine and its applications. Appl Microbiol Biotechnol. 2007;77:751–762. doi: 10.1007/s00253-007-1243-y. [DOI] [PubMed] [Google Scholar]
- 14.Patnaik R, Liao JC. Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield. Appl Environ Microbiol. 1994;60:3903–3908. doi: 10.1128/aem.60.11.3903-3908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patnaik R, Spitzer RG, Liao JC. Pathway engineering for production of aromatics in Escherichia coli: Confirmation of stoichiometric analysis by independent modulation of AroG, TktA, and Pps activities. Biotechnol Bioeng. 1995;46:361–370. doi: 10.1002/bit.260460409. [DOI] [PubMed] [Google Scholar]
- 16.Santos CNS, Stephanopoulos G. Melanin-based high-throughput screen for L-tyrosine production in Escherichia coli. Appl Environ Microbiol. 2008;74:1190–1197. doi: 10.1128/AEM.02448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lütke-Eversloh T, Stephanopoulos G. L-Tyrosine production by deregulated strains of Escherichia coli. Appl Microbiol Biotechnol. 2007;75:103–110. doi: 10.1007/s00253-006-0792-9. [DOI] [PubMed] [Google Scholar]
- 18.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 20.Murakami K, Fujita N, Ishihama A. Transcription factor recognition surface on the RNA polymerase alpha subunit is involved in contact with the DNA enhancer element. EMBO J. 1996;15:4358–4367. [PMC free article] [PubMed] [Google Scholar]
- 21.Gaal T, et al. DNA-binding determinants of the alpha subunit of RNA polymerase: Novel DNA-binding domain architecture. Genes Dev. 1996;10:16–26. doi: 10.1101/gad.10.1.16. [DOI] [PubMed] [Google Scholar]
- 22.Masuda N, Church GM. Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol. 2003;48:699–712. doi: 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- 23.Magnusson LU, Farewell A, Nystrom T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 25.Srivatsan A, Wang JD. Control of bacterial transcription, translation, and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber G, et al. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 27.Olson MM, et al. Production of tyrosine from sucrose or glucose achieved by rapid genetic changes to phenylalanine-producing Escherichia coli strains. Appl Microbiol Biotechnol. 2007;74:1031–1040. doi: 10.1007/s00253-006-0746-2. [DOI] [PubMed] [Google Scholar]
- 28.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riesenberg D, et al. High cell density cultivation of Escherichia coli at controlled specific growth rate. J Biotechnol. 1991;20:17–27. doi: 10.1016/0168-1656(91)90032-q. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda M, Ohnishi J, Hayashi M, Mitsuhashi S. A genome-based approach to create a minimally mutated Corynebacterium glutamicum strain for efficient L-lysine production. J Ind Microbiol Biotechnol. 2006;33:610–615. doi: 10.1007/s10295-006-0104-5. [DOI] [PubMed] [Google Scholar]
- 31.Mei B, Zalkin H. A cysteine-histidine-aspartate catalytic triad is involved in glutamine amide transfer function in purF-type glutamine amidotransferases. J Biol Chem. 1989;264:16613–16619. [PubMed] [Google Scholar]
- 32.Mei BG, Zalkin H. Amino-terminal deletions define a glutamine amide transfer domain in glutamine phosphoribosylpyrophosphate amidotransferase and other PurF-type amidotransferases. J Bacteriol. 1990;172:3512–3514. doi: 10.1128/jb.172.6.3512-3514.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Donoghue P, Amaro RE, Luthey-Schulten Z. On the structure of hisH: Protein structure prediction in the context of structural and functional genomics. J Struct Biol. 2001;134:257–268. doi: 10.1006/jsbi.2001.4390. [DOI] [PubMed] [Google Scholar]
- 34.Patnaik R, Zolandz RR, Green DA, Kraynie DF. L-tyrosine production by recombinant Escherichia coli: Fermentation optimization and recovery. Biotechnol Bioeng. 2008;99:741–752. doi: 10.1002/bit.21765. [DOI] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.