Abstract
Within the intestinal mucosa, epithelial cells serve multiple functions to partition the lumen from the lamina propria. As part of their natural function, intestinal epithelial cells actively transport electrolytes with passive water movement as a mechanism for mucosal hydration. Here, we hypothesized that electrogenic Cl- secretion, and associated mucosal hydration, influences bacterial-epithelial interactions and significantly influences the composition of the intestinal microbiota. An initial screen of different epithelial secretagogues identified lubiprostone as the most potent agonist for which to define these principles. In in vitro studies using cultured T84 cells, lubiprostone decreased E. coli translocation in a concentration-dependent manner (p < 0.001) and decreased S. typhimurium internalization and translocation by as much as 71 ± 6% (p < 0.01). Such decreases in bacterial translocation were abolished by inhibition of electrogenic Cl- secretion and water transport using the Na-K-Cl- antagonist bumetanide (p < 0.01). Extensions of these findings to microbiome analysis in vivo revealed that lubiprostone delivered orally to mice fundamentally shifted the intestinal microbiota, with notable changes within the Firmicutes and Bacteroidetes phyla of resident colonic bacteria. Such findings document a previously unappreciated role for epithelial Cl- secretion and water transport in influencing bacterial-epithelial interactions and suggest that active mucosal hydration functions as a primitive innate epithelial defense mechanism.
Keywords: Lactobacillus, commensal bacteria, epithelial transport, infection, innate immunity, microbiota, ribosomal RNA
Introduction
A primary physiologic function of epithelial cells is electrolyte transport. Mucosal tissues lined by epithelia, such as the lung and intestine, accomplish this function through active ion transport.1 As part of a tissue adaptive response, a number of endogenous molecules been shown to influence epithelial electrogenic chloride secretion, the transport event responsible for mucosal hydration.1 This aspect of epithelial function has been studied in detail utilizing models of intact epithelial cell layers coupled with electrophysiologic strategies.
The process of electrogenic chloride secretion occurs through a coordinated series of membrane transporters. In Cl- secreting epithelium, the rate limiting step is entry of Cl- via the Na-K-2Cl- cotransporters,2 a family of proteins that mediate the electroneutral transport of Na+, K+ and Cl- ions across cellular membranes.1 These cotransporters are distributed over a wide variety of tissues, and with the exception of renal epithelia, are localized along the basolateral membrane.1 Due to their particular ability to regulate both anionic and cationic fluxes, the Na-K-Cl- cotransporters serve a number of vital physiological functions. In the majority of cell types Na-K-Cl- cotransporters are involved in the regulation of cell volume and are functionally activated by cell shrinkage.1 In coordination with other solute transport pathways, Na-K-Cl- cotransporters play a key role in salt transport by secretory epithelia. Specifically, the apical/basolateral distribution determines whether Na-K-Cl- cotransporters participate in active fluid absorption or secretion depending on the particular organ and their localization.1
Innate immunity in the intestine includes a combination of chemical, mechanical and environmental barriers to the invasion of luminal microbes.3-5 It is currently assumed that water transport and mucosal hydration function as a necessary component to a normally protective barrier. However, this view of the normal mucosa is generally inferred from pathologic changes such as those elicited by decreased hydration (e.g., cystic fibrosis)6 or increased hydration (e.g., cholera and enterotoxigenic E. coli)7 and by observations that microbes have evolved mechanisms to overcome the normal clearance mechanisms of the mucosa.7,8 From this perspective, surprisingly little is known about the direct influence of water transport on bacterial-epithelial interactions.
Here, we hypothesized that epithelial fluid transport regulates bacterial interactions with intestinal epithelial cells. Studies using human in vitro models and murine in vivo approaches reveal that electrogenic Cl- secretion, reflected as fluid transport, significantly decreases bacterial uptake and translocation across cultured intestinal epithelial cells. Likewise, administration of the Cl- secretory agonist lubiprostone to mice resulted in a significant change in the microbiota, particularly Firmicutes and Bacteroidetes. These findings reveal active fluid transport as a primitive innate defense mechanism in the mucosa.
Results
Screen of epithelial chloride channel activators and resultant water transport.
Initially, we hypothesized that Cl- secretion and resultant water transport would influence bacterial translocation. As a starting point, epithelial electrogenic Cl- secretion was used to define responses to approximate EC50 concentrations (based on dose response curves, not shown) of the secretagogues forskolin (1 ∝M, Fsk), adenosine (10 ∝M, Ado), prostaglandin E2 (1 ∝M, PGE2) and lubiprostone (100 nM, Lub) relative to Hank’s buffer (HBSS) control. As shown in Figure 1A, of those agonists screened, lubiprostone elicited the highest maximal Cl- secretory response. Figure 1B shows the influence of individual secretagogues on water transport as a result of Cl- secretion. Monolayers of T84 cells were incubated with individual secretagogues for 24 h and examined for vectorial basolateral-to-apical fluid transport. T84 cells alone served as a negative control. Like Cl- secretion (Fig. 1A), lubiprostone elicited the highest degree of fluid transport of the secretagogues screened. Based on these results, we proceeded with lubiprostone to define the impact of fluid transport on bacterial translocation.
Figure 1.
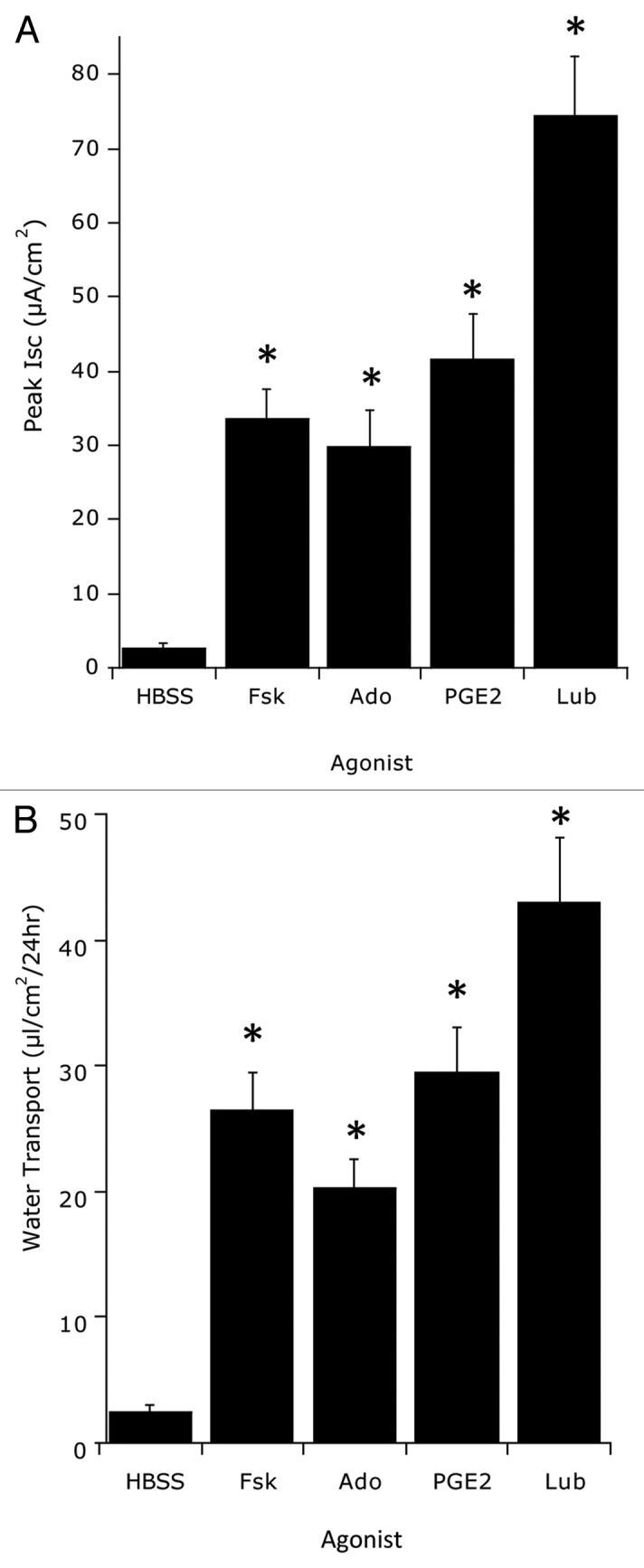
Screen of epithelial chloride channel activators and resultant water transport. (A) Epithelial electrogenic Cl- secretion was used to define responses to the secretagogues forskolin (1 μM, Fsk), adenosine (10 μM, Ado), prostaglandin E2 (1 μM, PGE2) and lubiprostone (100 nM, Lub) relative to Hank’s buffer (HBSS) control. Cells were exposed to each agonist for 1 h and Isc was monitored every 5 min. Results are pooled from eight monolayers in each condition and results are expressed as the mean ± SEM where * indicates p < 0.01. (B) Influence of individual secretagogues on water transport. Monolayers of T84 cells were incubated with individual secretagogues for 24 h and examined for vectorial basolateral-to-apical fluid transport. T84 cells alone served as a negative control. Data are pooled from eight monolayers in each condition and results are expressed as the mean ± SEM fluid movement over 24 h where * indicates p < 0.01.
Influence of epithelial electrogenic Cl- secretion on bacterial translocation.
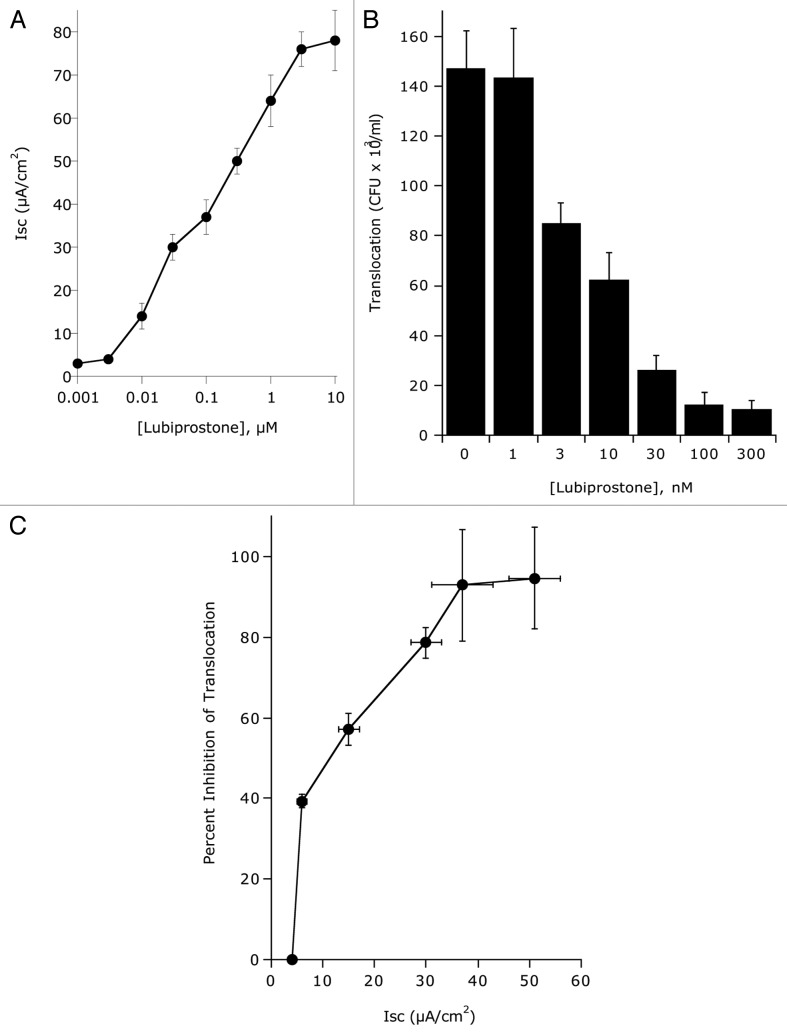
Epithelial electrogenic Cl- secretion was used to define the dose response to indicated concentrations of lubiprostone (0.001–10 ∝M) relative to Hank’s buffer (HBSS) control. As shown in Figure 2A, lubiprostone showed a concentration-dependent increase in Cl- secretion (p < 0.001 by ANOVA) with maximal increases at 3 ∝M (p < 0.001). In parallel, we addressed the influence of lubiprostone on bacterial translocation. As shown in Figure 2B, translocation of E. coli across T84 cell monolayers decreased with increasing concentrations of lubiprostone (p < 0.001 by ANOVA). Figure 2C shows the correlation between electrogenic Cl- secretion plotted with E. coli translocation and revealed that translocation significantly decreases with increasing concentrations of lubiprostone. A time course of lubiprostone (100 nM) on E. coli translocation relative to vehicle control revealed that lubiprostone significantly decreases bacterial translocation in a time-dependent manner (Fig. 3 and p < 0.01 by ANOVA). Importantly, at all concentrations tested (1 nM–1 ∝M), lubiprostone had no influence on E. coli growth compared with vehicle control (based on exponential growth rates monitored spectrophotometrically at 650 nm, data not shown).
Figure 2.
Influence of epithelial electrogenic Cl- secretion on E. coli translocation. (A) Epithelial electrogenic Cl- secretion was used to define the dose response to indicated concentrations of lubiprostone (0.001–10 μM) relative to Hank’s buffer (HBSS) control. Results are pooled from 6–8 monolayers in each condition and results are expressed as the mean ± SEM. (B) Translocation of E. coli across T84 cells (120 min time point) in the presence and absence of lubiprostone stimulation at indicated concentrations. Results are pooled from 6–8 monolayers in each condition and results are expressed as the mean ± SEM. (C) Correlation of electrogenic Cl- secretion (X-axis) with E. coli translocation (Y-axis, plotted as percent inhibition of translocation).
Figure 3.
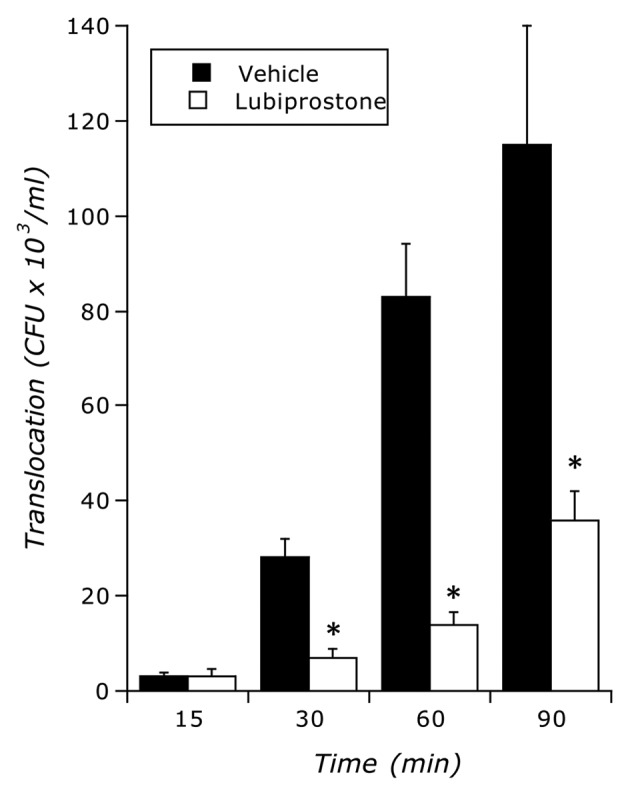
Influence of lubiprostone on E. coli translocation. Time course translocation of E. coli across T84 cells in the presence and absence of lubiprostone stimulation (100 nM). Results are pooled from 6–8 monolayers in each condition and results are expressed as the mean ± SEM where * indicates p < 0.01.
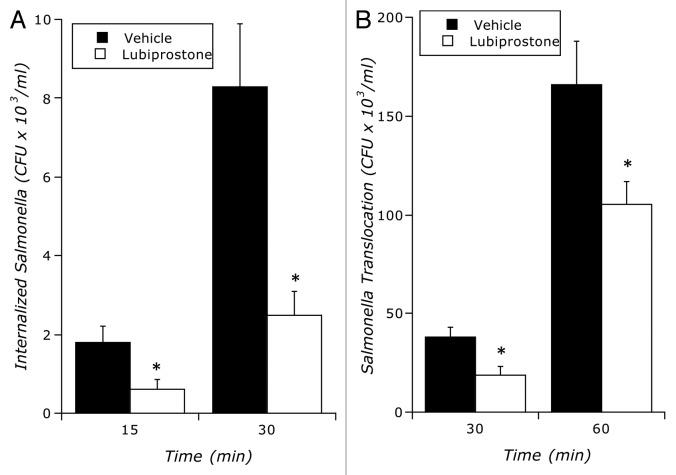
As shown in Figure 4, like our findings with E. coli, lubiprostone (100 nM) significantly decreased both internalization and translocation of S. typhimurium (p < 0.01 by ANOVA for both), suggesting that these findings are not bacterial species-specific.
Figure 4.
Influence of lubiprostone on S. typhimurium internalization and translocation: (A) Time course of lubiprostone (100 nM) on S. typhimurium internalization relative to vehicle control (p < 0.01 by ANOVA). (B) Time course of lubiprostone (100 nM) on S. typhimurium translocation across T84 epithelia relative to vehicle control. Results are pooled from 8 monolayers in each condition and results are expressed as the mean ± SEM where * indicates p < 0.01.
Influence of blocking electrogenic Cl- secretion on lubiprostone-mediated inhibition of E. coli translocation.
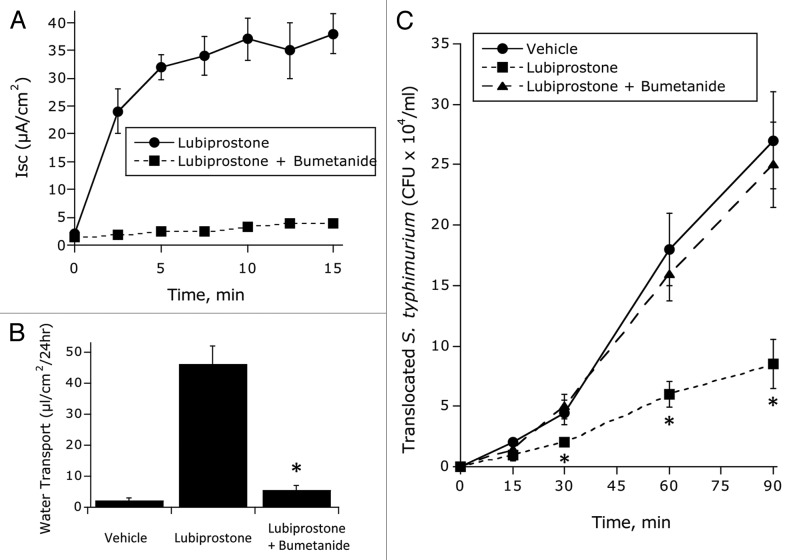
Having demonstrated that lubiprostone blocks bacterial translocation and invasion, we next addressed the relationship of water transport to bacterial-epithelial interactions. Epithelial electrogenic Cl- secretion was used to define responses to lubiprostone (100 nM, Lub) in the presence and absence of the NKCC1 inhibitor bumetanide (1 ∝M) relative to HBSS control. As shown in Figure 5, bumetanide blocked lubiprostone-induced Cl- secretion (Fig. 5A) and water transport (Fig. 5B) by as much as 95 ± 3% (p < 0.001). Using such conditions, we defined the relative importance of water transport to inhibition of bacterial translocation. As shown in Figure 5C, while lubiprostone significantly decreased S. typhimurium translocation (p < 0.001), such inhibition was fully blocked by the addition of bumetanide (p = not significant compared with vehicle). Such findings indicate that Cl- secretion and associated water transport explain the decrease in bacterial translocation associated with lubiprostone.
Figure 5.
Influence of blocking electrogenic Cl- secretion on lubiprostone-mediated inhibition of E. coli translocation: (A) Epithelial electrogenic Cl- secretion was used to define responses to lubiprostone (100 nM, Lub) in the presence and absence of the NKCC1 inhibitor bumetanide (1 μM) relative to Hank’s buffer (HBSS) control. Results are pooled from eight monolayers in each condition and results are expressed as the mean ± SEM (p < 0.001 by ANOVA). (B) Influence of bumetanide on lubiprostone-mediated on water transport in the presence and absence of the NKCC1 inhibitor bumetanide (1 μM) relative to Hank’s buffer (HBSS) control or Fsk control (where * indicates p < 0.01 in the comparison of lubiprostone vs. lubiprostone + bumetanide). (C) Influence of lubiprostone on E. coli translocation in the presence and absence of bumetanide (p < 0.01 by ANOVA in the comparison of lubiprostone vs. lubiprostone + bumetanide).
Influence of lubiprostone on murine colonic bacterial microfloramicrobiota.
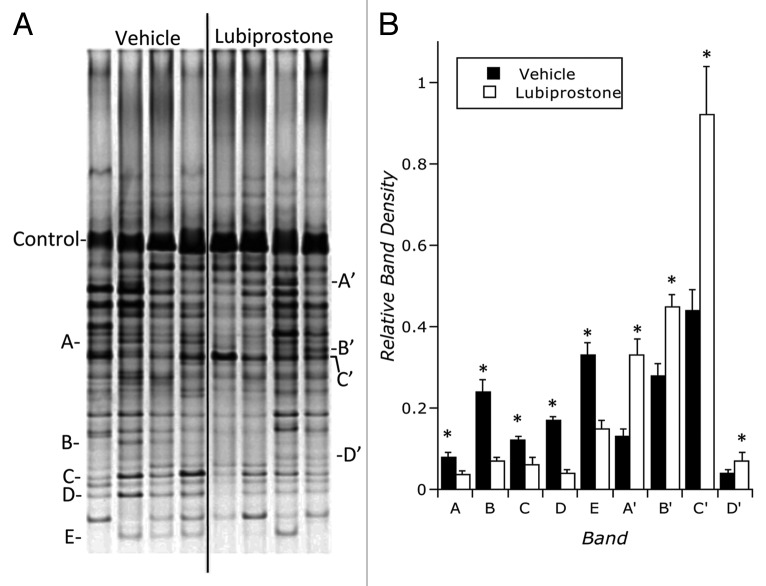
We next addressed the hypothesis that lubiprostone would influence bacterial colonic colonization in vivo. To do this, C57Bl/6 mice (n = 4 per group) were administered lubiprostone by oral gavage for (1 mg/kg/day) 7 d. Mice were sacrificed on day 7 and colonic stool and mucosal scrapings were harvested. Bacterial genomic DNA was isolated and samples were amplified using 16S rDNA-specific primers. Samples were resolved by denaturing gradient gel electrophoresis (DGGE). Figure 6 depicts a global example of stool DGGE from animals exposed to lubiprostone or vehicle for 7 d and revealed a significant change in microbiota with administration of lubiprostone. As can be seen, at least five bands are over-represented in vehicle control while at least four bands are over-represented in animals treated with lubiprostone. At a minimum, such results indicate that lubiprostone globally influences bacterial colonization in vivo.
Figure 6.
Influence of lubiprostone on murine colonic bacterial microbiota: C57Bl/6 mice (n = 4 per group) were administered lubiprostone (1 mg/kg/day) by oral gavage for 7 d. Mice were sacrificed on day 7 and colonic stool was harvested. Bacterial genomic DNA was isolated and samples were amplified using 16S rDNA-specific primers. Samples were resolved by denaturing gradient gel electrophoresis (DGGE). (A) is an example of stool DGGE from animals exposed to vehicle or lubiprostone for 7 d. Indicated bands (A–E) signify bands over-represented in vehicle exposed animals and bands (A’–D’) signify bands over-represented following 7 d of lubiprostone treatment. (B) represents densitometric analysis of indicated bands relative to the control band depicted in (A), where * over closed bars indicates significantly over-represented in control group (p < 0.05) and * over open bars indicates significantly over-represented in lubiprostone-treated group (p < 0.05).
Microbiome analysis of lubiprostone-mediated changes in in vivo.
We next extended these findings with DGGE to genus-level changes in bacterial colonization and mucosal-associated bacteria in response to orally administered lubiprostone in vivo. Bacterial genomic DNA was isolated from either stool or mucosal scrapings and samples were amplified using 16S rDNA-specific primers. Samples were analyzed by 454 next-generation sequencing. All results were analyzed between or within the treated and control groups using a rank based test (Wilcoxon).
As a starting point and as an important internal control, this analysis revealed that no differences were observable in stool microbiota between any of the mice at baseline (i.e., prior to administration of lubiprostone or vehicle, see Fig. S1). Likewise, no differences were observed between control and treated in mucosal-associated microbiota (i.e., from mucosal scrapings at day 7, see Fig. S2).
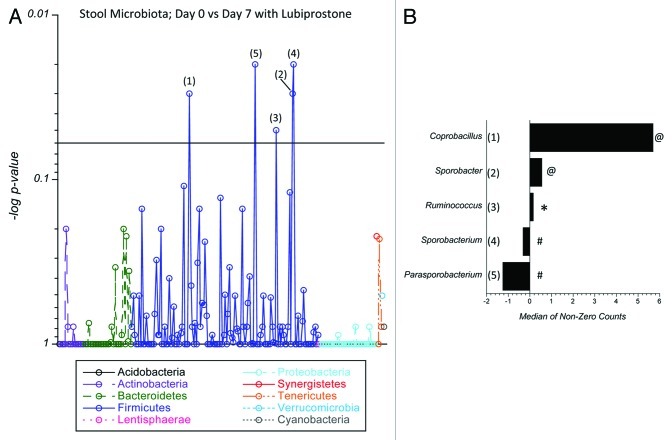
A comparison of the stool microbiota in lubiprostone-treated animals revealed significant shifts in bacterial genera, primarily within the Firmicutes and Bacteroidetes phyla. Coprobacillus, Parasporobacterium and Sporobacterium displayed interesting patterns across the tests performed and were further investigated. Coprobacillus significantly expanded between day 0 and day 7 in the treated mice (Fig. 7 and p = 0.03) and had a marginal difference between treated and control mice in stool at day 7 (p = 0.09), but did not differ between treated and control mice at day 0 (p = 0.33). Moreover, the differences between days 0 and 7 were significantly different between the treated and control groups (p = 0.02). Parasporobacterium differed significantly between day 0 and day 7 in the treated mice (Fig. 7 and p = 0.02) and had a marginal difference between treated and control mice in stool at day 7 (p = 0.11) but did not differ between treated and control mice at day 0 (p = 0.84). The differences between days 7 and 0 was not significantly different between the treated and control groups (p = 0.37). Sporobacterium differed significantly between day 0 and day 7 in the treated mice (p = 0.02) and had a marginal difference between treated and control mice in stool at day 7 (p = 0.05) but did not differ between treated and control mice at day 0 (p = 0.44). The differences between days 0 and 7 were significant between the treated and control groups (Fig. 7 and p = 0.04). As shown in Figure S3, some other differences were observed in the comparison of stool samples in control and treated on day 7, including Lachnobacterium (p = 0.04), Butyricoccous (p = 0.02), Dorea (p = 0.04), Acetanaerobacterium (p = 0.04) and Hydrogenoanaerobacterium (p = 0.04).
Figure 7.
Analysis of lubiprostone-mediated changes on murine colonic stool microbiota: C57Bl/6 mice (n = 13 per group) were administered lubiprostone (1 mg/kg/day) by oral gavage for 7 d. Stool was harvested on days 0 and 7 following treatment. Bacterial genomic DNA was isolated and samples were amplified using 16S rDNA-specific primers. Shown here a comparison of genus level groups identified in day 0 and day 7 samples of lubiprostone treated mice. In (A), the Manhattan plot displays results of statistical significance testing (p-value plotted on the y-axis) for the 230 genera identified in the two groups. Numbers corresponding to individual bacterial genera are depicted in (B) as the change in relative abundance between the two groups, plotted as the median of non-zero sequencing counts. Values > 1 indicate enrichment with lubiprostone treatment and values < 1 indicate enrichment with vehicle treatment, where # indicates p = 0.02, @ indicates p = 0.03 and * indicates p = 0.05.
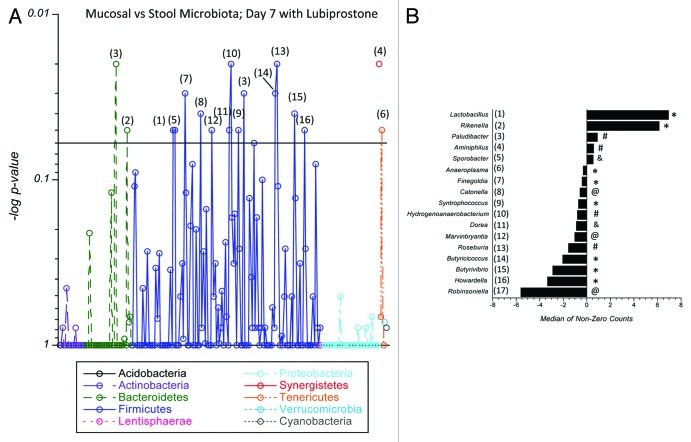
Figure 8 depicts differences in the microbiota between the stool and mucosa in lubiprostone treated mice only (i.e., differences not reflected in vehicle-treatment). Notable was the over-representation of Lactobacillus in lubiprostone treated mice (Fig. 8). Lactobacillus differed significantly between day 0 and day 7 in the treated mice (p = 0.05), was different between treated and control mice in stool at day 7 (p = 0.04) and was associated only with the stool and not mucosa (Fig. 8). Some differences other than Lactobacillus were also observed within the Firmicutes between the mucosa and the stool in lubiprostone treated mice (Fig. 8), including Sporobacter (p = 0.04), Finegoldia (p = 0.05), Catonella (p = 0.03), Syntrophococcous (p = 0.05), Dorea (p = 0.04), Marvinbryantia (p = 0.03), Butyricicoccus (p = 0.05) and Howardella (p = 0.05) as well as one Tenericute (Anaeroplasma, p = 0.05). Other bacterial genera differences were accounted for as baseline differences between the stool and mucosa (ie. also different in vehicle-treated mice, see Fig. S4), including Rikenella, Paludibacter, Aminiphilus, Coprococcus, Roseburia, Butyrivibrio, Hydrogenoanaeroobacterium and Robinsoniella.
Figure 8.
Comparison of mucosal and stool microbiota in lubiprostone-treated mice: C57Bl/6 mice (n = 13 per group) were administered lubiprostone (1 mg/kg/day) by oral gavage for 7 d. Mice were sacrificed on day 7 and colonic stool and mucosal scrapings were harvested. Bacterial genomic DNA was isolated and samples were amplified using 16S rDNA-specific primers. Samples were analyzed by next-generation sequencing (230 genus level taxa identified). Shown here a comparison of differences in the microbiota between the mucosa and the stool of lubiprostone treated mice on day 7. In (A), the Manhattan plot displays results of statistical significance testing (p-value plotted on the y-axis) for the 230 genera identified in the two groups. Numbers corresponding to individual bacterial genera are depicted in (B) as the change in relative abundance between the two groups, plotted as the median of non-zero sequencing counts. Values > 1 indicate enrichment in the stool of mice with lubiprostone treatment and values < 1 indicate enrichment in the mucosa of mice with lubiprostone treatment, where # indicates p = 0.02, @ indicates p = 0.03 and indicates p = 0.04, and * indicates p = 0.05.
Discussion
The intestinal epithelium normally functions to provide a selective barrier to luminal contents and to provide vectoral ion transport, the basis of water movement across the epithelium.9 In this study, we sought to determine whether the activation of intestinal epithelial chloride channels might influenced bacterial translocation in vitro and the composition of the commensal microbiota inhabiting the murine GI tract in vivo.
We demonstrate here that the potent Cl- secretagogue lubiprostone provides a strong driving force for water movement across the apical membrane of the epithelium. Indeed, using an in vitro water transport assay in conjunction with an established electrogenic Cl- secretion model (T84 cells) 10 revealed that the activation of water movement significantly diminishes E. coli and S. typhimurium internalization and translocation. Such inhibition of bacterial translocation strongly correlated with electrogenic Cl- secretion over a broad range of lubiprostone concentrations (1–300 nM).
While inhibition of bacterial infection by fluid transport could occur through a number of mechanisms, our results with bumetanide strongly implicate Cl- secretion and associated water movement as the mechanism for inhibition. While the decrease in bacterial translocation afforded by bumetanide could result from a change in membrane potential, more likely this change reflects a decrease in fluid transport. Indeed, concentrations of bumetanide that blocked Cl- secretion also decreased fluid transport by > 90% and normalized bacterial translocation in this in vitro model. From this observation, we surmise that in addition to mucosal hydration, a physiological function for fluid transport is the “flushing” of potentially noxious components from the luminal surface during an ongoing inflammatory response. Some evidence suggests that such a mechanism may be protective. For example, Asfaha et al. used a murine model of colitis and demonstrated that as late as six weeks post-induction of colitis, barrier function defects had resolved but colonic secretory dysfunction persisted.11 These findings paralleled increased bacterial translocation and increased colonic aerobes, which were subsequently shown to involve chronic expression of cyclooxygenase-2.12 Such findings suggest that secretory impairments may contribute to bacterial translocation independent of defects in epithelial barrier function.
Turning our attention to profiling of resident bacteria, mice were administered lubiprostone via oral gavage for 1 week and the stool and colonic mucosal microbiota was profiled by DGGE and/or 454 NextGen sequencing. Lubiprostone has been demonstrated to be a potent epithelial secretagogue in mouse ileal tissue ex vivo13 and has been shown to increase gastric emptying and small intestinal transit in vivo.14 We did not observe obvious signs of diarrhea or significant differences in tissue or fecal wet:dry ratios (not shown). We did, however, observe a significant shift in the intestinal microbiota of mice administered lubiprostone orally. In particular, members of the Firmicutes and Bacteroidetes phyla changed in fundamental ways. Somewhat surprising was the finding that lubiprostone-associated differences were observed only in stool samples and not in mucosal-associated populations. Given the findings of our in vitro model, we presumed that the microbiota closest to epithelium (e.g., at the mucosa or within the mucus gel layer) would shift with active water movement. More likely, our results reflect a change associated with water transport proximal to the colon (e.g., small intestine or cecum) and that such changes proximally, condition the stool microbiota distally. This exemplifies one limitation of our study, namely that we could not sample the microbiota of the small intestine in real-time. Likely, changes within the small intestine during active water transport are important. Given the necessity to internally control our analysis, it was not possible to sample the small intestine in a non-invasive manner.
Notable from these microbiota results was the over-representation of the genus Coprobacillus with lubiprostone administration to mice. Coprobacillus, which was isolated from human feces within the past decade,15 is an intestinal resident commensal bacteria that has been strongly associated with irritable bowel syndrome (IBS),16 particularly diarrhea predominant IBS.17 Indeed, using intestinal microbial fecal cloning, Kassinen et al. recently showed that Coprobacillus was significantly increased in patients with IBS.16 In this regard, there is significant interest in understanding how lubiprostone functions in the treatment of IBS.18,19 While these studies do not reveal a direct mechanism for IBS, they provide initial clues to potential changes that reflect disease status.
Also notable from our analysis was the increased association of Lactobacillus in stool samples of lubiprostone-treated mice. The beneficial influences of Lactobacillus are exemplified by their common use as probiotic agents.20,21 Numerous studies have shown health benefits of Lactobacillus and that species within this genera harbor anti-inflammatory properties as well as beneficial influences for the host across a range of functions, including colonization resistance, increased availability of nutrients to the intestine and improved digestion.21 The anti-inflammatory properties of Lactobacillus in the colon have been studied in some detail, and while the mechanisms are incompletely understood, much of the activity is attributable to cell surface proteins interacting with the host immune response.22 Original reports in the IL-10-/- spontaneous colitis mouse model revealed that abnormal colonization of Lactobacillus sp, that when normalized, reduced levels of mucosal adherent bacteria and attenuated the development of colitis.23 Of interest for the current work, colons from IL-10-/- mice show significant defects in activated Cl- secretion linked to decreased expression of the cystic fibrosis transmembrane regulator.24 From such observations, it is possible that water transport promotes the colonization of Lactobacillus and promotes colonic homeostasis.
In summary, these results define a previously unappreciated role for ion secretion and water transport in the prevention of bacterial invasion at the mucosal interface. We show here that agents which selectively activate epithelial ion transport also diminish bacterial infection. Central to this pathway is a fundamental change in the microbiota of the fecal stream of mice subjected to such conditions.
Materials and Methods
Cell culture.
T84 intestinal epithelial cells were grown and maintained as confluent monolayers in 1:1 Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F12 medium with 10% FBS at 37°C in 5% CO2 in room air. For electrophysiological measurements described below, T84 cells were plated on collagen coated permeable supports and grown to high resistance (> 1,000 ohm · cm2) as previously described in detail in reference 10. Monolayers were grown on 0.33-cm2 ring-supported polycarbonate filters (Costar Corp., Cambridge MA) unless otherwise noted, and they were used 6 to 12 d after plating as described previously in reference 25.
Electrophysiological measurements.
To measure agonist stimulated short-circuit current (Isc), transepithelial potential and resistance, we use a commercially available voltage clamp (Iowa Dual Voltage Clamps, Bioengineering, University of Iowa) interfaced with an equilibrated pair of calomel electrodes and a pair of Ag-AgCl electrodes, as described in detail in reference 10. Cl- secretory responses are expressed as peak Isc (peak Isc) designated the Isc necessary to maintain zero potential difference across the monolayer.
Fluid transport assay.
T84 cells growing on inserts net fluid movement was measured as described previously in reference 26, with minor modifications. In brief, the apical solution of confluent T84 cell monolayers grown on 0.33 cm2 permeable supports was replaced with 30 ∝l of media and layered with 60 ∝l of warm, sterile mineral oil to minimize evaporation as previously described in reference 26. In some monolayers, lubiprostone (a kind gift from Takeda Pharmaceuticals) was added at indicated concentrations to the apical solution to promote fluid movement. After 24 h, the apical solution was collected, centrifuged at 10,000× g, and fluid transport quantified with a calibrated pipette and weighed on a balance (Sartorius, Inc.).
Bacterial cultures. Primary bacterial cultures of E. coli (ATCC# 33694)and S. typhimurium (ATCC# 700408) were streaked on LB agar plates and incubated overnight at 37°C. Colonies were removed from the plate and inoculated into 5 ml of LB broth. This broth was incubated, shaking (250 rpm), overnight at 37°C.
Infection studies.
E. coli and S. typhimurium was cultured in LB broth overnight. Fifty ∝l of the resulting culture was inoculated into 5 ml of fresh LB broth and incubated static for 3 h at 37°C.27 The resulting culture (~1 × 108 CFU/ml) was centrifuged at 3,000× g for 10 min. The pellet was resuspended in HBSS (supplemented with magnesium and calcium) buffer to give a concentration of 1 × 107 CFU/ml. T84 cells were seeded on 0.33 cm2, 3 ∝m pore transwell inserts and maintained until consistent epithelial resistances formed. Monolayers were pre-incubated with either vehicle or lubiprostone at indicated concentrations on the apical surface for 30 min. Monolayers were then apically challenged with indicated bacteria at a multiplicity of infection of 100:1 and incubated at 37°C. Samples of 100 ∝l in volume were taken from the basolateral chamber 15, 30, 60 and 120 min after challenge. Basolateral samples were diluted 1/100 and 50 ∝l samples were spot plated on LB agar plates. Plates were incubated overnight at 37°C and plate counts were performed to determine bacterial translocation.
Denaturing gradient gel electrophoresis.
Age and weight matched female C57/B6 mice were purchased from (The Jackson Laboratory). All procedures were performed with approval of the Animal Care and Use Committee of the University of Colorado Denver. Animals (n = 4 per group) were administered either vehicle (10 ∝l/g/day, medium chain triglyceride) or lubiprostone (1 mg/kg/day) by oral gavage daily for 7 d. Fecal pellets were harvested and DNA Extraction was performed using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol with the addition of chicken egg white lysozyme (0.36 mg/sample, Sigma-Aldrich) to the first step of the protocol and incubation at 37° for 1 h before proceeding to step 2. PCR amplification of the bacterial 16S ribosome gene was performed in preparation for DGGE profiling. Briefly, 500 ng of DNA isolated from colonic stool and mucosal scrapings was used in a 50 ∝l reaction with 1ul of the AccuPrime Pfx DNA polymerase (Invitrogen), 5 ∝l of the corresponding 10× reaction mix, and 3 ∝l of 10 ∝M HDA-1 GC-clamped (5'-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3') and HDA-2 (5'-GTA TTA CCG CGG CTG CTG GCA C-3') primers that target the V2-V3 hypervariable region. Initial denaturing was at 95° for 3 min, followed by 40 cycles of 30 sec at 95°, 30 sec at 56° and 45 sec at 70°. Final extension was 10 min at 72°. Following visualization of the expected 240 bp product on a 2% agarose gel, 20 ∝l of PCR product was used for DGGE profiling with the DCode Universal Mutation Detection System (BioRad). Samples were loaded in 16 cm polyacrylamide gradient gels (30–50% urea-formamide) prepared according to the mutation detection system manual. Electrophoresis was performed for 4.5 h at 130 V in a TRIS-acetate-EDTA buffer. Gels were stained with ethidium bromide (1 ∝g/ml) for 10 min before imaging. Differential banding was quantified using Image J (NIH).
Microbial diversity analysis.
Age and weight matched female C57/B6 mice were purchased from (The Jackson Laboratory). The microbiota was allowed to establish in our animal facility for 2 weeks prior to experiementation. At that point, animals (n = 13 per group) were administered either vehicle (10 ∝l/g/day, medium chain triglyceride) or lubiprostone (1 mg/kg/day) by oral gavage daily for 7 d.
Fecal samples from the colon (Day 0 and Day 7) and the scraped mucosa (Day 7) were collected and genomic DNA was isolated using the DNeasy Blood and Tissue Kit with the aforementioned modifications (Qiagen). Amplicons were generated for each sample with a unique barcoded primer compatible with the Roche 454 FLX system.28 DNA concentration for each amplicon was normalized using the Invitrogen SequalPrep kit, and mixed in equal volume to construct the amplicon pool for sequencing.29 The amplicon pool was concentrated by evaporation and gel purified using the Montage Gel Purification kit prior to sequencing. Sequencing was performed per manufacturer’s instructions.
Sequence analysis consisted of sample assignment and low-level quality filtering (length, quality score) using BARTAB.30 Matching to the bacterial SSU-rRNA secondary structure was checked using Infernal,31 and chimera detection was done with ChimeraSlayer.32 Sequences were classified using the RDP classifier and taxonomy lines were used to assemble sequence clusters.33 Changes in relative abundance between treated and untreated groups for genera were analyzed, statistically compared for relative abundance (Student’s t-test, p < 0.05 level of significance) and expressed as the median of non-zero sequencing counts.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants (R37-DK50189, RO1-HL60569), by support from the Crohn’s and Colitis Foundation of America and by an Investigator Initiated Preclinical Study from Takeda Pharmaceuticals.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Matieral
Supplemental materials may be found here: http://www.landesbioscience.com/journals/gutmicrobes/article/20529/
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/20529
References
- 1.Gamba G. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev. 2005;85:423–93. doi: 10.1152/physrev.00011.2004. [DOI] [PubMed] [Google Scholar]
- 2.Barrett KE, Keely SJ. Chloride secretion by the intestinal epithelium: molecular basis and regulatory aspects. Annu Rev Physiol. 2000;62:535–72. doi: 10.1146/annurev.physiol.62.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Campbell EL, Serhan CN, Colgan SP. Antimicrobial aspects of inflammatory resolution in the mucosa: a role for proresolving mediators. J Immunol. 2011;187:3475–81. doi: 10.4049/jimmunol.1100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanov AI, Parkos CA, Nusrat A. Cytoskeletal regulation of epithelial barrier function during inflammation. Am J Pathol. 2010;177:512–24. doi: 10.2353/ajpath.2010.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 6.Clunes MT, Boucher RC. Introduction to section I: overview of approaches to study cystic fibrosis pathophysiology. Methods Mol Biol. 2011;742:3–14. doi: 10.1007/978-1-61779-120-8_1. [DOI] [PubMed] [Google Scholar]
- 7.Glenn GM, Francis DH, Danielsen EM. Toxin-mediated effects on the innate mucosal defenses: implications for enteric vaccines. Infect Immun. 2009;77:5206–15. doi: 10.1128/IAI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallstrom K, McCormick BA. Salmonella Interaction with and Passage through the Intestinal Mucosa: Through the Lens of the Organism. Front Microbiol. 2011;2:88. doi: 10.3389/fmicb.2011.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCole DF, Barrett KE. Varied role of the gut epithelium in mucosal homeostasis. Curr Opin Gastroenterol. 2007;23:647–54. doi: 10.1097/MOG.0b013e3282f0153b. [DOI] [PubMed] [Google Scholar]
- 10.Dharmsathaphorn K, Madara JL. Established intestinal cell lines as model systems for electrolyte transport studies. Methods Enzymol. 1990;192:354–89. doi: 10.1016/0076-6879(90)92082-O. [DOI] [PubMed] [Google Scholar]
- 11.Asfaha S, MacNaughton WK, Appleyard CB, Chadee K, Wallace JL. Persistent epithelial dysfunction and bacterial translocation after resolution of intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2001;281:G635–44. doi: 10.1152/ajpgi.2001.281.3.G635. [DOI] [PubMed] [Google Scholar]
- 12.Zamuner SR, Warrier N, Buret AG, MacNaughton WK, Wallace JL. Cyclooxygenase 2 mediates post-inflammatory colonic secretory and barrier dysfunction. Gut. 2003;52:1714–20. doi: 10.1136/gut.52.12.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bijvelds MJ, Bot AG, Escher JC, De Jonge HR. Activation of intestinal Cl- secretion by lubiprostone requires the cystic fibrosis transmembrane conductance regulator. Gastroenterology. 2009;137:976–85. doi: 10.1053/j.gastro.2009.05.037. [DOI] [PubMed] [Google Scholar]
- 14.De Lisle RC, Mueller R, Roach E. Lubiprostone ameliorates the cystic fibrosis mouse intestinal phenotype. BMC Gastroenterol. 2010;10:107. doi: 10.1186/1471-230X-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kageyama A, Benno Y. Coprobacillus catenaformis gen. nov., sp. nov., a new genus and species isolated from human feces. Microbiol Immunol. 2000;44:23–8. doi: 10.1111/j.1348-0421.2000.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 16.Kassinen A, Krogius-Kurikka L, Mäkivuokko H, Rinttilä T, Paulin L, Corander J, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Lyra A, Rinttilä T, Nikkilä J, Krogius-Kurikka L, Kajander K, Malinen E, et al. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol. 2009;15:5936–45. doi: 10.3748/wjg.15.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukudo S, Hongo M, Kaneko H, Ueno R. Efficacy and safety of oral lubiprostone in constipated patients with or without irritable bowel syndrome: a randomized, placebo-controlled and dose-finding study. Neurogastroenterol Motil. 2011;23:544–e205. doi: 10.1111/j.1365-2982.2011.01668.x. [DOI] [PubMed] [Google Scholar]
- 19.Saad RJ. Peripherally acting therapies for the treatment of irritable bowel syndrome. Gastroenterol Clin North Am. 2011;40:163–82. doi: 10.1016/j.gtc.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Boesten RJ, de Vos WM. Interactomics in the human intestine: Lactobacilli and Bifidobacteria make a difference. J Clin Gastroenterol. 2008;42(Suppl 3 Pt 2):S163–7. doi: 10.1097/MCG.0b013e31817dbd62. [DOI] [PubMed] [Google Scholar]
- 21.Reading NC, Kasper DL. The starting lineup: key microbial players in intestinal immunity and homeostasis. Front Microbiol. 2011;2:148. doi: 10.3389/fmicb.2011.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sánchez B, Urdaci MC, Margolles A. Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiology. 2010;156:3232–42. doi: 10.1099/mic.0.044057-0. [DOI] [PubMed] [Google Scholar]
- 23.Madsen KL, Doyle JS, Jewell LD, Tavernini MM, Fedorak RN. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116:1107–14. doi: 10.1016/S0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- 24.Walker J, Jijon HB, Churchill T, Kulka M, Madsen KL. Activation of AMP-activated protein kinase reduces cAMP-mediated epithelial chloride secretion. Am J Physiol Gastrointest Liver Physiol. 2003;285:G850–60. doi: 10.1152/ajpgi.00077.2003. [DOI] [PubMed] [Google Scholar]
- 25.Madara JL, Parkos CA, Colgan SP, MacLeod RJ, Nash S, Matthews J, et al. Cl- secretion in a model intestinal epithelium induced by a neutrophil-derived secretagogue. J Clin Invest. 1992;89:1938–44. doi: 10.1172/JCI115800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blume ED, Taylor CT, Lennon PF, Stahl GL, Colgan SP. Activated endothelial cells elicit paracrine induction of epithelial chloride secretion. 6-Keto-PGF1alpha is an epithelial secretagogue. J Clin Invest. 1998;102:1161–72. doi: 10.1172/JCI3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keely S, Rawlinson LA, Haddleton DM, Brayden DJ. A tertiary amino-containing polymethacrylate polymer protects mucus-covered intestinal epithelial monolayers against pathogenic challenge. Pharm Res. 2008;25:1193–201. doi: 10.1007/s11095-007-9501-3. [DOI] [PubMed] [Google Scholar]
- 28.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–7. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris JK, Sahl JW, Castoe TA, Wagner BD, Pollock DD, Spear JR. Comparison of normalization methods for construction of large, multiplex amplicon pools for next-generation sequencing. Appl Environ Microbiol. 2010;76:3863–8. doi: 10.1128/AEM.02585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank DN. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–7. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Human Microbiome Consortium Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.